

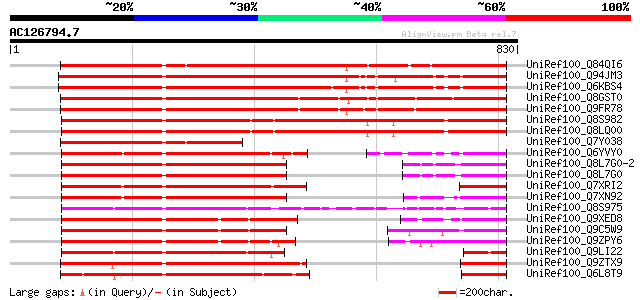
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126794.7 - phase: 1 /pseudo
(830 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84QI6 Auxin response factor-like protein [Mangifera i... 1003 0.0
UniRef100_Q94JM3 Auxin response factor 2 [Arabidopsis thaliana] 974 0.0
UniRef100_Q6KBS4 Putative auxin response factor [Brassica napus] 936 0.0
UniRef100_Q8GST0 Auxin response factor 1 [Oryza sativa] 790 0.0
UniRef100_Q9FR78 Putative auxin response factor 1 [Oryza sativa] 788 0.0
UniRef100_Q8S982 Auxin response factor 2 [Oryza sativa] 780 0.0
UniRef100_Q8LQ00 Auxin response factor 2 [Oryza sativa] 780 0.0
UniRef100_Q7Y038 Auxin response factor-like protein [Mangifera i... 533 e-150
UniRef100_Q6YVY0 Putative auxin-responsive factor [Oryza sativa] 515 e-144
UniRef100_Q8L7G0-2 Splice isoform 2 of Q8L7G0 [Arabidopsis thali... 513 e-144
UniRef100_Q8L7G0 Auxin response factor 1 [Arabidopsis thaliana] 513 e-144
UniRef100_Q7XRI2 P0076O17.10 protein [Oryza sativa] 488 e-136
UniRef100_Q7XN92 OSJNBa0064D20.11 protein [Oryza sativa] 488 e-136
UniRef100_Q8S975 Auxin response factor 16 [Oryza sativa] 461 e-128
UniRef100_Q9XED8 Auxin response factor 9 [Arabidopsis thaliana] 457 e-127
UniRef100_Q9C5W9 Auxin response factor 18 [Arabidopsis thaliana] 453 e-126
UniRef100_Q9ZPY6 Auxin response factor 11 [Arabidopsis thaliana] 452 e-125
UniRef100_Q9LI22 EST C26826(C50159) corresponds to a region of t... 420 e-115
UniRef100_Q9ZTX9 Auxin response factor 4 [Arabidopsis thaliana] 402 e-110
UniRef100_Q6L8T9 Auxin response factor 5 [Cucumis sativus] 398 e-109
>UniRef100_Q84QI6 Auxin response factor-like protein [Mangifera indica]
Length = 840
Score = 1003 bits (2594), Expect = 0.0
Identities = 519/762 (68%), Positives = 592/762 (77%), Gaps = 43/762 (5%)
Query: 83 EAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILC 142
E ALY+ELWHACAGPLVTVPR+GE V+YFPQGHIEQVEASTNQ ++Q MP+YDLR KILC
Sbjct: 31 ETALYKELWHACAGPLVTVPRQGERVYYFPQGHIEQVEASTNQFADQQMPIYDLRSKILC 90
Query: 143 RVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTS 202
RVINV LKA+PDTDEVFAQ+TL+PEPNQDENAVEKE PP PRFHVHSFCKTLTASDTS
Sbjct: 91 RVINVQLKAKPDTDEVFAQITLLPEPNQDENAVEKEPPPPLLPRFHVHSFCKTLTASDTS 150
Query: 203 THGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLL 262
THGGFSVLRRHA+ECLP LDMS+QPPTQ+LVAKDLHGNEWRFRHIFRG QPRRHLL
Sbjct: 151 THGGFSVLRRHAEECLPELDMSQQPPTQDLVAKDLHGNEWRFRHIFRG-----QPRRHLL 205
Query: 263 QSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLAT 322
QSGWSVFVSSKRLVAGDAFIFLR E ELRVGVRRAMRQQGNVPSSVISSHSMHLGVLAT
Sbjct: 206 QSGWSVFVSSKRLVAGDAFIFLRCEK-ELRVGVRRAMRQQGNVPSSVISSHSMHLGVLAT 264
Query: 323 AWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFT 382
AWHAV TGTMFTVYYKPR SPAEFIVP+DQYMES+K+NY+IGM F+MRFEGEEAPEQR+T
Sbjct: 265 AWHAVSTGTMFTVYYKPRISPAEFIVPFDQYMESVKSNYSIGMGFEMRFEGEEAPEQRYT 324
Query: 383 GTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRP 442
GTIVGIED+D +RWP SKWRCLKVRWDETS +PRPERVSPWKIEPALAP ALNPLP+ RP
Sbjct: 325 GTIVGIEDADPQRWPDSKWRCLKVRWDETSTVPRPERVSPWKIEPALAPLALNPLPLSRP 384
Query: 443 KRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLRGNLAE--SNDSY 500
KRPR+N+VPSSPDSSVLTRE S KV++DP +GF RVLQGQE STLRGN AE SN+
Sbjct: 385 KRPRSNMVPSSPDSSVLTREGSFKVNVDPSSATGFSRVLQGQEFSTLRGNFAERDSNEFD 444
Query: 501 TAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFGS---------- 550
TAEKSV + D++K+D V SRRYG EN +P R EP +DLLSG G+
Sbjct: 445 TAEKSVVRPSSLDDKKIDVVFASRRYGFENCVPAGRSEPMCTDLLSGLGTNSDSVHGYSP 504
Query: 551 -----------------TREGKHNML-TQWPVMPPGLSLNFLHSNMKGSAQGSDNATYQA 592
++EGK NML + W +MP LSL +N K QG D Y
Sbjct: 505 SIDQSLASAVPVRKSLLSQEGKFNMLGSPWSLMPSSLSLKMPETNAKVQVQGGD-INYLV 563
Query: 593 QGNMRYSAFGDYSVLHGHKVENPHGNFLMPPPPPTQYES-PHSRELSQKQMSAKISEAAK 651
QGN RY DY L H+V +GN+ MPP + +E+ SREL +K +S + EA K
Sbjct: 564 QGNARYGGLSDYPTLQSHRVGPSNGNWFMPPLVSSHFENLVPSRELMEKPISVQHHEAGK 623
Query: 652 PKDSDCKLFGFSLLSSPTML-EPSLSQRNATSETSSHMQISSQHHTFENDQKSEHSKSSK 710
K DCKLFG L+SS + EP L +N+ +E HM + Q E+D KSE SKS
Sbjct: 624 TK--DCKLFGIPLVSSSCVTPEPILLHQNSMNEPVGHM--NHQLGVLESDPKSEQSKSPT 679
Query: 711 PADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYD 770
A+ V E K QT QPHVKDV KPQSGS+RSCTKVHK+GIALGRSVDL+KF++Y+
Sbjct: 680 LANDSNCVSEQGKPSQTCQPHVKDVHSKPQSGSSRSCTKVHKQGIALGRSVDLSKFNNYE 739
Query: 771 ELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
EL AELD+LFEF GEL++P+K+WL+++TD+EGD+MLVGDDPW
Sbjct: 740 ELIAELDRLFEFGGELMTPKKNWLIIYTDDEGDIMLVGDDPW 781
>UniRef100_Q94JM3 Auxin response factor 2 [Arabidopsis thaliana]
Length = 859
Score = 974 bits (2518), Expect = 0.0
Identities = 504/771 (65%), Positives = 585/771 (75%), Gaps = 62/771 (8%)
Query: 81 EAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKI 140
+ EAALYRELWHACAGPLVTVPR+ + VFYFPQGHIEQVEASTNQA+EQ MP+YDL K+
Sbjct: 53 DPEAALYRELWHACAGPLVTVPRQDDRVFYFPQGHIEQVEASTNQAAEQQMPLYDLPSKL 112
Query: 141 LCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASD 200
LCRVINV LKAE DTDEV+AQ+TL+PE NQDENA+EKEAP PPPRF VHSFCKTLTASD
Sbjct: 113 LCRVINVDLKAEADTDEVYAQITLLPEANQDENAIEKEAPLPPPPRFQVHSFCKTLTASD 172
Query: 201 TSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRH 260
TSTHGGFSVLRRHADECLPPLDMS+QPPTQELVAKDLH NEWRFRHIFRG QPRRH
Sbjct: 173 TSTHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHANEWRFRHIFRG-----QPRRH 227
Query: 261 LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL 320
LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL
Sbjct: 228 LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL 287
Query: 321 ATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQR 380
ATAWHA+ TGTMFTVYYKPRTSP+EFIVP+DQYMES+KNNY+IGMRFKMRFEGEEAPEQR
Sbjct: 288 ATAWHAISTGTMFTVYYKPRTSPSEFIVPFDQYMESVKNNYSIGMRFKMRFEGEEAPEQR 347
Query: 381 FTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMP 440
FTGTIVGIE+SD RWP SKWR LKVRWDETS+IPRP+RVSPWK+EPALAPPAL+P+PMP
Sbjct: 348 FTGTIVGIEESDPTRWPKSKWRSLKVRWDETSSIPRPDRVSPWKVEPALAPPALSPVPMP 407
Query: 441 RPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLRGNLAESNDSY 500
RPKRPR+N+ PSSPDSS+LTRE ++K +MDPLP SG RVLQGQE STLR ES +
Sbjct: 408 RPKRPRSNIAPSSPDSSMLTREGTTKANMDPLPASGLSRVLQGQEYSTLRTKHTESVECD 467
Query: 501 TAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFG----------- 549
E SV W + D++K+D VS SRRYGSENWM +R EPTY+DLLSGFG
Sbjct: 468 APENSVVWQSSADDDKVDVVSGSRRYGSENWMSSARHEPTYTDLLSGFGTNIDPSHGQRI 527
Query: 550 -----------------STREGKHNML-TQWPVMPPGLSLNFLHSNMKGSAQGSDNATYQ 591
S EGK + L QW ++ GLSL LH + K A + +A+ Q
Sbjct: 528 PFYDHSSSPSMPAKRILSDSEGKFDYLANQWQMIHSGLSLK-LHESPKVPA--ATDASLQ 584
Query: 592 AQGNMRYSAFGDYSVLHGHKVENPHGNFLMPPPPPTQYE-------SPHSRELSQKQMSA 644
+ N++YS +Y VL+G EN GN+ + P YE +RE KQ
Sbjct: 585 GRCNVKYS---EYPVLNGLSTENAGGNWPIRPRALNYYEEVVNAQAQAQAREQVTKQPFT 641
Query: 645 KISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFENDQKSE 704
E AK ++ +C+LFG L ++ + ++SQRN ++ + QI+S + S+
Sbjct: 642 IQEETAKSREGNCRLFGIPLTNNMNGTDSTMSQRNNLNDAAGLTQIASP----KVQDLSD 697
Query: 705 HSKSSKPADKLVIVDEHEKQ---LQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSV 761
SK SK ++H +Q QT+ PH KD Q K + S+RSCTKVHK+GIALGRSV
Sbjct: 698 QSKGSKS------TNDHREQGRPFQTNNPHPKDAQTK--TNSSRSCTKVHKQGIALGRSV 749
Query: 762 DLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
DL+KF +Y+EL AELD+LFEF GEL++P+KDWL+V+TD E DMMLVGDDPW
Sbjct: 750 DLSKFQNYEELVAELDRLFEFNGELMAPKKDWLIVYTDEENDMMLVGDDPW 800
>UniRef100_Q6KBS4 Putative auxin response factor [Brassica napus]
Length = 848
Score = 936 bits (2419), Expect = 0.0
Identities = 488/767 (63%), Positives = 572/767 (73%), Gaps = 60/767 (7%)
Query: 81 EAEAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKI 140
+ EAALYRELWHACAGPLVTVPR+ + VFYFPQGHIEQVEASTNQA+EQ MP+YDL KI
Sbjct: 48 DPEAALYRELWHACAGPLVTVPRQDDRVFYFPQGHIEQVEASTNQAAEQQMPLYDLPSKI 107
Query: 141 LCRVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASD 200
LCRVINV LKAE DTDEV+AQ+TL+PEP QDEN++EKEAPP PPPRF VHSFCKTLTASD
Sbjct: 108 LCRVINVDLKAEADTDEVYAQITLLPEPVQDENSIEKEAPPPPPPRFQVHSFCKTLTASD 167
Query: 201 TSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRH 260
TSTHGGFSVLRRHADECLPPLDMS+QPPTQELVAKDLH +EWRFRHIFRG QPRRH
Sbjct: 168 TSTHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHASEWRFRHIFRG-----QPRRH 222
Query: 261 LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL 320
LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL
Sbjct: 223 LLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVL 282
Query: 321 ATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQR 380
ATAWHA+ TGTMFTVYYKPRTSP+EFIVP+DQY ES+K NY+IGMRFKMRFEGEEAPEQR
Sbjct: 283 ATAWHAISTGTMFTVYYKPRTSPSEFIVPFDQYTESVKINYSIGMRFKMRFEGEEAPEQR 342
Query: 381 FTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMP 440
FTGTIVGIEDSD RW SKWR LKVRWDET++IPRP+RVSPWKIEPAL+PPAL+P+PMP
Sbjct: 343 FTGTIVGIEDSDPTRWAKSKWRSLKVRWDETTSIPRPDRVSPWKIEPALSPPALSPVPMP 402
Query: 441 RPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLRGNLAESNDSY 500
RPKRPR+N+ S+PDSS+ RE SSK +MDPLP SG RVLQGQE TLR ES +
Sbjct: 403 RPKRPRSNLASSTPDSSMRIREGSSKANMDPLPASGLSRVLQGQEYPTLRTKHVESVECD 462
Query: 501 TAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFG----------- 549
E SV W +TD++K+D +S SRRY ENW+ R PT +DLLSGFG
Sbjct: 463 APENSVVWQSSTDDDKVDVISASRRY--ENWISSGRHGPTCTDLLSGFGTNIEPPHGHQI 520
Query: 550 ------------------STREGKHNMLTQWPVMPPGLSLNFLHSNMKGSAQGSDNATYQ 591
S ++GK L +M GLSL LH + K A + +A++Q
Sbjct: 521 PFYDRLSSPPSVAARKILSDQDGKFEYLANQWMMHSGLSLK-LHESPKVPA--ASDASFQ 577
Query: 592 AQGNMRYSAFGDYSVLHGHKVENPHGNFLMPPPPPTQYES---PHSRELSQKQMSAKISE 648
GN Y G+Y++ EN GN+ + P +E +RE K+ + E
Sbjct: 578 GIGNPNY---GEYALPRAVTTENAAGNWPIRPRALNYFEEAVHAQAREHVTKRPAVVQEE 634
Query: 649 AAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFENDQKSEHSKS 708
AAKP+D +C+LFG L+++ + +LSQRN ++ + Q++S + S+ SK
Sbjct: 635 AAKPRDGNCRLFGIPLVNNVNGTDTTLSQRNNLNDPAGPTQMASP----KVQDLSDQSKG 690
Query: 709 SKPADKLVIVDEHEKQ---LQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTK 765
SK ++H +Q S+PH KDVQ K + S RSCTKV K+GIALGRSVDL+K
Sbjct: 691 SKS------TNDHREQGRPFPVSKPHPKDVQTK--TNSCRSCTKVQKQGIALGRSVDLSK 742
Query: 766 FSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
F +Y+EL ELD+LFEF GEL++P+KDWL+V+TD+E DMMLVGDDPW
Sbjct: 743 FQNYEELVTELDRLFEFNGELMAPKKDWLIVYTDDENDMMLVGDDPW 789
>UniRef100_Q8GST0 Auxin response factor 1 [Oryza sativa]
Length = 836
Score = 790 bits (2041), Expect = 0.0
Identities = 435/772 (56%), Positives = 516/772 (66%), Gaps = 56/772 (7%)
Query: 83 EAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILC 142
E AL+ ELW ACAGPLVTVPR GE VFYFPQGHIEQVEASTNQ EQ M +Y+L KILC
Sbjct: 18 EDALFTELWSACAGPLVTVPRVGEKVFYFPQGHIEQVEASTNQVGEQRMQLYNLPWKILC 77
Query: 143 RVINVMLKAEPDTDEVFAQVTLVPEPNQDEN--AVEKEAPPAPPP---RFHVHSFCKTLT 197
V+NV LKAEPDTDEV+AQ+TL+PE Q E+ + E+E P AP R VHSFCKTLT
Sbjct: 78 EVMNVELKAEPDTDEVYAQLTLLPESKQQEDNGSTEEEVPSAPAAGHVRPRVHSFCKTLT 137
Query: 198 ASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQP 257
ASDTSTHGGFSVLRRHADECLPPLDMS+QPPTQELVAKDLHG EWRFRHIFRG QP
Sbjct: 138 ASDTSTHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHGVEWRFRHIFRG-----QP 192
Query: 258 RRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHL 317
RRHLLQSGWSVFVS+KRLVAGDAFIFLRGENGELRVGVRRAMRQQ NVPSSVISSHSMHL
Sbjct: 193 RRHLLQSGWSVFVSAKRLVAGDAFIFLRGENGELRVGVRRAMRQQTNVPSSVISSHSMHL 252
Query: 318 GVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAP 377
GVLATAWHAV TGTMFTVYYKPRTSPAEF+VPYD+YMESLK NY+IGMRFKMRFEGEEAP
Sbjct: 253 GVLATAWHAVNTGTMFTVYYKPRTSPAEFVVPYDRYMESLKQNYSIGMRFKMRFEGEEAP 312
Query: 378 EQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPL 437
EQRFTGTIVG+ DSD WP SKWR LKVRWDE S+IPRPERVSPW+IEPA++PP +NPL
Sbjct: 313 EQRFTGTIVGMGDSDPAGWPESKWRSLKVRWDEASSIPRPERVSPWQIEPAVSPPPVNPL 372
Query: 438 PMPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLRGNLAESN 497
P+PR KR R N DSS + +EA++KV ++ P +G QR Q QE++T + S+
Sbjct: 373 PVPRTKRLRPNATALPADSSAIAKEAATKVVVESEP-NGTQRTFQTQENATPKSGFGNSS 431
Query: 498 DSYTAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFGSTRE---- 553
+ +A+KS+ D EK + + GS+ M MS+ E +YS++LSGF ++
Sbjct: 432 ELESAQKSIMRPSGFDREKNNT-PIQWKLGSDGRMQMSKPE-SYSEMLSGFQPPKDVQIP 489
Query: 554 -----------------------------GKHNML-TQWPVMPPGLSLNFLHSNMKGSAQ 583
HNM + W MPP L N +
Sbjct: 490 QGFWSLPEQITAGHSNFWHTVNAQYQDQQSNHNMFPSSWSFMPPNTRLGLNKQNYSMIQE 549
Query: 584 GSDNATYQAQGNMRYSAFGDYSVLHGHKVENPHGNFLMPPPPPTQYESPHSRELSQKQMS 643
Q GN ++ G Y+ L G E G + P + + R + K +
Sbjct: 550 AG--VLSQRPGNTKFGN-GVYAALPGRGTEQYSGGWFGHMMPNSHMDDTQPRLIKPKPLV 606
Query: 644 AKISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHH---TFEND 700
+ K K + CKLFG L SP EP S + + + +++ E +
Sbjct: 607 VAHGDVQKAKGASCKLFGIHL-DSPAKSEPLKSPSSVVYDGTPQTPGATEWRRPDVTEVE 665
Query: 701 QKSEHSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRS 760
+ S+ SK+ KP D EK +SQ +++ K Q S RSC KVHK+GIALGRS
Sbjct: 666 KCSDPSKAMKPLDTPQPDSVPEKP--SSQQASRNMSCKSQGVSTRSCKKVHKQGIALGRS 723
Query: 761 VDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
VDLTKF+ Y+EL AELD +F+F GEL P+K+W+VV+TDNEGDMMLVGDDPW
Sbjct: 724 VDLTKFNGYEELIAELDDMFDFNGELKGPKKEWMVVYTDNEGDMMLVGDDPW 775
>UniRef100_Q9FR78 Putative auxin response factor 1 [Oryza sativa]
Length = 857
Score = 788 bits (2034), Expect = 0.0
Identities = 433/775 (55%), Positives = 516/775 (65%), Gaps = 57/775 (7%)
Query: 83 EAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILC 142
E AL+ ELW ACAGPLVTVPR GE FYFPQGHIEQVEASTNQ EQ M +Y+L KILC
Sbjct: 34 EDALFTELWSACAGPLVTVPRVGEKEFYFPQGHIEQVEASTNQVGEQRMQLYNLPWKILC 93
Query: 143 RVINVMLKAEPDTDEVFAQVTLVPE--PNQDENAVEKEAPPAPPP---RFHVHSFCKTLT 197
V+NV LKAEPDTDEV+AQ+TL+PE +D + E+E P AP R VHSFCKTLT
Sbjct: 94 EVMNVELKAEPDTDEVYAQLTLLPELKRQEDNGSTEEEVPSAPAAGHVRPRVHSFCKTLT 153
Query: 198 ASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQP 257
ASDTSTHGGFSVLRRHADECLPPLDMS+QPPTQELVAKDLHG EWRFRHIFRG QP
Sbjct: 154 ASDTSTHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHGVEWRFRHIFRG-----QP 208
Query: 258 RRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHL 317
RRHLLQSGWSVFVS+KRLVAGDAFIFLRGENGELRVGVRRAMRQQ NVPSSVISSHSMHL
Sbjct: 209 RRHLLQSGWSVFVSAKRLVAGDAFIFLRGENGELRVGVRRAMRQQTNVPSSVISSHSMHL 268
Query: 318 GVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAP 377
GVLATAWHAV TGTMFTVYYKPRTSPAEF+VPYD+YMESLK NY+IGMRFKMRFE EEAP
Sbjct: 269 GVLATAWHAVNTGTMFTVYYKPRTSPAEFVVPYDRYMESLKRNYSIGMRFKMRFESEEAP 328
Query: 378 EQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPL 437
EQRFTGTIVG+ DSD WP SKWR LKVRWDE S+IPRPERVSPW+IEPA++PP +NPL
Sbjct: 329 EQRFTGTIVGMGDSDPAGWPESKWRSLKVRWDEASSIPRPERVSPWQIEPAVSPPPVNPL 388
Query: 438 PMPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLRGNLAESN 497
P+PR KR R N DSS + +EA++KV ++ P +G QR Q QE++T + S+
Sbjct: 389 PVPRTKRLRPNATALPADSSAIAKEAATKVVVESEP-NGTQRTFQTQENATPKSGFGNSS 447
Query: 498 DSYTAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFG-------- 549
+ +A+KS+ D EK + + GS+ M MS+ E +YS++LSGF
Sbjct: 448 ELESAQKSIMRPSGFDREK-NNTPIQWKLGSDGRMQMSKPE-SYSEMLSGFQPPKDVQTP 505
Query: 550 -------------------------STREGKHNML-TQWPVMPPGLSLNFLHSNMKGSAQ 583
++ HNM + W MPP L N S
Sbjct: 506 QGFCSLPEQITAGHSNFWHTVNAQYQDQQSNHNMFPSSWSFMPPNTRLGLNKQNY--SMI 563
Query: 584 GSDNATYQAQGNMRYSAFGDYSVLHGHKVENPHGNFLMPPPPPTQYESPHSRELSQKQMS 643
Q GN ++ G Y+ L G E G + P + + R + K +
Sbjct: 564 QEAGVLSQRPGNTKFGN-GVYAALPGRGTEQYSGGWFGLMMPNSHMDDTQPRLIKPKPLV 622
Query: 644 AKISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHH---TFEND 700
+ K K + CKLFG L SP EPS S + + + +++ E +
Sbjct: 623 VAHGDVQKAKGASCKLFGIH-LDSPAKSEPSKSPSSVVYDGTPQTPGATEWRRPDVTEVE 681
Query: 701 QKSEHSKSSKPADKL---VIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIAL 757
+ S+ SK+ KP D + ++ Q +SQ +++ K Q S RSC KVHK+GIAL
Sbjct: 682 KCSDPSKAMKPLDTPQPDSVPEKPSSQQASSQQASRNMSCKSQGVSTRSCKKVHKQGIAL 741
Query: 758 GRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
GRSVDLTKF+ Y+EL AELD +F+F GEL P+K+W+VV+TDNEGDMMLVGDDPW
Sbjct: 742 GRSVDLTKFNGYEELIAELDDMFDFNGELKGPKKEWMVVYTDNEGDMMLVGDDPW 796
>UniRef100_Q8S982 Auxin response factor 2 [Oryza sativa]
Length = 791
Score = 780 bits (2014), Expect = 0.0
Identities = 426/752 (56%), Positives = 510/752 (67%), Gaps = 38/752 (5%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRVI 145
LY ELWHACAGPLVTVPR G+LVFYFPQGHIEQVEAS NQ ++ M +YDL K+LCRV+
Sbjct: 4 LYDELWHACAGPLVTVPRVGDLVFYFPQGHIEQVEASMNQVADSQMRLYDLPSKLLCRVL 63
Query: 146 NVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPP--PRFHVHSFCKTLTASDTST 203
NV LKAE DTDEV+AQV L+PEP Q+E AVEK P + P R V SFCKTLTASDTST
Sbjct: 64 NVELKAEQDTDEVYAQVMLMPEPEQNEMAVEKTTPTSGPVQARPPVRSFCKTLTASDTST 123
Query: 204 HGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQ 263
HGGFSVLRRHADECLPPLDM++ PPTQELVAKDLH +WRFRHIFRG QPRRHLLQ
Sbjct: 124 HGGFSVLRRHADECLPPLDMTQSPPTQELVAKDLHSMDWRFRHIFRG-----QPRRHLLQ 178
Query: 264 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATA 323
SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQ NVPSSVISS SMHLGVLATA
Sbjct: 179 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQLSNVPSSVISSQSMHLGVLATA 238
Query: 324 WHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTG 383
WHA+ T +MFTVYYKPRTSP+EFI+PYDQYMES+KNNY++GMRF+MRFEGEEAPEQRFTG
Sbjct: 239 WHAINTKSMFTVYYKPRTSPSEFIIPYDQYMESVKNNYSVGMRFRMRFEGEEAPEQRFTG 298
Query: 384 TIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPK 443
TI+G E+ D WP S WR LKVRWDE S IPRP+RVSPWKIEPA +PP +NPLP+ R K
Sbjct: 299 TIIGSENLDPV-WPESSWRSLKVRWDEPSTIPRPDRVSPWKIEPASSPP-VNPLPLSRVK 356
Query: 444 RPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQR--VLQGQESSTLRGNLAESNDS-Y 500
RPR N P+SP+S +LT+EA++KV DP Q VLQGQE TLR NL ESNDS
Sbjct: 357 RPRPNAPPASPESPILTKEAATKVDTDPAQAQRSQNSTVLQGQEQMTLRSNLTESNDSDV 416
Query: 501 TAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFGSTREGKHNMLT 560
TA K + W+P+ + K ++ +R +NWM + R+E + D+ SG S + +
Sbjct: 417 TAHKPMMWSPSPNAAKAHPLTFQQRPPMDNWMQLGRRETDFKDVRSGSQSFGDSPGFFMQ 476
Query: 561 QWPVMPPGLSLNFLHSNMKGSAQ---------------GSDNATYQAQGNMRYSAFGDYS 605
+ P L+ +GSA+ +++T + + S
Sbjct: 477 NFDEAPNRLTSFKNQFQDQGSARHFSDPYYYVSPQPSLTVESSTQMHTDSKELHFWNGQS 536
Query: 606 VLHGHKVENPHGNFLMPPPPPT----QYESPHSRELSQKQMSAKISEAAKPKDSDCKLFG 661
++G+ + P NF + + P + + S E K + S K+FG
Sbjct: 537 TVYGNSRDRPQ-NFRFEQNSSSWLNQSFARPEQPRVIRPHASIAPVELEKTEGSGFKIFG 595
Query: 662 FSLLSSPTMLEPSLSQRNATSETSSHMQIS-SQHHTFENDQKSEHSKSSKPADKLVIVDE 720
F + ++ S AT E S +Q + D E S S+ E
Sbjct: 596 FKVDTTNAPNNHLSSPMAATHEPMLQTPSSLNQLQPVQTDCIPEVSVSTAGT-----ATE 650
Query: 721 HEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLF 780
+EK Q +Q KDVQ K Q S RSCTKVHK+G+ALGRSVDL+KFS+YDEL AELD++F
Sbjct: 651 NEKSGQQAQQSSKDVQSKTQVASTRSCTKVHKQGVALGRSVDLSKFSNYDELKAELDKMF 710
Query: 781 EFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
EF GEL+S K+W +V+TDNEGDMMLVGDDPW
Sbjct: 711 EFDGELVSSNKNWQIVYTDNEGDMMLVGDDPW 742
>UniRef100_Q8LQ00 Auxin response factor 2 [Oryza sativa]
Length = 826
Score = 780 bits (2014), Expect = 0.0
Identities = 426/752 (56%), Positives = 510/752 (67%), Gaps = 38/752 (5%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRVI 145
LY ELWHACAGPLVTVPR G+LVFYFPQGHIEQVEAS NQ ++ M +YDL K+LCRV+
Sbjct: 16 LYDELWHACAGPLVTVPRVGDLVFYFPQGHIEQVEASMNQVADSQMRLYDLPSKLLCRVL 75
Query: 146 NVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPP--PRFHVHSFCKTLTASDTST 203
NV LKAE DTDEV+AQV L+PEP Q+E AVEK P + P R V SFCKTLTASDTST
Sbjct: 76 NVELKAEQDTDEVYAQVMLMPEPEQNEMAVEKTTPTSGPVQARPPVRSFCKTLTASDTST 135
Query: 204 HGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQ 263
HGGFSVLRRHADECLPPLDM++ PPTQELVAKDLH +WRFRHIFRG QPRRHLLQ
Sbjct: 136 HGGFSVLRRHADECLPPLDMTQSPPTQELVAKDLHSMDWRFRHIFRG-----QPRRHLLQ 190
Query: 264 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATA 323
SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQ NVPSSVISS SMHLGVLATA
Sbjct: 191 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQLSNVPSSVISSQSMHLGVLATA 250
Query: 324 WHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTG 383
WHA+ T +MFTVYYKPRTSP+EFI+PYDQYMES+KNNY++GMRF+MRFEGEEAPEQRFTG
Sbjct: 251 WHAINTKSMFTVYYKPRTSPSEFIIPYDQYMESVKNNYSVGMRFRMRFEGEEAPEQRFTG 310
Query: 384 TIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPK 443
TI+G E+ D WP S WR LKVRWDE S IPRP+RVSPWKIEPA +PP +NPLP+ R K
Sbjct: 311 TIIGSENLDPV-WPESSWRSLKVRWDEPSTIPRPDRVSPWKIEPASSPP-VNPLPLSRVK 368
Query: 444 RPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQR--VLQGQESSTLRGNLAESNDS-Y 500
RPR N P+SP+S +LT+EA++KV DP Q VLQGQE TLR NL ESNDS
Sbjct: 369 RPRPNAPPASPESPILTKEAATKVDTDPAQAQRSQNSTVLQGQEQMTLRSNLTESNDSDV 428
Query: 501 TAEKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFGSTREGKHNMLT 560
TA K + W+P+ + K ++ +R +NWM + R+E + D+ SG S + +
Sbjct: 429 TAHKPMMWSPSPNAAKAHPLTFQQRPPMDNWMQLGRRETDFKDVRSGSQSFGDSPGFFMQ 488
Query: 561 QWPVMPPGLSLNFLHSNMKGSAQ---------------GSDNATYQAQGNMRYSAFGDYS 605
+ P L+ +GSA+ +++T + + S
Sbjct: 489 NFDEAPNRLTSFKNQFQDQGSARHFSDPYYYVSPQPSLTVESSTQMHTDSKELHFWNGQS 548
Query: 606 VLHGHKVENPHGNFLMPPPPPT----QYESPHSRELSQKQMSAKISEAAKPKDSDCKLFG 661
++G+ + P NF + + P + + S E K + S K+FG
Sbjct: 549 TVYGNSRDRPQ-NFRFEQNSSSWLNQSFARPEQPRVIRPHASIAPVELEKTEGSGFKIFG 607
Query: 662 FSLLSSPTMLEPSLSQRNATSETSSHMQIS-SQHHTFENDQKSEHSKSSKPADKLVIVDE 720
F + ++ S AT E S +Q + D E S S+ E
Sbjct: 608 FKVDTTNAPNNHLSSPMAATHEPMLQTPSSLNQLQPVQTDCIPEVSVSTAGT-----ATE 662
Query: 721 HEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLF 780
+EK Q +Q KDVQ K Q S RSCTKVHK+G+ALGRSVDL+KFS+YDEL AELD++F
Sbjct: 663 NEKSGQQAQQSSKDVQSKTQVASTRSCTKVHKQGVALGRSVDLSKFSNYDELKAELDKMF 722
Query: 781 EFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
EF GEL+S K+W +V+TDNEGDMMLVGDDPW
Sbjct: 723 EFDGELVSSNKNWQIVYTDNEGDMMLVGDDPW 754
>UniRef100_Q7Y038 Auxin response factor-like protein [Mangifera indica]
Length = 326
Score = 533 bits (1373), Expect = e-150
Identities = 261/298 (87%), Positives = 277/298 (92%), Gaps = 6/298 (2%)
Query: 83 EAALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILC 142
E ALY+ELWHACAGPLVTVPR+GE V+YFPQGHIEQVEASTNQ ++Q MP+YDLR KILC
Sbjct: 31 ETALYKELWHACAGPLVTVPRQGERVYYFPQGHIEQVEASTNQFADQQMPIYDLRSKILC 90
Query: 143 RVINVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTS 202
RVINV LKA+PDTDEVFAQ+TL+PEPNQDENAVEKE PP PRFHVHSFCKTLTASDTS
Sbjct: 91 RVINVQLKAKPDTDEVFAQITLLPEPNQDENAVEKEPPPPLLPRFHVHSFCKTLTASDTS 150
Query: 203 THGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLL 262
THGGFSVLRRHA+ECLP LDMS+QPPTQ+LVAKDLHGNEWRFRHIFR GQPRRHLL
Sbjct: 151 THGGFSVLRRHAEECLPVLDMSQQPPTQDLVAKDLHGNEWRFRHIFR-----GQPRRHLL 205
Query: 263 QSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLAT 322
QSGWSVFVSSKRLVAGDAFIFLR E ELRVGVRRAMRQQGNVPSSVISSHSMHLGVLAT
Sbjct: 206 QSGWSVFVSSKRLVAGDAFIFLRCEK-ELRVGVRRAMRQQGNVPSSVISSHSMHLGVLAT 264
Query: 323 AWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQR 380
AWHAV TGTMFTVYYKPR SPAEFIVP+DQYMES+K+NY+IGMRFKMRFEGEEAPEQR
Sbjct: 265 AWHAVSTGTMFTVYYKPRISPAEFIVPFDQYMESVKSNYSIGMRFKMRFEGEEAPEQR 322
>UniRef100_Q6YVY0 Putative auxin-responsive factor [Oryza sativa]
Length = 678
Score = 515 bits (1327), Expect = e-144
Identities = 259/409 (63%), Positives = 314/409 (76%), Gaps = 15/409 (3%)
Query: 85 ALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRV 144
ALYRELWHACAGPLVTVPR+GELV+YFPQGH+EQ+EAST+Q +QH+P+++L KILC+V
Sbjct: 22 ALYRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFNLPSKILCKV 81
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
+NV L+AE D+DEV+AQ+ L PE +Q+E K P P + +VHSFCKTLTASDTSTH
Sbjct: 82 VNVELRAETDSDEVYAQIMLQPEADQNELTSPKPEPHEPE-KCNVHSFCKTLTASDTSTH 140
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLRRHA+ECLPPLDM++ PP QELVA+DLHGNEW FRHIFRG QPRRHLL +
Sbjct: 141 GGFSVLRRHAEECLPPLDMTQNPPWQELVARDLHGNEWHFRHIFRG-----QPRRHLLTT 195
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRR MRQ N+PSSVISSHSMHLGVLATA
Sbjct: 196 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRLMRQLNNMPSSVISSHSMHLGVLATAS 255
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HA+ TGT+F+V+YKPRTS +EF+V ++Y+E+ + ++GMRFKMRFEG+EAPE+RF+GT
Sbjct: 256 HAISTGTLFSVFYKPRTSQSEFVVSANKYLEAKNSKISVGMRFKMRFEGDEAPERRFSGT 315
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPKR 444
I+G+ + W S WR LKV+WDE S +PRP+RVSPW++EP LA P P P +
Sbjct: 316 IIGVGSMSTSPWANSDWRSLKVQWDEPSVVPRPDRVSPWELEP-LAVSNSQPSPQPPARN 374
Query: 445 PRA------NVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESS 487
RA ++ P P L + SS S SG QR + SS
Sbjct: 375 KRARPPASNSIAPELPPVFGLWK--SSAESTQGFSFSGLQRTQELYPSS 421
Score = 102 bits (255), Expect = 4e-20
Identities = 70/228 (30%), Positives = 112/228 (48%), Gaps = 33/228 (14%)
Query: 585 SDNATYQAQGNMRYSAFGDYSVLHGHKVENPHGNFLMPPPPPTQYESPHSRELSQKQMSA 644
S N + N+ +S + S L N H + M Y + S+ S+K+
Sbjct: 421 SPNPIFSTSLNVGFSTKNEPSAL-----SNKHFYWPMRETRANSYSASISKVPSEKKQ-- 473
Query: 645 KISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFENDQKSE 704
+P + C+LFG + S+ P L+ + + + + E+DQ S+
Sbjct: 474 ------EPSSAGCRLFGIEISSAVEATSP-LAAVSGVGQDQPAASVDA-----ESDQLSQ 521
Query: 705 HSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLT 764
S ++K + +S+P + Q S RSCTKV +G+A+GR+VDLT
Sbjct: 522 PSHANKS----------DAPAASSEPSPHETQ----SRQVRSCTKVIMQGMAVGRAVDLT 567
Query: 765 KFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
+ YD+L +L+++F+ +GEL + K W VV+TD+E DMMLVGDDPW
Sbjct: 568 RLHGYDDLRCKLEEMFDIQGELSASLKKWKVVYTDDEDDMMLVGDDPW 615
>UniRef100_Q8L7G0-2 Splice isoform 2 of Q8L7G0 [Arabidopsis thaliana]
Length = 662
Score = 513 bits (1322), Expect = e-144
Identities = 251/370 (67%), Positives = 302/370 (80%), Gaps = 8/370 (2%)
Query: 85 ALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRV 144
AL RELWHACAGPLVT+PREGE V+YFP+GH+EQ+EAS +Q EQ MP ++L KILC+V
Sbjct: 18 ALCRELWHACAGPLVTLPREGERVYYFPEGHMEQLEASMHQGLEQQMPSFNLPSKILCKV 77
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
IN+ +AEP+TDEV+AQ+TL+PE +Q E +AP P + VHSFCKTLTASDTSTH
Sbjct: 78 INIQRRAEPETDEVYAQITLLPELDQSE-PTSPDAPVQEPEKCTVHSFCKTLTASDTSTH 136
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLRRHAD+CLPPLDMS+QPP QELVA DLH +EW FRHIFRG QPRRHLL +
Sbjct: 137 GGFSVLRRHADDCLPPLDMSQQPPWQELVATDLHNSEWHFRHIFRG-----QPRRHLLTT 191
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWSVFVSSK+LVAGDAFIFLRGEN ELRVGVRR MRQQ N+PSSVISSHSMH+GVLATA
Sbjct: 192 GWSVFVSSKKLVAGDAFIFLRGENEELRVGVRRHMRQQTNIPSSVISSHSMHIGVLATAA 251
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HA+ TGT+F+V+YKPRTS +EFIV ++Y+E+ ++GMRFKMRFEGEEAPE+RF+GT
Sbjct: 252 HAITTGTIFSVFYKPRTSRSEFIVSVNRYLEAKTQKLSVGMRFKMRFEGEEAPEKRFSGT 311
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALA--PPALNPLPMPRP 442
IVG++++ S W S+WR LKV+WDE S++ RPERVSPW++EP +A P+ P P R
Sbjct: 312 IVGVQENKSSVWHDSEWRSLKVQWDEPSSVFRPERVSPWELEPLVANSTPSSQPQPPQRN 371
Query: 443 KRPRANVVPS 452
KRPR +PS
Sbjct: 372 KRPRPPGLPS 381
Score = 100 bits (249), Expect = 2e-19
Identities = 59/174 (33%), Positives = 99/174 (55%), Gaps = 14/174 (8%)
Query: 643 SAKISEAAKPKDSD---CKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFEN 699
SA +E+ + K ++ C+LFGF L+ + + E S + + + + S F++
Sbjct: 445 SAFNNESTEKKQTNGNVCRLFGFELVENVNVDE-CFSAASVSGAVAVDQPVPSNE--FDS 501
Query: 700 DQKSEHSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGR 759
Q+SE ++ D + L++ Q + QS RSCTKVH +G A+GR
Sbjct: 502 GQQSEPLNINQSDIPSGSGDPEKSSLRSPQ--------ESQSRQIRSCTKVHMQGSAVGR 553
Query: 760 SVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPWH 813
++DLT+ Y++L +L+++F+ +GEL+ K W VV+TD+E DMM+VGDDPW+
Sbjct: 554 AIDLTRSECYEDLFKKLEEMFDIKGELLESTKKWQVVYTDDEDDMMMVGDDPWN 607
>UniRef100_Q8L7G0 Auxin response factor 1 [Arabidopsis thaliana]
Length = 665
Score = 513 bits (1322), Expect = e-144
Identities = 251/370 (67%), Positives = 302/370 (80%), Gaps = 8/370 (2%)
Query: 85 ALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRV 144
AL RELWHACAGPLVT+PREGE V+YFP+GH+EQ+EAS +Q EQ MP ++L KILC+V
Sbjct: 18 ALCRELWHACAGPLVTLPREGERVYYFPEGHMEQLEASMHQGLEQQMPSFNLPSKILCKV 77
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
IN+ +AEP+TDEV+AQ+TL+PE +Q E +AP P + VHSFCKTLTASDTSTH
Sbjct: 78 INIQRRAEPETDEVYAQITLLPELDQSE-PTSPDAPVQEPEKCTVHSFCKTLTASDTSTH 136
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLRRHAD+CLPPLDMS+QPP QELVA DLH +EW FRHIFRG QPRRHLL +
Sbjct: 137 GGFSVLRRHADDCLPPLDMSQQPPWQELVATDLHNSEWHFRHIFRG-----QPRRHLLTT 191
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWSVFVSSK+LVAGDAFIFLRGEN ELRVGVRR MRQQ N+PSSVISSHSMH+GVLATA
Sbjct: 192 GWSVFVSSKKLVAGDAFIFLRGENEELRVGVRRHMRQQTNIPSSVISSHSMHIGVLATAA 251
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HA+ TGT+F+V+YKPRTS +EFIV ++Y+E+ ++GMRFKMRFEGEEAPE+RF+GT
Sbjct: 252 HAITTGTIFSVFYKPRTSRSEFIVSVNRYLEAKTQKLSVGMRFKMRFEGEEAPEKRFSGT 311
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALA--PPALNPLPMPRP 442
IVG++++ S W S+WR LKV+WDE S++ RPERVSPW++EP +A P+ P P R
Sbjct: 312 IVGVQENKSSVWHDSEWRSLKVQWDEPSSVFRPERVSPWELEPLVANSTPSSQPQPPQRN 371
Query: 443 KRPRANVVPS 452
KRPR +PS
Sbjct: 372 KRPRPPGLPS 381
Score = 100 bits (249), Expect = 2e-19
Identities = 59/174 (33%), Positives = 99/174 (55%), Gaps = 14/174 (8%)
Query: 643 SAKISEAAKPKDSD---CKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFEN 699
SA +E+ + K ++ C+LFGF L+ + + E S + + + + S F++
Sbjct: 448 SAFNNESTEKKQTNGNVCRLFGFELVENVNVDE-CFSAASVSGAVAVDQPVPSNE--FDS 504
Query: 700 DQKSEHSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGR 759
Q+SE ++ D + L++ Q + QS RSCTKVH +G A+GR
Sbjct: 505 GQQSEPLNINQSDIPSGSGDPEKSSLRSPQ--------ESQSRQIRSCTKVHMQGSAVGR 556
Query: 760 SVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPWH 813
++DLT+ Y++L +L+++F+ +GEL+ K W VV+TD+E DMM+VGDDPW+
Sbjct: 557 AIDLTRSECYEDLFKKLEEMFDIKGELLESTKKWQVVYTDDEDDMMMVGDDPWN 610
>UniRef100_Q7XRI2 P0076O17.10 protein [Oryza sativa]
Length = 1673
Score = 488 bits (1256), Expect = e-136
Identities = 253/408 (62%), Positives = 306/408 (74%), Gaps = 17/408 (4%)
Query: 85 ALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRV 144
AL+RELWHACAGPLVTVP+ GE V+YFPQGH+EQ+EASTNQ +Q++P+++L KILC V
Sbjct: 16 ALFRELWHACAGPLVTVPKRGERVYYFPQGHMEQLEASTNQQLDQYLPMFNLPSKILCSV 75
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDE-NAVEKEAPPAPPPRFHVHSFCKTLTASDTST 203
+NV L+AE D+DEV+AQ+ L PE +Q E +++ E + HSFCKTLTASDTST
Sbjct: 76 VNVELRAEADSDEVYAQIMLQPEADQSELTSLDPELQDLE--KCTAHSFCKTLTASDTST 133
Query: 204 HGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQ 263
HGGFSVLRRHA+ECLP LDMS+ PP QELVAKDLHG EW FRHIFRG QPRRHLL
Sbjct: 134 HGGFSVLRRHAEECLPQLDMSQNPPCQELVAKDLHGTEWHFRHIFRG-----QPRRHLLT 188
Query: 264 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATA 323
+GWSVFVSSKRLVAGDAFIFLRGE+GELRVGVRR MRQ N+PSSVISSHSMHLGVLATA
Sbjct: 189 TGWSVFVSSKRLVAGDAFIFLRGESGELRVGVRRLMRQVNNMPSSVISSHSMHLGVLATA 248
Query: 324 WHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTG 383
HA+ TGT+F+V+YKPRTS +EF+V ++Y+E+ K N ++GMRFKMRFEG+EAPE+RF+G
Sbjct: 249 SHAISTGTLFSVFYKPRTSRSEFVVSVNKYLEAKKQNLSVGMRFKMRFEGDEAPERRFSG 308
Query: 384 TIVGIED---SDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMP 440
TI+GI W S W+ LKV+WDE S I RP+RVSPW++EP A NP P
Sbjct: 309 TIIGIGSVPAMSKSPWADSDWKSLKVQWDEPSAIVRPDRVSPWELEPL---DASNPQPPQ 365
Query: 441 RPKRPRANVVPSSPDSSVLTREASSKVSMDPL--PTSGFQRVLQGQES 486
P R + P+SP S V S V D + P++G + G S
Sbjct: 366 PPLRNKRARPPASP-SVVAELPPSFDVDSDQISQPSNGNKSDAPGTSS 412
Score = 91.7 bits (226), Expect = 8e-17
Identities = 41/76 (53%), Positives = 58/76 (75%)
Query: 737 LKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVV 796
L+ QS RSCTKV +G+A+GR+VDLTK + Y +L ++L+++F+ +G+L K W VV
Sbjct: 417 LESQSRQVRSCTKVIMQGMAVGRAVDLTKLNGYGDLRSKLEEMFDIQGDLCPTLKRWQVV 476
Query: 797 FTDNEGDMMLVGDDPW 812
+TD+E DMMLVGDDPW
Sbjct: 477 YTDDEDDMMLVGDDPW 492
>UniRef100_Q7XN92 OSJNBa0064D20.11 protein [Oryza sativa]
Length = 669
Score = 488 bits (1255), Expect = e-136
Identities = 244/372 (65%), Positives = 291/372 (77%), Gaps = 11/372 (2%)
Query: 85 ALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCRV 144
AL+RELWHACAGPLVTVP+ GE V+YFPQGH+EQ+EASTNQ +Q++P+++L KILC V
Sbjct: 16 ALFRELWHACAGPLVTVPKRGERVYYFPQGHMEQLEASTNQQLDQYLPMFNLPSKILCSV 75
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDE-NAVEKEAPPAPPPRFHVHSFCKTLTASDTST 203
+NV L+AE D+DEV+AQ+ L PE +Q E +++ E + HSFCKTLTASDTST
Sbjct: 76 VNVELRAEADSDEVYAQIMLQPEADQSELTSLDPELQDLE--KCTAHSFCKTLTASDTST 133
Query: 204 HGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQ 263
HGGFSVLRRHA+ECLP LDMS+ PP QELVAKDLHG EW FRHIFRG QPRRHLL
Sbjct: 134 HGGFSVLRRHAEECLPQLDMSQNPPCQELVAKDLHGTEWHFRHIFRG-----QPRRHLLT 188
Query: 264 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATA 323
+GWSVFVSSKRLVAGDAFIFLRGE+GELRVGVRR MRQ N+PSSVISSHSMHLGVLATA
Sbjct: 189 TGWSVFVSSKRLVAGDAFIFLRGESGELRVGVRRLMRQVNNMPSSVISSHSMHLGVLATA 248
Query: 324 WHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTG 383
HA+ TGT+F+V+YKPRTS +EF+V ++Y+E+ K N ++GMRFKMRFEG+EAPE+RF+G
Sbjct: 249 SHAISTGTLFSVFYKPRTSRSEFVVSVNKYLEAKKQNLSVGMRFKMRFEGDEAPERRFSG 308
Query: 384 TIVGIED---SDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMP 440
TI+GI W S W+ LKV+WDE S I RP+RVSPW++EP A P P
Sbjct: 309 TIIGIGSVPAMSKSPWADSDWKSLKVQWDEPSAIVRPDRVSPWELEPLDASNPQPPQPPL 368
Query: 441 RPKRPRANVVPS 452
R KR R PS
Sbjct: 369 RNKRARPPASPS 380
Score = 95.1 bits (235), Expect = 7e-18
Identities = 57/168 (33%), Positives = 87/168 (50%), Gaps = 20/168 (11%)
Query: 645 KISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFENDQKSE 704
++ +P C+LFG + S+ +E +L + +S + + Q S
Sbjct: 463 RVERKQEPTTMGCRLFGIEISSA---VEEALPAATVSGVGYDQTVLSVDVDSDQISQPSN 519
Query: 705 HSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLT 764
+KS P TS L+ QS RSCTKV +G+A+GR+VDLT
Sbjct: 520 GNKSDAPG--------------TSSERSP---LESQSRQVRSCTKVIMQGMAVGRAVDLT 562
Query: 765 KFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
K + Y +L ++L+++F+ +G+L K W VV+TD+E DMMLVGDDPW
Sbjct: 563 KLNGYGDLRSKLEEMFDIQGDLCPTLKRWQVVYTDDEDDMMLVGDDPW 610
>UniRef100_Q8S975 Auxin response factor 16 [Oryza sativa]
Length = 695
Score = 461 bits (1186), Expect = e-128
Identities = 287/730 (39%), Positives = 406/730 (55%), Gaps = 89/730 (12%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQAS-EQHMPVYDLRPKILCRV 144
L+ ELW ACAGPLV VP+ E VFYF QGH+EQ++ T+ A + + ++ + KILC+V
Sbjct: 11 LFAELWRACAGPLVEVPQRDERVFYFLQGHLEQLQEPTDPALLAEQIKMFQVPYKILCKV 70
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPP-PRFHVHSFCKTLTASDTST 203
+NV LKAE +TDEVFAQ+TL P+P+Q EN PP P PR VHSFCK LT SDTST
Sbjct: 71 VNVELKAETETDEVFAQITLQPDPDQ-ENLPTLPDPPLPEQPRPVVHSFCKILTPSDTST 129
Query: 204 HGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQ 263
HGGFSVLRRHA+ECLPPLDMS PTQEL+ KDLHG+EWRF+HI+RG QPRRHLL
Sbjct: 130 HGGFSVLRRHANECLPPLDMSMATPTQELITKDLHGSEWRFKHIYRG-----QPRRHLLT 184
Query: 264 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATA 323
+GWS FV+SK+L++GDAF++LR E GE RVGVRR +++Q +P+SVISS SMHLGVLA+A
Sbjct: 185 TGWSTFVTSKKLISGDAFVYLRSETGEQRVGVRRLVQKQSTMPASVISSQSMHLGVLASA 244
Query: 324 WHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTG 383
HA+ T ++F VYY+PR S +++IV ++Y+ + K + +GMRFKM FEGE+ P ++F+G
Sbjct: 245 SHAIKTNSIFLVYYRPRLSQSQYIVSVNKYLAASKVGFNVGMRFKMSFEGEDVPVKKFSG 304
Query: 384 TIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPK 443
TIVG E S +W S+W+ LKV+WDE +N+ PERVSPW+IE
Sbjct: 305 TIVG-EGDLSLQWSGSEWKSLKVQWDEVTNVNGPERVSPWEIETC------------DGT 351
Query: 444 RPRANVVPSSPDSSVLTREASSKVSMDPL-PTSGFQRVLQGQESSTLRGNLAESNDSYTA 502
P NV S + RE S + + L P F L G + + S + +
Sbjct: 352 APAINVPLQSATKNKRPREPSETIDLQSLEPAQEFW--LSGMPQQHEKTGIGSSEPNCIS 409
Query: 503 EKSVAWTPATDEEKMDAVSTSRRYGSENWMPMSRQEPTYSDLLSGFGSTREGKHNMLTQW 562
V W + AVS+S + + L F S+ +G L++
Sbjct: 410 GHQVVW--PGEHPGYGAVSSS----------VCQNPLVLESWLKDFNSSNKGVSPTLSE- 456
Query: 563 PVMPPGLSLNFLHSNMKGSAQGSDNATYQAQGNMRYSAFGDYSVLHGHKVENPHGNFLMP 622
+ + SN A + YQA+ S + G++ E N
Sbjct: 457 ----ISQKIFQVTSNEARIATWPARSAYQAEEPT--SKLSSNTAACGYRTEEVAPN---- 506
Query: 623 PPPPTQYESPHSRELSQKQMSAKISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATS 682
++K+ E K + + +LFG L+ T + + +++
Sbjct: 507 --------------------ASKVVE-GKKEPAMFRLFGVDLMKC-TSISTTTDDKSSVG 544
Query: 683 ETSSHMQISSQHHTFENDQKSEHSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSG 742
+ + + H ++ Q S SK +K + + DE +++Q+ Q +
Sbjct: 545 AGEASAKGTGSHE--DSGQLSAFSKVTK---EHIAADESPQEIQSHQNY----------- 588
Query: 743 SARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEG 802
+AR+ KV G A+GR+VDL Y++L EL+++F + ++ W V FTD+EG
Sbjct: 589 TARTRIKVQMHGNAVGRAVDLANLDGYEQLMNELEEMFNIK----DLKQKWKVAFTDDEG 644
Query: 803 DMMLVGDDPW 812
D M VGDDPW
Sbjct: 645 DTMEVGDDPW 654
>UniRef100_Q9XED8 Auxin response factor 9 [Arabidopsis thaliana]
Length = 638
Score = 457 bits (1177), Expect = e-127
Identities = 229/387 (59%), Positives = 286/387 (73%), Gaps = 9/387 (2%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHM-PVYDLRPKILCRV 144
LY ELW CAGPLV VP+ E V+YFPQGH+EQ+EAST Q M P++ L PKILC V
Sbjct: 9 LYDELWKLCAGPLVDVPQAQERVYYFPQGHMEQLEASTQQVDLNTMKPLFVLPPKILCNV 68
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
+NV L+AE DTDEV+AQ+TL+P + + + + P R VHSF K LTASDTSTH
Sbjct: 69 MNVSLQAEKDTDEVYAQITLIPVGTEVDEPMSPDPSPPELQRPKVHSFSKVLTASDTSTH 128
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLR+HA ECLPPLDM++Q PTQELVA+D+HG +W+F+HIFR GQPRRHLL +
Sbjct: 129 GGFSVLRKHATECLPPLDMTQQTPTQELVAEDVHGYQWKFKHIFR-----GQPRRHLLTT 183
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWS FV+SKRLVAGD F+FLRGENGELRVGVRRA QQ ++PSSVISSHSMHLGVLATA
Sbjct: 184 GWSTFVTSKRLVAGDTFVFLRGENGELRVGVRRANLQQSSMPSSVISSHSMHLGVLATAR 243
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HA T TMF VYYKPRTS +FI+ ++Y+E++ N +++GMRFKMRFEGE++PE+R++GT
Sbjct: 244 HATQTKTMFIVYYKPRTS--QFIISLNKYLEAMSNKFSVGMRFKMRFEGEDSPERRYSGT 301
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLPMPRPKR 444
++G++D S W SKWRCL+V WDE ++I RP +VSPW+IEP + + M + KR
Sbjct: 302 VIGVKDC-SPHWKDSKWRCLEVHWDEPASISRPNKVSPWEIEPFVNSENVPKSVMLKNKR 360
Query: 445 PRANVVPSSPDSSVLTREASSKVSMDP 471
PR S+ D + S V P
Sbjct: 361 PRQVSEVSALDVGITASNLWSSVLTQP 387
Score = 102 bits (253), Expect = 6e-20
Identities = 65/172 (37%), Positives = 96/172 (55%), Gaps = 19/172 (11%)
Query: 641 QMSAKISEAAKPKDSDCKLFGFSLLSSPTMLEPSLSQRNATSETSSHMQISSQHHTFEND 700
QM + + + ++ +LFG L+SS SL+ + + IS +D
Sbjct: 437 QMVSPVEQKKPETTANYRLFGIDLMSS------SLAVPEEKTAPMRPINISKPTMDSHSD 490
Query: 701 QKSEHSKSSKPADKLVIVDEHEKQLQTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRS 760
KSE SK S+ EK+ + ++ K+VQ K QS S RS TKV +G+ +GR+
Sbjct: 491 PKSEISKVSE-----------EKKQEPAEGSPKEVQSK-QSSSTRSRTKVQMQGVPVGRA 538
Query: 761 VDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGDMMLVGDDPW 812
VDL Y+EL ++++LF+ +GEL S + W +VFTD+EGDMMLVGDDPW
Sbjct: 539 VDLNALKGYNELIDDIEKLFDIKGELRS-RNQWEIVFTDDEGDMMLVGDDPW 589
>UniRef100_Q9C5W9 Auxin response factor 18 [Arabidopsis thaliana]
Length = 602
Score = 453 bits (1166), Expect = e-126
Identities = 231/370 (62%), Positives = 280/370 (75%), Gaps = 11/370 (2%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQA-SEQHMPVYDLRPKILCRV 144
LY ELW CAGPLV VPR E VFYFPQGH+EQ+ ASTNQ + + +PV+DL PKILCRV
Sbjct: 22 LYTELWKVCAGPLVEVPRAQERVFYFPQGHMEQLVASTNQGINSEEIPVFDLPPKILCRV 81
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
++V LKAE +TDEV+AQ+TL PE +Q E + P P + HSF K LTASDTSTH
Sbjct: 82 LDVTLKAEHETDEVYAQITLQPEEDQSE-PTSLDPPIVGPTKQEFHSFVKILTASDTSTH 140
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLR+HA ECLP LDM++ PTQELV +DLHG EWRF+HIFRG QPRRHLL +
Sbjct: 141 GGFSVLRKHATECLPSLDMTQATPTQELVTRDLHGFEWRFKHIFRG-----QPRRHLLTT 195
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWS FVSSKRLVAGDAF+FLRGENG+LRVGVRR R Q +P+SVISS SMHLGVLATA
Sbjct: 196 GWSTFVSSKRLVAGDAFVFLRGENGDLRVGVRRLARHQSTMPTSVISSQSMHLGVLATAS 255
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HAV T T+F V+YKPR S +FIV ++YME++K+ +++G RF+MRFEGEE+PE+ FTGT
Sbjct: 256 HAVRTTTIFVVFYKPRIS--QFIVGVNKYMEAIKHGFSLGTRFRMRFEGEESPERIFTGT 313
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALA-PPALNPLPMPRPK 443
IVG D S +WP SKWR L+V+WDE + + RP++VSPW+IEP LA P P P+ K
Sbjct: 314 IVGSGDL-SSQWPASKWRSLQVQWDEPTTVQRPDKVSPWEIEPFLATSPISTPAQQPQSK 372
Query: 444 RPRANVVPSS 453
R+ + S
Sbjct: 373 CKRSRPIEPS 382
Score = 92.4 bits (228), Expect = 5e-17
Identities = 65/208 (31%), Positives = 95/208 (45%), Gaps = 20/208 (9%)
Query: 619 FLMPPPPPTQYESPHSRELSQKQMSAKISEAAKP-------KDSDCKLFGFSLLSSPTM- 670
FL P T + P S+ + + + A P + D L P++
Sbjct: 356 FLATSPISTPAQQPQSKCKRSRPIEPSVKTPAPPSFLYSLPQSQDSINASLKLFQDPSLE 415
Query: 671 -LEPSLSQRNATSETSSHMQISSQHHTFENDQKSEHS----KSSKPADKLVIVDEHEKQL 725
+ S N+ + + + F D S + +P D E
Sbjct: 416 RISGGYSSNNSFKPETPPPPTNCSYRLFGFDLTSNSPAPIPQDKQPMDTCGAAKCQEPIT 475
Query: 726 QTSQPHVKDVQLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGE 785
TS K Q ++RS TKV +GIA+GR+VDLT YDEL EL+++FE +G+
Sbjct: 476 PTSMSEQKKQQ------TSRSRTKVQMQGIAVGRAVDLTLLKSYDELIDELEEMFEIQGQ 529
Query: 786 LISPQKDWLVVFTDNEGDMMLVGDDPWH 813
L++ K W+VVFTD+EGDMML GDDPW+
Sbjct: 530 LLARDK-WIVVFTDDEGDMMLAGDDPWN 556
>UniRef100_Q9ZPY6 Auxin response factor 11 [Arabidopsis thaliana]
Length = 601
Score = 452 bits (1164), Expect = e-125
Identities = 235/389 (60%), Positives = 290/389 (74%), Gaps = 16/389 (4%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQAS-EQHMPVYDLRPKILCRV 144
LY ELW ACAGPLV VPR GE VFYFPQGH+EQ+ ASTNQ +Q +PV++L PKILCRV
Sbjct: 18 LYTELWKACAGPLVEVPRYGERVFYFPQGHMEQLVASTNQGVVDQEIPVFNLPPKILCRV 77
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPPPRFHVHSFCKTLTASDTSTH 204
++V LKAE +TDEV+AQ+TL PE +Q E + P P + V SF K LTASDTSTH
Sbjct: 78 LSVTLKAEHETDEVYAQITLQPEEDQSE-PTSLDPPLVEPAKPTVDSFVKILTASDTSTH 136
Query: 205 GGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQS 264
GGFSVLR+HA ECLP LDM++ PTQELVA+DLHG EWRF+HIFRG QPRRHLL +
Sbjct: 137 GGFSVLRKHATECLPSLDMTQPTPTQELVARDLHGYEWRFKHIFRG-----QPRRHLLTT 191
Query: 265 GWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAW 324
GWS FV+SKRLVAGDAF+FLRGE G+LRVGVRR +QQ +P+SVISS SM LGVLATA
Sbjct: 192 GWSTFVTSKRLVAGDAFVFLRGETGDLRVGVRRLAKQQSTMPASVISSQSMRLGVLATAS 251
Query: 325 HAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGT 384
HAV T T+F V+YKPR S +FI+ ++YM ++KN +++GMR++MRFEGEE+PE+ FTGT
Sbjct: 252 HAVTTTTIFVVFYKPRIS--QFIISVNKYMMAMKNGFSLGMRYRMRFEGEESPERIFTGT 309
Query: 385 IVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPALNPLP-----M 439
I+G D S +WP SKWR L+++WDE S+I RP +VSPW+IEP +P AL P P
Sbjct: 310 IIGSGDL-SSQWPASKWRSLQIQWDEPSSIQRPNKVSPWEIEP-FSPSALTPTPTQQQSK 367
Query: 440 PRPKRPRANVVPSSPDSSVLTREASSKVS 468
+ RP + + S SS L+ + S S
Sbjct: 368 SKRSRPISEITGSPVASSFLSSFSQSHES 396
Score = 97.4 bits (241), Expect = 2e-18
Identities = 74/209 (35%), Positives = 109/209 (51%), Gaps = 23/209 (11%)
Query: 620 LMPPPPPTQYESPHSRELSQKQMSAKISEAAKPKDSDCKLFGFSLLSSPTML----EPSL 675
L P P Q +S SR +S+ S S S F S S+P++ +P+
Sbjct: 357 LTPTPTQQQSKSKRSRPISEITGSPVAS-------SFLSSFSQSHESNPSVKLLFQDPAT 409
Query: 676 SQRNATSETSSHMQ-------ISSQHHTFENDQKSEHSKSSKPADK-LVIVDEH--EKQL 725
+ + S SS +Q ++S F D S+ + ++ P DK L+ VD + +
Sbjct: 410 ERNSNKSVFSSGLQCKITEAPVTSSCRLFGFDLTSKPASATIPHDKQLISVDSNISDSTT 469
Query: 726 QTSQPHVKDV-QLKPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRG 784
+ P+ + + + Q S RS KV +G A+GR+VDLT YDEL EL+++FE G
Sbjct: 470 KCQDPNSSNSPKEQKQQTSTRSRIKVQMQGTAVGRAVDLTLLRSYDELIKELEKMFEIEG 529
Query: 785 ELISPQKDWLVVFTDNEGDMMLVGDDPWH 813
EL SP+ W +VFTD+EGD MLVGDDPW+
Sbjct: 530 EL-SPKDKWAIVFTDDEGDRMLVGDDPWN 557
>UniRef100_Q9LI22 EST C26826(C50159) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 613
Score = 420 bits (1079), Expect = e-115
Identities = 210/372 (56%), Positives = 273/372 (72%), Gaps = 15/372 (4%)
Query: 86 LYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQAS-EQHMPVYDLRPKILCRV 144
L+ ELW ACAGPLV VP+ E VFYF QGH+EQ++ T+ A + + ++ + KILC+V
Sbjct: 15 LFAELWRACAGPLVEVPQRDERVFYFLQGHLEQLQEPTDPALLAEQIKMFQVPYKILCKV 74
Query: 145 INVMLKAEPDTDEVFAQVTLVPEPNQDENAVEKEAPPAPP-PRFHVHSFCKTLTASDTST 203
+NV LKAE +TDEVFAQ+TL P+P+Q EN PP P PR VHSFCK LT SDTST
Sbjct: 75 VNVELKAETETDEVFAQITLQPDPDQ-ENLPTLPDPPLPEQPRPVVHSFCKILTPSDTST 133
Query: 204 HGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELISGQPRRHLLQ 263
HGGFSVLRRHA+ECLPPLDMS PTQEL+ KDLHG+EWRF+HI+RG QPRRHLL
Sbjct: 134 HGGFSVLRRHANECLPPLDMSMATPTQELITKDLHGSEWRFKHIYRG-----QPRRHLLT 188
Query: 264 SGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHLGVLATA 323
+GWS FV+SK+L++GDAF++LR E GE RVGVRR +++Q +P+SVISS SMHLGVLA+A
Sbjct: 189 TGWSTFVTSKKLISGDAFVYLRSETGEQRVGVRRLVQKQSTMPASVISSQSMHLGVLASA 248
Query: 324 WHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTG 383
HA+ T ++F VYY+PR S +++IV ++Y+ + K + +GMRFKM FEGE+ P ++F+G
Sbjct: 249 SHAIKTNSIFLVYYRPRLSQSQYIVSVNKYLAASKVGFNVGMRFKMSFEGEDVPVKKFSG 308
Query: 384 TIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIE------PALAPPALNPL 437
TIVG E S +W S+W+ LKV+WDE +N+ PERVSPW+IE PA+ P +
Sbjct: 309 TIVG-EGDLSLQWSGSEWKSLKVQWDEVTNVNGPERVSPWEIETCDGTAPAINVPLQSAT 367
Query: 438 PMPRPKRPRANV 449
RP+ P +
Sbjct: 368 KNKRPREPSETI 379
Score = 62.8 bits (151), Expect = 4e-08
Identities = 30/69 (43%), Positives = 43/69 (61%), Gaps = 4/69 (5%)
Query: 744 ARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTDNEGD 803
A + +KV G A+GR+VDL Y++L EL+++F + ++ W V FTD+EGD
Sbjct: 508 APNASKVQMHGNAVGRAVDLANLDGYEQLMNELEEMFNIK----DLKQKWKVAFTDDEGD 563
Query: 804 MMLVGDDPW 812
M VGDDPW
Sbjct: 564 TMEVGDDPW 572
>UniRef100_Q9ZTX9 Auxin response factor 4 [Arabidopsis thaliana]
Length = 788
Score = 402 bits (1033), Expect = e-110
Identities = 202/413 (48%), Positives = 279/413 (66%), Gaps = 20/413 (4%)
Query: 84 AALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCR 143
+++Y ELWHACAGPL +P++G +V YFPQGH+EQ +A + +S +P +DL P+I+CR
Sbjct: 60 SSIYSELWHACAGPLTCLPKKGNVVVYFPQGHLEQ-DAMVSYSSPLEIPKFDLNPQIVCR 118
Query: 144 VINVMLKAEPDTDEVFAQVTLVP----------EPNQDENAVEKEAPPAPPPRFHVHSFC 193
V+NV L A DTDEV+ QVTL+P E E+E + + H FC
Sbjct: 119 VVNVQLLANKDTDEVYTQVTLLPLQEFSMLNGEGKEVKELGGEEERNGSSSVKRTPHMFC 178
Query: 194 KTLTASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRGELI 253
KTLTASDTSTHGGFSV RR A++C PLD +Q P+QEL+AKDLHG EW+FRHI+RG
Sbjct: 179 KTLTASDTSTHGGFSVPRRAAEDCFAPLDYKQQRPSQELIAKDLHGVEWKFRHIYRG--- 235
Query: 254 SGQPRRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSH 313
QPRRHLL +GWS+FVS K LV+GDA +FLR E GELR+G+RRA R + +P S+I +
Sbjct: 236 --QPRRHLLTTGWSIFVSQKNLVSGDAVLFLRDEGGELRLGIRRAARPRNGLPDSIIEKN 293
Query: 314 SMHLGVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEG 373
S +L+ +AV T +MF V+Y PR + AEF++PY++Y+ S+++ IG RF+MRFE
Sbjct: 294 SCS-NILSLVANAVSTKSMFHVFYSPRATHAEFVIPYEKYITSIRSPVCIGTRFRMRFEM 352
Query: 374 EEAPEQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALAPPA 433
+++PE+R G + G+ D D RWP SKWRCL VRWDE+ ERVSPW+I+P+++ P
Sbjct: 353 DDSPERRCAGVVTGVCDLDPYRWPNSKWRCLLVRWDESFVSDHQERVSPWEIDPSVSLPH 412
Query: 434 LNPLPMPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQES 486
L+ PRPKRP A ++ ++P + +T+ + + S +VLQGQE+
Sbjct: 413 LSIQSSPRPKRPWAGLLDTTPPGNPITKRGGFLDFEESVRPS---KVLQGQEN 462
Score = 100 bits (250), Expect = 1e-19
Identities = 42/76 (55%), Positives = 58/76 (76%)
Query: 738 KPQSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVF 797
KPQS S R CTKVHK+G +GR++DL++ + YD+L EL++LF G L P+K W +++
Sbjct: 658 KPQSSSKRICTKVHKQGSQVGRAIDLSRLNGYDDLLMELERLFNMEGLLRDPEKGWRILY 717
Query: 798 TDNEGDMMLVGDDPWH 813
TD+E DMM+VGDDPWH
Sbjct: 718 TDSENDMMVVGDDPWH 733
>UniRef100_Q6L8T9 Auxin response factor 5 [Cucumis sativus]
Length = 733
Score = 398 bits (1023), Expect = e-109
Identities = 204/420 (48%), Positives = 279/420 (65%), Gaps = 25/420 (5%)
Query: 84 AALYRELWHACAGPLVTVPREGELVFYFPQGHIEQVEASTNQASEQHMPVYDLRPKILCR 143
++ Y ELWHACAGPL ++P++G +V YFPQGH+EQ+ AS + S M +DL+P ILCR
Sbjct: 49 SSTYLELWHACAGPLTSLPKKGNVVVYFPQGHLEQI-ASASPFSPMEMRTFDLQPHILCR 107
Query: 144 VINVMLKAEPDTDEVFAQVTLVPEPNQ----------DE---NAVEKEAPPAPPPRFHVH 190
VINV L A + DEV+ Q+TL P P +E N + + P + H
Sbjct: 108 VINVHLLANKENDEVYTQLTLRPLPELLGTGVAGKELEELALNGADGDGSGGSPTKSTPH 167
Query: 191 SFCKTLTASDTSTHGGFSVLRRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFRG 250
FCKTLTASDTSTHGGFSV RR A++C PPLD ++ P+QEL+AKDLHG EWRFRHI+RG
Sbjct: 168 MFCKTLTASDTSTHGGFSVPRRAAEDCFPPLDYTQLRPSQELIAKDLHGVEWRFRHIYRG 227
Query: 251 ELISGQPRRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVI 310
QPRRHLL +GWS+FVS K L++GDA +FLRGENGELR+G+RRA+R + +P S++
Sbjct: 228 -----QPRRHLLTTGWSIFVSQKNLISGDAVLFLRGENGELRLGIRRAVRPRNGLPDSIV 282
Query: 311 SSHSMHLGVLATAWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMR 370
+ + LA A+ T + F V+Y PR A+FI+ +Y++S+ N ++G RFKMR
Sbjct: 283 GNQNSCANDLARVVKAISTKSTFDVFYNPRAYHAQFIISCQKYVKSINNPVSVGTRFKMR 342
Query: 371 FEGEEAPEQRFTGTIVGIEDSDSKRWPTSKWRCLKVRWDETSNIPRPERVSPWKIEPALA 430
FE +++PE++F G +VGI D DS RWP SKWRCL VRWD+ S+ ERVSPW+I+P+++
Sbjct: 343 FEMDDSPERKFNGVVVGISDMDSFRWPNSKWRCLTVRWDKDSD--HQERVSPWEIDPSVS 400
Query: 431 PPALNPLPMPRPKRPRANVVPSSPDSSVLTREASSKVSMDPLPTSGFQRVLQGQESSTLR 490
P L+ PR K+ R ++ + P+++ R MD + +VLQGQE +LR
Sbjct: 401 LPPLSVQSSPRLKKLRTSLQAAPPNNAFTGRGG----FMDFEDSVRSSKVLQGQEICSLR 456
Score = 91.7 bits (226), Expect = 8e-17
Identities = 39/74 (52%), Positives = 54/74 (72%)
Query: 740 QSGSARSCTKVHKKGIALGRSVDLTKFSDYDELTAELDQLFEFRGELISPQKDWLVVFTD 799
QS RSCTKVHK+G +GR++DL++ + Y +L +EL++LF G L P K W V++TD
Sbjct: 600 QSSGKRSCTKVHKQGSLVGRAIDLSRLNGYTDLISELERLFSMEGLLKDPDKGWRVLYTD 659
Query: 800 NEGDMMLVGDDPWH 813
NE D+M+VGD PWH
Sbjct: 660 NENDVMVVGDYPWH 673
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,426,587,971
Number of Sequences: 2790947
Number of extensions: 62358716
Number of successful extensions: 217895
Number of sequences better than 10.0: 455
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 376
Number of HSP's that attempted gapping in prelim test: 216512
Number of HSP's gapped (non-prelim): 1133
length of query: 830
length of database: 848,049,833
effective HSP length: 136
effective length of query: 694
effective length of database: 468,481,041
effective search space: 325125842454
effective search space used: 325125842454
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC126794.7


