

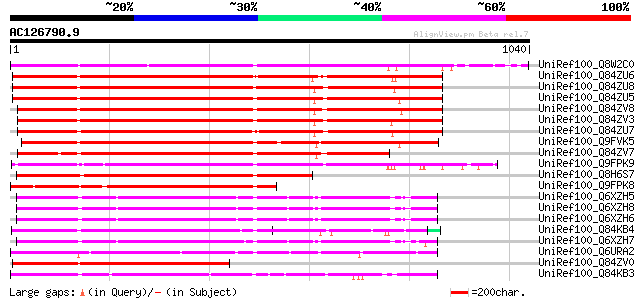
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.9 + phase: 0 /pseudo
(1040 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Gl... 884 0.0
UniRef100_Q84ZU6 R 1 protein [Glycine max] 815 0.0
UniRef100_Q84ZU8 R 10 protein [Glycine max] 806 0.0
UniRef100_Q84ZU5 R 8 protein [Glycine max] 803 0.0
UniRef100_Q84ZV8 R 3 protein [Glycine max] 796 0.0
UniRef100_Q84ZV3 R 4 protein [Glycine max] 795 0.0
UniRef100_Q84ZU7 R 5 protein [Glycine max] 780 0.0
UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max] 763 0.0
UniRef100_Q84ZV7 R 12 protein [Glycine max] 716 0.0
UniRef100_Q9FPK9 Putative resistance protein [Glycine max] 704 0.0
UniRef100_Q8H6S7 Resistance protein KR3 [Glycine max] 579 e-163
UniRef100_Q9FPK8 Putative resistance protein [Glycine max] 505 e-141
UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuber... 498 e-139
UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum] 493 e-138
UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuber... 492 e-137
UniRef100_Q84KB4 MRGH5 [Cucumis melo] 487 e-136
UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuber... 483 e-134
UniRef100_Q6URA2 TIR-NBS-LRR type R protein 7 [Malus baccata] 482 e-134
UniRef100_Q84ZV0 R 14 protein [Glycine max] 478 e-133
UniRef100_Q84KB3 MRGH63 [Cucumis melo] 475 e-132
>UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Glycine max]
Length = 1124
Score = 884 bits (2285), Expect = 0.0
Identities = 528/1137 (46%), Positives = 686/1137 (59%), Gaps = 134/1137 (11%)
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEIT 60
M SSS+S SY + VFLSFRG DTR GFTGNLYKAL+D+GI+TF+D + RGD+IT
Sbjct: 1 MAKQSSSSSFSYRFSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKKIPRGDQIT 60
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHR 120
L KAIEESRIFI V S NYASSSFCL+EL +I+ K KG L+LPVF+ V+P+ VR+
Sbjct: 61 SGLEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKVDPSDVRNH 120
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPG--YEYKLIGKIVK 178
GS+G+AL HEK+F++ +ME+L+ WK AL++ ANLSGYH G YEY+ I +IV+
Sbjct: 121 TGSFGKALTNHEKKFKST-NDMEKLETWKMALNKVANLSGYHHFKHGEEYEYEFIQRIVE 179
Query: 179 YISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIY 238
+S KI+R PLHVA YPVGL+SR+Q+VK+LLD GSDD VHM+GI+G+GG+GK+TLA +Y
Sbjct: 180 LVSKKINRAPLHVADYPVGLESRIQEVKALLDVGSDDVVHMLGIHGLGGVGKTTLAAAVY 239
Query: 239 NFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRK 298
N +AD FE CFL +VRE S ++ L++LQ LL + G E KL V +GI +I+ RL +K
Sbjct: 240 NSIADHFEALCFLQNVRETSKKHGLQHLQRNLLSEMAG-EDKLIGVKQGISIIEHRLRQK 298
Query: 299 KILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEA 358
K+LLILDDVD +QL ALAG D FG GSRVIITTR+K LL+ HG+E T+ V LNE A
Sbjct: 299 KVLLILDDVDKREQLQALAGRPDLFGPGSRVIITTRDKQLLACHGVERTYEVNELNEEYA 358
Query: 359 LELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDR 418
LELL W AFK +KV Y+D+LNRA YA GLPL LEV+GSNL GK+IE W LD Y R
Sbjct: 359 LELLNWKAFKLEKVDPFYKDVLNRAATYASGLPLALEVIGSNLSGKNIEQWISALDRYKR 418
Query: 419 IPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLA 478
IPNKEIQ+ILKVSYDALEE+EQS+FLDIACCFK Y E +DIL AH+ HC+ HH+GVL
Sbjct: 419 IPNKEIQEILKVSYDALEEDEQSIFLDIACCFKKYDLAEVQDILHAHHGHCMKHHIGVLV 478
Query: 479 GKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKEN 538
KSL+KIS V LHDLI+DMGKE+VR+ESP+EPG+RSRLW DI+ VL+EN
Sbjct: 479 EKSLIKISL------DGYVTLHDLIEDMGKEIVRKESPQEPGKRSRLWLPTDIVQVLEEN 532
Query: 539 TGTSKIEMIYMNLHSM--ESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLK 596
GTS I +I MN +S E I G AFKKM LKTLII +G FS G K+ P SLRVL+
Sbjct: 533 KGTSHIGIICMNFYSSFEEVEIQWDGDAFKKMKNLKTLIIRSGHFSKGPKHFPKSLRVLE 592
Query: 597 WKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPD 656
W S + I + C ++ + +KF N+ L D C++LT IPD
Sbjct: 593 WWRYPSHYFPYDFQMEKLAIFNLPDCGFTSRELAAMLKKKFVNLTSLNFDSCQHLTLIPD 652
Query: 657 VSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYE 716
VS + +L+KLSF CDNL IH S+G L KL L A GC +L++F P+ L SL++L L
Sbjct: 653 VSCVPHLQKLSFKDCDNLYAIHPSVGFLEKLRILDAEGCSRLKNFPPIKLTSLEQLKLGF 712
Query: 717 CECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELH------------------- 757
C L+NFPE+L KM +I E+D+ T + + P SFQNL+ L
Sbjct: 713 CHSLENFPEILGKMENITELDLEQTPVKKFPLSFQNLTRLETVLLCFPRNQANGCTGIFL 772
Query: 758 ----------ELTVTSGMKFPKIVF---------------SNMTKLSLSFFNLSDECLPI 792
EL G+ + +F SN+ L L NLSD+ PI
Sbjct: 773 SNICPMQESPELINVIGVGWEGCLFRKEDEGAENVSLTTSSNVQFLDLRNCNLSDDFFPI 832
Query: 793 VLKWCVNMTHLDLSFSNFKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYC 852
L N+ L+LS +NF ++PEC++EC L + + CE L EIRGIPPNLK A C
Sbjct: 833 ALPCFANVMELNLSGNNFTVIPECIKECRFLTTLYLNYCERLREIRGIPPNLKYFYAEEC 892
Query: 853 KSLSSSSRRMLMSQ--------------------------------------PSITCIFI 874
SL+SS R ML+SQ P+I I
Sbjct: 893 LSLTSSCRSMLLSQELHEAGRTFFYLPGAKIPEWFDFQTSEFPISFWFRNKFPAIAICHI 952
Query: 875 LPKGNEYATS----------VNVFVNGYEIEIGCYWSLFFFTDHTTLFHTSKLNELIKTQ 924
+ + E+++S V +NG + S+ +D T LF +L +
Sbjct: 953 IKRVAEFSSSRGWTFRPNIRTKVIING---NANLFNSVVLGSDCTCLF------DLRGER 1003
Query: 925 CEYNIEKGLLKNEWIYVEFKLKDHENS---VYAQRGIHVWNEKSNTEEENVVFTDPCITK 981
N+++ LL+NEW + E + + + G+HV ++SN E+ + F+DPC K
Sbjct: 1004 VTDNLDEALLENEWNHAEVTCPGFTFTFAPTFIKTGLHVLKQESNMED--IRFSDPC-RK 1060
Query: 982 TKSDEYLNQSNNTSLSQFEPPLKKQRLVEVGVSETEEDINASLQQQELMKEEQRKTW 1038
TK D N S K QR V V++T+ +QQQ+LM + W
Sbjct: 1061 TKLDNDFNSSKP----------KNQRWVGNDVAKTQ-----VVQQQQLMGSFLSRMW 1102
>UniRef100_Q84ZU6 R 1 protein [Glycine max]
Length = 902
Score = 815 bits (2105), Expect = 0.0
Identities = 465/908 (51%), Positives = 594/908 (65%), Gaps = 65/908 (7%)
Query: 6 SSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLK 65
++T+ S Y VFL+FRG DTRYGFTGNLYKAL DKGI+TF D++ L GD+ITP+L K
Sbjct: 2 AATTRSLASIYDVFLNFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGDDITPALSK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYG 125
AI+ESRI I V S NYASSSFCLDELV I+HC K +G LV+PVF V+P+ VRH KGSYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KREGLLVIPVFHNVDPSAVRHLKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
EA+A+H+KRF+ E+LQ W+ AL Q A+LSGYH YEYK IG IV+ +S KI
Sbjct: 121 EAMAKHQKRFK---AKKEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIGNIVEEVSRKI 177
Query: 185 SRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQ 244
+ PLHVA YPVGL S+V +V LLD GSDD VH++GI+G+GGLGK+TLA +YNF+A
Sbjct: 178 NCAPLHVADYPVGLGSQVIEVMKLLDVGSDDLVHIIGIHGMGGLGKTTLALAVYNFIALH 237
Query: 245 FEGSCFLHDVRENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLI 303
F+ SCFL +VRE S ++ LK+ Q LL K G +I L EG +I+ RL RKK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHFQSILLSKLLGEKDITLTSWQEGASMIQHRLRRKKVLLI 297
Query: 304 LDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLR 363
LDDVD +QL A+ G DWFG GSRVIITTR+K LL H +E T+ V+ LN AL+LL
Sbjct: 298 LDDVDKREQLEAIVGRSDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNHNAALQLLT 357
Query: 364 WMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKE 423
W AFK +K+ Y+D+LNR V YA GLPL LEV+GS+LFGK++ +W+ ++ Y RIP+ E
Sbjct: 358 WNAFKREKIDPIYDDVLNRVVTYASGLPLALEVIGSDLFGKTVAEWESAVEHYKRIPSDE 417
Query: 424 IQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLV 483
I KILKVS+DAL EE+++VFLDIACCFKGY+W E +DIL A Y +C HH+GVL KSL+
Sbjct: 418 ILKILKVSFDALGEEQKNVFLDIACCFKGYKWTEVDDILRAFYGNCKKHHIGVLVEKSLI 477
Query: 484 KISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSK 543
K++ Y SG+ V +HDLI+DMG+E+ RQ SP+EP + RLW +DI VLK NTGTSK
Sbjct: 478 KLNCY-DSGT---VEMHDLIQDMGREIERQRSPEEPWKCKRLWSPKDIFQVLKHNTGTSK 533
Query: 544 IEMIYMN--LHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCL 601
IE+I ++ + E ++ AF KM LK LII NG FS G Y P L VL+W
Sbjct: 534 IEIICLDFSISDKEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPEGLTVLEWHRYP 593
Query: 602 SKCL------SSSILNKA--SEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTH 653
S CL ++ ++ K S ITS F S KF ++ VL D CE+LT
Sbjct: 594 SNCLPYNFHPNNLLICKLPDSSITS----FELHGPS------KFWHLTVLNFDQCEFLTQ 643
Query: 654 IPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLI 713
IPDVS L NL++LSF C++LI + +SIG LNKL+ LSAYGCRKL F PL L SL+ L
Sbjct: 644 IPDVSDLPNLKELSFDWCESLIAVDDSIGFLNKLKKLSAYGCRKLRSFPPLNLTSLETLQ 703
Query: 714 LYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSG--------- 764
L C L+ FPE+L +M +IK +D+ I ELPFSFQNL L LT+ S
Sbjct: 704 LSGCSSLEYFPEILGEMENIKALDLDGLPIKELPFSFQNLIGLCRLTLNSCGIIQLPCSL 763
Query: 765 MKFPKI----------------------VFSNMTKLSLSFF----NLSDECLPIVLKWCV 798
P++ V S ++ L F NL D+ K
Sbjct: 764 AMMPELSVFRIENCNRWHWVESEEGEEKVGSMISSKELWFIAMNCNLCDDFFLTGSKRFT 823
Query: 799 NMTHLDLSFSNFKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSS 858
+ +LDLS +NF ILPE +E L + V CE L+EIRG+PPNL+ AR C SL+SS
Sbjct: 824 RVEYLDLSGNNFTILPEFFKELQFLRALMVSDCEHLQEIRGLPPNLEYFDARNCASLTSS 883
Query: 859 SRRMLMSQ 866
++ ML++Q
Sbjct: 884 TKSMLLNQ 891
>UniRef100_Q84ZU8 R 10 protein [Glycine max]
Length = 901
Score = 806 bits (2083), Expect = 0.0
Identities = 450/906 (49%), Positives = 590/906 (64%), Gaps = 61/906 (6%)
Query: 6 SSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLK 65
++T+ S Y VFL+FRG DTRYGFTGNLY+AL DKGI+TF D+ L RG+EITP+LLK
Sbjct: 2 AATTRSRASIYDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYG 125
AI+ESRI I V S NYASSSFCLDELV I+HC K++G LV+PVF+ V+P+ VRH+KGSYG
Sbjct: 62 AIQESRIAITVLSKNYASSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDVRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
+A+H+KRF+ E+LQ W+ AL Q A+L GYH YEYK I IV+ +S +I
Sbjct: 121 VEMAKHQKRFK---AKKEKLQKWRIALKQVADLCGYHFKDGDAYEYKFIQSIVEQVSREI 177
Query: 185 SRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQ 244
+R PLHVA YPVGL S+V +V+ LLD GS D VH++GI+G+GGLGK+TLA +YN +A
Sbjct: 178 NRAPLHVADYPVGLGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVYNLIALH 237
Query: 245 FEGSCFLHDVRENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLI 303
F+ SCFL +VRE S ++ LK+LQ LL K G +I L EG +I+ RL RKK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLI 297
Query: 304 LDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLR 363
LDDVD +QL A+ G DWFG GSRVIITTR+K LL H +E T+ V+ LN++ AL+LL+
Sbjct: 298 LDDVDKREQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLK 357
Query: 364 WMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKE 423
W AFK +K+ YED+LNR V YA GLPL LEV+GSNLFGK++ +W+ ++ Y RIP+ E
Sbjct: 358 WNAFKREKIDPSYEDVLNRVVTYASGLPLALEVIGSNLFGKTVAEWESAMEHYKRIPSDE 417
Query: 424 IQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLV 483
I +ILKVS+DAL EE+++VFLDIACCF+GY+W E +DIL A Y +C HH+GVL KSL+
Sbjct: 418 ILEILKVSFDALGEEQKNVFLDIACCFRGYKWTEVDDILRALYGNCKKHHIGVLVEKSLI 477
Query: 484 KISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSK 543
K++ Y + V +HDLI+DM +E+ R+ SP+EPG+ RLW +DII V K+NTGTSK
Sbjct: 478 KLNCY----GTDTVEMHDLIQDMAREIERKRSPQEPGKCKRLWLPKDIIQVFKDNTGTSK 533
Query: 544 IEMIYMN--LHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCL 601
IE+I ++ + E ++ AF KM LK LII N FS G Y P LRVL+W
Sbjct: 534 IEIICLDSSISDKEETVEWNENAFMKMENLKILIIRNDKFSKGPNYFPEGLRVLEWHRYP 593
Query: 602 SKCLSSS------ILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIP 655
S CL S+ ++ K + F F K F ++ VL D C++LT IP
Sbjct: 594 SNCLPSNFHPNNLVICKLPDSCMTSFEFHGPSK--------FGHLTVLKFDNCKFLTQIP 645
Query: 656 DVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILY 715
DVS L NL +LSF C++L+ + +SIG LNKL+ LSAYGC KL+ F PL L SL+ L L
Sbjct: 646 DVSDLPNLRELSFEECESLVAVDDSIGFLNKLKKLSAYGCSKLKSFPPLNLTSLQTLELS 705
Query: 716 ECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSG--MKFP----- 768
+C L+ FPE++ +M +IK + + I EL FSFQNL L LT+ S +K P
Sbjct: 706 QCSSLEYFPEIIGEMENIKHLFLYGLPIKELSFSFQNLIGLRWLTLRSCGIVKLPCSLAM 765
Query: 769 ----------------------------KIVFSNMTKLSLSFFNLSDECLPIVLKWCVNM 800
I S + S NL D+ K +
Sbjct: 766 MPELFEFHMEYCNRWQWVESEEGEKKVGSIPSSKAHRFSAKDCNLCDDFFLTGFKTFARV 825
Query: 801 THLDLSFSNFKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSSSR 860
HL+LS +NF ILPE +E L + V CE L+EIRG+PPNL+ AR C SL+SSS+
Sbjct: 826 GHLNLSGNNFTILPEFFKELQLLRSLMVSDCEHLQEIRGLPPNLEYFDARNCASLTSSSK 885
Query: 861 RMLMSQ 866
ML++Q
Sbjct: 886 NMLLNQ 891
>UniRef100_Q84ZU5 R 8 protein [Glycine max]
Length = 892
Score = 803 bits (2075), Expect = 0.0
Identities = 457/903 (50%), Positives = 591/903 (64%), Gaps = 59/903 (6%)
Query: 6 SSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLK 65
++T+ S Y Y VFLSF G DTR GFTG LYKAL D+GI TFID L+RGDEI P+L
Sbjct: 2 AATTRSLAYNYDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSN 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYG 125
AI+ESRI I V S NYASSSFCLDELV I+HC K++G LV+PVF+ V+P+ VRH+KGSYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
EA+A+H+KRF+ N E+LQ W+ AL Q A+LSGYH YEY+ IG IV+ IS K
Sbjct: 121 EAMAKHQKRFK---ANKEKLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRKF 177
Query: 185 SRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQ 244
SR LHVA YPVGL+S V +V LLD GS D VH++GI+G+GGLGK+TLA ++NF+A
Sbjct: 178 SRASLHVADYPVGLESEVTEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALH 237
Query: 245 FEGSCFLHDVRENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLI 303
F+ SCFL +VRE S ++ LK+LQ LL K G +I L EG +I+ RL RKK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLI 297
Query: 304 LDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLR 363
LDDVD +QL A+ G DWFG GSRVIITTR+K LL H +E T+ V+ LN++ AL+LL
Sbjct: 298 LDDVDKRQQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLT 357
Query: 364 WMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKE 423
W AFK +K+ YED+LNR V YA GLPL LEV+GSNLF K++ +W+ ++ Y RIP+ E
Sbjct: 358 WNAFKREKIDPSYEDVLNRVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDE 417
Query: 424 IQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLV 483
IQ+ILKVS+DAL EE+++VFLDIACCFKGY+W E ++IL Y +C HH+GVL KSLV
Sbjct: 418 IQEILKVSFDALGEEQKNVFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLV 477
Query: 484 KISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSK 543
K+S + V +HD+I+DMG+E+ RQ SP+EPG+ RL +DII VLK+NTGTSK
Sbjct: 478 KVSC------CDTVEMHDMIQDMGREIERQRSPEEPGKCKRLLLPKDIIQVLKDNTGTSK 531
Query: 544 IEMIYMN--LHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCL 601
IE+I ++ + E ++ AF KM LK LII N FS G Y P LRVL+W
Sbjct: 532 IEIICLDFSISDKEETVEWNENAFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYP 591
Query: 602 SKCLSSS------ILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIP 655
S CL S+ ++ K + + F F K K ++ VL D CE+LT IP
Sbjct: 592 SNCLPSNFDPINLVICKLPDSSITSFEFHGSSK-------KLGHLTVLNFDRCEFLTKIP 644
Query: 656 DVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILY 715
DVS L NL++LSF C++L+ + +SIG LNKL+ LSAYGCRKL F PL L SL+ L L
Sbjct: 645 DVSDLPNLKELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLTSFPPLNLTSLETLNLG 704
Query: 716 ECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTS-GMKFPKIVFSN 774
C L+ FPE+L +M +I + + + I ELPFSFQNL L L + S G+ + +
Sbjct: 705 GCSSLEYFPEILGEMKNITVLALHDLPIKELPFSFQNLIGLLFLWLDSCGIVQLRCSLAT 764
Query: 775 MTKLS---------------------------LSF----FNLSDECLPIVLKWCVNMTHL 803
M KL LSF NL D+ I K ++ +L
Sbjct: 765 MPKLCEFCITDSCNRWQWVESEEGEEKVVGSILSFEATDCNLCDDFFFIGSKRFAHVGYL 824
Query: 804 DLSFSNFKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSSSRRML 863
+L +NF ILPE +E L + V C+ L+EIRG+PPNLK AR C SL+SSS+ ML
Sbjct: 825 NLPGNNFTILPEFFKELQFLTTLVVHDCKHLQEIRGLPPNLKHFDARNCASLTSSSKSML 884
Query: 864 MSQ 866
++Q
Sbjct: 885 LNQ 887
>UniRef100_Q84ZV8 R 3 protein [Glycine max]
Length = 897
Score = 796 bits (2056), Expect = 0.0
Identities = 447/900 (49%), Positives = 583/900 (64%), Gaps = 68/900 (7%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLYKAL D+GI TFID L RGDEITP+L KAI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRF 135
V S NYASSSFCLDELV ++ C K KG LV+PVF+ V+P+ VR +KGSYGEA+A+H+KRF
Sbjct: 72 VLSQNYASSSFCLDELVTVLLC-KRKGLLVIPVFYNVDPSDVRQQKGSYGEAMAKHQKRF 130
Query: 136 QNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISRQPLHVATY 194
+ E+LQ W+ AL Q A+LSGYH YEYK I IV+ +S +I+R PLHVA Y
Sbjct: 131 K---AKKEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIQSIVEQVSREINRTPLHVADY 187
Query: 195 PVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDV 254
PVGL S+V +V+ LLD GS D VH++GI+G+GGLGK+TLA +YN +A F+ SCFL +V
Sbjct: 188 PVGLGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVYNLIALHFDESCFLQNV 247
Query: 255 RENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQL 313
RE S ++ LK+LQ +L K G +I L EG +I+ RL RKK+LLILDDVD +QL
Sbjct: 248 REESNKHGLKHLQSIILSKLLGEKDINLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQL 307
Query: 314 HALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVP 373
A+ G DWFG GSRVIITTR+K +L H +E T+ V+ LN++ AL+LL+W AFK +K
Sbjct: 308 KAIVGRPDWFGPGSRVIITTRDKHILKYHEVERTYEVKVLNQSAALQLLKWNAFKREKND 367
Query: 374 SGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSYD 433
YED+LNR V YA GLPL LE++GSNLFGK++ +W+ ++ Y RIP+ EI +ILKVS+D
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEIIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFD 427
Query: 434 ALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSGS 493
AL EE+++VFLDIACC KG + E E +L YD+C+ HH+ VL KSL K+
Sbjct: 428 ALGEEQKNVFLDIACCLKGCKLTEVEHMLRGLYDNCMKHHIDVLVDKSLTKVRH------ 481
Query: 494 INDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYM--NL 551
V +HDLI+DMG+E+ RQ SP+EPG+R RLW +DII VLK NTGTSKIE+IY+ ++
Sbjct: 482 -GIVEMHDLIQDMGREIERQRSPEEPGKRKRLWSPKDIIQVLKHNTGTSKIEIIYVDFSI 540
Query: 552 HSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSS--- 608
E ++ AF KM LK LII NG FS G Y P LRVL+W S CL S+
Sbjct: 541 SDKEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPQGLRVLEWHRYPSNCLPSNFDP 600
Query: 609 ---ILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLSNLEK 665
++ K + + F F K ++ VL D+C++LT IPDVS L NL +
Sbjct: 601 INLVICKLPDSSMTSFEFHGS--------SKLGHLTVLKFDWCKFLTQIPDVSDLPNLRE 652
Query: 666 LSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECLDNFPE 725
LSF C++L+ + +SIG LNKL+ L+AYGCRKL F PL L SL+ L L C L+ FPE
Sbjct: 653 LSFQWCESLVAVDDSIGFLNKLKKLNAYGCRKLTSFPPLHLTSLETLELSHCSSLEYFPE 712
Query: 726 LLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTS-GMKFPKIVFSNMTKLSLSFF- 783
+L +M +I+ +D+ I ELPFSFQNL L +L++ G+ + + M KLS F
Sbjct: 713 ILGEMENIERLDLHGLPIKELPFSFQNLIGLQQLSMFGCGIVQLRCSLAMMPKLSAFKFV 772
Query: 784 -------------------------------------NLSDECLPIVLKWCVNMTHLDLS 806
NL D+ K ++ +L+LS
Sbjct: 773 NCNRWQWVESEEAEEKVGSIISSEARFWTHSFSAKNCNLCDDFFLTGFKKFAHVGYLNLS 832
Query: 807 FSNFKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSSSRRMLMSQ 866
+NF ILPE +E L +NV C+ L+EIRGIP NL+ AR C SL+SSS+ ML++Q
Sbjct: 833 RNNFTILPEFFKELQFLGSLNVSHCKHLQEIRGIPQNLRLFNARNCASLTSSSKSMLLNQ 892
>UniRef100_Q84ZV3 R 4 protein [Glycine max]
Length = 895
Score = 795 bits (2053), Expect = 0.0
Identities = 446/896 (49%), Positives = 580/896 (63%), Gaps = 63/896 (7%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLYKAL D+GI T ID L RGDEITP+L KAI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRF 135
V S NYASSSFCLDELV I+HC K++G LV+PVF+ V+P+ VRH+KGSYGEA+A+H+KRF
Sbjct: 72 VLSQNYASSSFCLDELVTILHC-KSEGLLVIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 130
Query: 136 QNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISRQPLHVATY 194
+ E+LQ W+ AL Q A+LSGYH + YEYK IG IV+ +S KISR LHVA Y
Sbjct: 131 K---AKKEKLQKWRMALKQVADLSGYHFEDGDAYEYKFIGSIVEEVSRKISRASLHVADY 187
Query: 195 PVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDV 254
PVGL+S+V +V LLD GSDD VH++GI+G+GGLGK+TLA ++YN +A F+ SCFL +V
Sbjct: 188 PVGLESQVTEVMKLLDVGSDDLVHIIGIHGMGGLGKTTLALEVYNLIALHFDESCFLQNV 247
Query: 255 RENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQL 313
RE S ++ LK+LQ LL K G +I L EG I+ RL RKK+LLILDDV+ +QL
Sbjct: 248 REESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASTIQHRLQRKKVLLILDDVNKREQL 307
Query: 314 HALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVP 373
A+ G DWFG GSRVIITTR+K LL H +E T+ V+ LN AL+LL W AFK +K+
Sbjct: 308 KAIVGRPDWFGPGSRVIITTRDKHLLKCHEVERTYEVKVLNHNAALQLLTWNAFKREKID 367
Query: 374 SGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSYD 433
YED+LNR V YA GLPL LE++GSN+FGKS+ W+ ++ Y RIPN EI +ILKVS+D
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEIIGSNMFGKSVAGWESAVEHYKRIPNDEILEILKVSFD 427
Query: 434 ALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSGS 493
AL EE+++VFLDIA C KG + E E +LC+ YD+C+ HH+ VL KSL+K+
Sbjct: 428 ALGEEQKNVFLDIAFCLKGCKLTEVEHMLCSLYDNCMKHHIDVLVDKSLIKVKH------ 481
Query: 494 INDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYMN--L 551
V +HDLI+ +G+E+ RQ SP+EPG+R RLW +DIIHVLK+NTGTSKIE+I ++ +
Sbjct: 482 -GIVEMHDLIQVVGREIERQRSPEEPGKRKRLWLPKDIIHVLKDNTGTSKIEIICLDFSI 540
Query: 552 HSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSS--- 608
E ++ AF KM LK LII NG FS G Y P LRVL+W S L S+
Sbjct: 541 SYKEETVEFNENAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSNFLPSNFDP 600
Query: 609 ---ILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLSNLEK 665
++ K + + + F F K K ++ VL D C++LT IPDVS L NL +
Sbjct: 601 INLVICKLPDSSIKSFEFHGSSK-------KLGHLTVLKFDRCKFLTQIPDVSDLPNLRE 653
Query: 666 LSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECLDNFPE 725
LSF C++L+ + +SIG L KL+ LSAYGCRKL F PL L SL+ L L C L+ FPE
Sbjct: 654 LSFEDCESLVAVDDSIGFLKKLKKLSAYGCRKLTSFPPLNLTSLETLQLSSCSSLEYFPE 713
Query: 726 LLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTS---------------------- 763
+L +M +I+E+ ++ I ELPFSFQNL+ L L ++
Sbjct: 714 ILGEMENIRELRLTGLYIKELPFSFQNLTGLRLLALSGCGIVQLPCSLAMMPELSSFYTD 773
Query: 764 -------------GMKFPKIVFSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDLSFSNF 810
K I+ S + NL D+ K ++ +L+LS +NF
Sbjct: 774 YCNRWQWIELEEGEEKLGSIISSKAQLFCATNCNLCDDFFLAGFKRFAHVGYLNLSGNNF 833
Query: 811 KILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSSSRRMLMSQ 866
ILPE +E L ++V CE L+EIRG+PP L+ AR C S +SSS ML++Q
Sbjct: 834 TILPEFFKELQFLRTLDVSDCEHLQEIRGLPPILEYFDARNCVSFTSSSTSMLLNQ 889
>UniRef100_Q84ZU7 R 5 protein [Glycine max]
Length = 907
Score = 780 bits (2014), Expect = 0.0
Identities = 436/897 (48%), Positives = 579/897 (63%), Gaps = 67/897 (7%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIP 75
Y VFLSFRG DTRYGFTGNLYKAL DKGI+TF D++ L G+EITP+LLKAI++SRI I
Sbjct: 12 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRF 135
V S ++ASSSFCLDEL I+ C + G +V+PVF+ V P VRH+KG+YGEALA+H+KRF
Sbjct: 72 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 131
Query: 136 QNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISRQPLHVATY 194
++LQ W++AL Q ANLSG H YEYK IG+IV +S KI+ LHVA
Sbjct: 132 P------DKLQKWERALRQVANLSGLHFKDRDEYEYKFIGRIVASVSEKINPASLHVADL 185
Query: 195 PVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYN--FVADQFEGSCFLH 252
PVGL+S+VQ+V+ LLD G+ DGV M+GI+G+GG+GKSTLAR +YN + + F+G CFL
Sbjct: 186 PVGLESKVQEVRKLLDVGNHDGVCMIGIHGMGGIGKSTLARAVYNDLIITENFDGLCFLE 245
Query: 253 DVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQ 312
+VRE+S + L++LQ LL + G +IK+ +GI I+ L KK+LLILDDVD +Q
Sbjct: 246 NVRESSNNHGLQHLQSILLSEILGEDIKVRSKQQGISKIQSMLKGKKVLLILDDVDKPQQ 305
Query: 313 LHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKV 372
L +AG DWFG GS +IITTR+K LL+ HG++ + VE LN+ AL+LL W AFK +K+
Sbjct: 306 LQTIAGRRDWFGPGSIIIITTRDKQLLAPHGVKKRYEVEVLNQNAALQLLTWNAFKREKI 365
Query: 373 PSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSY 432
YED+LNR V YA GLPL LEV+GSN+FGK + +WK ++ Y RIPN EI +ILKVS+
Sbjct: 366 DPSYEDVLNRVVTYASGLPLALEVIGSNMFGKRVAEWKSAVEHYKRIPNDEILEILKVSF 425
Query: 433 DALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSG 492
DAL EE+++VFLDIACCFKG + E E +L Y++C+ HH+ VL KSL+K+ G
Sbjct: 426 DALGEEQKNVFLDIACCFKGCKLTEVEHMLRGLYNNCMKHHIDVLVDKSLIKVR----HG 481
Query: 493 SINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYMN-- 550
++N +HDLI+ +G+E+ RQ SP+EPG+ RLW +DII VLK NTGTSKIE+I ++
Sbjct: 482 TVN---MHDLIQVVGREIERQISPEEPGKCKRLWLPKDIIQVLKHNTGTSKIEIICLDFS 538
Query: 551 LHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSS-- 608
+ E ++ AF KM LK LII NG FS G Y P LRVL+W SKCL S+
Sbjct: 539 ISDKEQTVEWNQNAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSKCLPSNFH 598
Query: 609 ----ILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLSNLE 664
++ K + + F F K F ++ VL D C++LT IPDVS L NL
Sbjct: 599 PNNLLICKLPDSSMASFEFHGSSK--------FGHLTVLKFDNCKFLTQIPDVSDLPNLR 650
Query: 665 KLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECLDNFP 724
+LSF C++L+ + +SIG LNKL+ L+AYGCRKL F PL L SL+ L L C L+ FP
Sbjct: 651 ELSFKGCESLVAVDDSIGFLNKLKKLNAYGCRKLTSFPPLNLTSLETLQLSGCSSLEYFP 710
Query: 725 ELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGM------------------- 765
E+L +M +IK++ + + I ELPFSFQNL L L + S +
Sbjct: 711 EILGEMENIKQLVLRDLPIKELPFSFQNLIGLQVLYLWSCLIVELPCRLVMMPELFQLHI 770
Query: 766 ----------------KFPKIVFSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDLSFSN 809
K I+ S NL D+ K ++ +LDLS +N
Sbjct: 771 EYCNRWQWVESEEGEEKVGSILSSKARWFRAMNCNLCDDFFLTGSKRFTHVEYLDLSGNN 830
Query: 810 FKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSSSRRMLMSQ 866
F ILPE +E L ++V CE L++IRG+PPNLK+ A C SL+SSS+ ML++Q
Sbjct: 831 FTILPEFFKELKFLRTLDVSDCEHLQKIRGLPPNLKDFRAINCASLTSSSKSMLLNQ 887
>UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max]
Length = 863
Score = 763 bits (1969), Expect = 0.0
Identities = 442/879 (50%), Positives = 567/879 (64%), Gaps = 62/879 (7%)
Query: 24 GSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIPVFSINYAS 83
G DTR GFTG LYKAL D+GI TFID L+RGDEI P+L AI+ESRI I V S NYAS
Sbjct: 3 GQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAITVLSQNYAS 62
Query: 84 SSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRFQNDPKNME 143
SSFCLDELV I+HC K++G LV+PVF+ V+P+ VRH+KGSYGEA+A+H+KRF+ N E
Sbjct: 63 SSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFK---ANKE 118
Query: 144 RLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISRQPLHVATYPVGLQSRV 202
+LQ W+ AL Q A+LSGYH YEY+ IG IV+ IS K SR LHVA YPVGL+S V
Sbjct: 119 KLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRKFSRASLHVADYPVGLESEV 178
Query: 203 QQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDVRENSAQNN 262
+V LLD GS D VH++GI+G+GGLGK+TLA ++NF+A F+ SCFL +VRE S ++
Sbjct: 179 TEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALHFDESCFLQNVREESNKHG 238
Query: 263 LKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQLHALAGGLD 321
LK+LQ LL K G +I L EG +I+ RL RKK+LLILDDVD +QL A+ G D
Sbjct: 239 LKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAIVGRPD 298
Query: 322 WFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVPSGYEDILN 381
WFG GSRVIITTR+K LL H +E T+ V+ LN++ AL+LL W AFK +K+ YED+LN
Sbjct: 299 WFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLTWNAFKREKIDPSYEDVLN 358
Query: 382 RAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSYDALEEEEQS 441
R V YA GLPL LEV+GSNLF K++ +W+ ++ Y RIP+ EIQ+ILKVS+DAL EE+++
Sbjct: 359 RVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDEIQEILKVSFDALGEEQKN 418
Query: 442 VFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYYPSGSINDVRLHD 501
VFLDIACCFKGY+W E ++IL Y +C HH+GVL KSLVK+S + V +HD
Sbjct: 419 VFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLVKVSC------CDTVEMHD 472
Query: 502 LIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYMN--LHSMESVID 559
+I+DMG+E+ RQ SP+EPG+ RL +DII V K IE+I ++ + E ++
Sbjct: 473 MIQDMGREIERQRSPEEPGKCKRLLLPKDIIQVFK-------IEIICLDFSISDKEETVE 525
Query: 560 KKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSSI--LNKA---- 613
AF KM LK LII N FS G Y P LRVL+W S CL S+ +N
Sbjct: 526 WNENAFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSNCLPSNFDPINLVICKL 585
Query: 614 --SEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLSNLEKLSFTCC 671
S ITS F K S +QK ++ VL D CE+LT IPDVS L NL++LSF C
Sbjct: 586 PDSSITSFEF-HGSSKASLKSSLQKLGHLTVLNFDRCEFLTKIPDVSDLPNLKELSFNWC 644
Query: 672 DNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECLDNFPELLCKMA 731
++L+ + +SIG LNKL+ LSAYGCRKL F PL L SL+ L L C L+ FPE+L +M
Sbjct: 645 ESLVAVDDSIGFLNKLKTLSAYGCRKLTSFPPLNLTSLETLNLGGCSSLEYFPEILGEMK 704
Query: 732 HIKEIDISNTSIGELPFSFQNLSELHELTVTS-GMKFPKIVFSNMTKLS----------- 779
+I + + + I ELPFSFQNL L L + S G+ + + M KL
Sbjct: 705 NITVLALHDLPIKELPFSFQNLIGLLFLWLDSCGIVQLRCSLATMPKLCEFCITDSCNRW 764
Query: 780 ----------------LSF----FNLSDECLPIVLKWCVNMTHLDLSFSNFKILPECLRE 819
LSF NL D+ I K ++ +L+L +NF ILPE +E
Sbjct: 765 QWVESEEGEEKVVGSILSFEATDCNLCDDFFFIGSKRFAHVGYLNLPGNNFTILPEFFKE 824
Query: 820 CHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSSS 858
L + V C+ L+EIRG+PPNLK AR C SL+SS
Sbjct: 825 LQFLTTLVVHDCKHLQEIRGLPPNLKHFDARNCASLTSS 863
>UniRef100_Q84ZV7 R 12 protein [Glycine max]
Length = 893
Score = 716 bits (1848), Expect = 0.0
Identities = 399/759 (52%), Positives = 502/759 (65%), Gaps = 38/759 (5%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFIP 75
Y VFLSF DT GFT LYKAL D+GI TF L R E+TP L KAI SR+ I
Sbjct: 12 YDVFLSFIREDTHRGFTFYLYKALNDRGIYTFFYDQELPRETEVTPGLYKAILASRVAII 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKRF 135
V S NYA SSFCLDELV I+HC R V+PVF V+P+ VRH+KGSYGEA+A+H+KRF
Sbjct: 72 VLSENYAFSSFCLDELVTILHCE----REVIPVFHNVDPSDVRHQKGSYGEAMAKHQKRF 127
Query: 136 QNDPKNMERLQGWKKALSQAANLSGYHDSPPG-YEYKLIGKIVKYISNKISRQPLHVATY 194
+ ++LQ W+ AL Q ANL GYH G YEY LIG+IVK +S LHVA Y
Sbjct: 128 K-----AKKLQKWRMALKQVANLCGYHFKDGGSYEYMLIGRIVKQVSRMFGLASLHVADY 182
Query: 195 PVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFLHDV 254
PVGL+S+V +V LLD GSDD VH++GI+G+GGLGK+TLA +YNF+A F+ SCFL +V
Sbjct: 183 PVGLESQVTEVMKLLDVGSDDVVHIIGIHGMGGLGKTTLAMAVYNFIAPHFDESCFLQNV 242
Query: 255 RENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLKQL 313
RE S ++ LK+LQ LL K G +I L EG +I+ RL KKILLILDDVD +QL
Sbjct: 243 REESNKHGLKHLQSVLLSKLLGEKDITLTSWQEGASMIQHRLRLKKILLILDDVDKREQL 302
Query: 314 HALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKSDKVP 373
A+ G DWFG GSRVIITTR+K LL H +E T+ V LN +A +LL W AFK +K+
Sbjct: 303 KAIVGKPDWFGPGSRVIITTRDKHLLKYHEVERTYEVNVLNHDDAFQLLTWNAFKREKID 362
Query: 374 SGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILKVSYD 433
Y+D+LNR V YA GLPL LEV+GSNL+GK++ +W+ L+ Y RIP+ EI KIL+VS+D
Sbjct: 363 PSYKDVLNRVVTYASGLPLALEVIGSNLYGKTVAEWESALETYKRIPSNEILKILEVSFD 422
Query: 434 ALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKS-LVKISTYYPSG 492
ALEEE+++VFLDIACCFKGY+W E DI A Y +C HH+GVL KS L+K+S
Sbjct: 423 ALEEEQKNVFLDIACCFKGYKWTEVYDIFRALYSNCKMHHIGVLVEKSLLLKVSWR---- 478
Query: 493 SINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYM--N 550
++V +HDLI+DMG+++ RQ SP+EPG+ RLW +DII VLK NTGTSK+E+I + +
Sbjct: 479 --DNVEMHDLIQDMGRDIERQRSPEEPGKCKRLWSPKDIIQVLKHNTGTSKLEIICLDSS 536
Query: 551 LHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSSI- 609
+ E ++ AF KM LK LII NG FS G Y P LRVL+W S CL S+
Sbjct: 537 ISDKEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSNCLPSNFD 596
Query: 610 -LNKA------SEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLSN 662
+N S ITS F S K ++ VL D C++LT IPDVS L N
Sbjct: 597 PINLVICKLPDSSITSLEFHGS----------SKLGHLTVLKFDKCKFLTQIPDVSDLPN 646
Query: 663 LEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECLDN 722
L +LSF C++L+ I +SIG LNKLE L+A GCRKL F PL L SL+ L L C L+
Sbjct: 647 LRELSFVGCESLVAIDDSIGFLNKLEILNAAGCRKLTSFPPLNLTSLETLELSHCSSLEY 706
Query: 723 FPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTV 761
FPE+L +M +I + + I ELPFSFQNL L E+T+
Sbjct: 707 FPEILGEMENITALHLERLPIKELPFSFQNLIGLREITL 745
>UniRef100_Q9FPK9 Putative resistance protein [Glycine max]
Length = 1093
Score = 704 bits (1816), Expect = 0.0
Identities = 452/1112 (40%), Positives = 630/1112 (56%), Gaps = 169/1112 (15%)
Query: 5 SSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLL 64
S ST I Y VFLSFRG DTR FTGNLY L +GI+TFI + G+EI SL
Sbjct: 6 SESTDIRV---YDVFLSFRGEDTRRSFTGNLYNCLEKRGIHTFIGDYDFESGEEIKASLS 62
Query: 65 KAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSY 124
+AIE SR+F+ VFS NYASSS+CLD LV I+ + R V+PVFF VEP+ VRH+KG Y
Sbjct: 63 EAIEHSRVFVIVFSENYASSSWCLDGLVRILDFTEDNHRPVIPVFFDVEPSHVRHQKGIY 122
Query: 125 GEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGY-HDSPPGYEYKLIGKIVKYISNK 183
GEALA HE+R +P++ + ++ W+ AL QAANLSGY GYEYKLI KIV+ ISNK
Sbjct: 123 GEALAMHERRL--NPESYKVMK-WRNALRQAANLSGYAFKHGDGYEYKLIEKIVEDISNK 179
Query: 184 IS-RQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVA 242
I +P V PVGL+ R+ +V LLD S GVHM+GI GIGG+GK+TLAR +Y+ A
Sbjct: 180 IKISRP--VVDRPVGLEYRMLEVDWLLDATSLAGVHMIGICGIGGIGKTTLARAVYHSAA 237
Query: 243 DQFEGSCFLHDVRENSAQNNLKYLQEKLLLKT-TGLEIKLDHVSEGIPVIKERLCRKKIL 301
F+ SCFL +VREN+ ++ L +LQ+ LL + I+L V +GI +IK+ L RK++L
Sbjct: 238 GHFDTSCFLGNVRENAMKHGLVHLQQTLLAEIFRENNIRLTSVEQGISLIKKMLPRKRLL 297
Query: 302 LILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALEL 361
L+LDDV L L AL G DWFG GSRVIITTR++ LL +HG++ + VE L EALEL
Sbjct: 298 LVLDDVCELDDLRALVGSPDWFGPGSRVIITTRDRHLLKAHGVDKVYEVEVLANGEALEL 357
Query: 362 LRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPN 421
L W AF++D+V + + LNRA+ +A G+PL LE++GS+L+G+ IE+W+ TLD Y++ P
Sbjct: 358 LCWKAFRTDRVHPDFINKLNRAITFASGIPLALELIGSSLYGRGIEEWESTLDQYEKNPP 417
Query: 422 KEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKS 481
++I LK+S+DAL E+ VFLDIAC F G++ E E IL AH+ C+ H+G L KS
Sbjct: 418 RDIHMALKISFDALGYLEKEVFLDIACFFNGFELAEIEHILGAHHGCCLKFHIGALVEKS 477
Query: 482 LVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGT 541
L+ I + V++HDLI+ MG+E+VRQESP+ PG+RSRLW EDI+HVL++NTGT
Sbjct: 478 LIMIDEH------GRVQMHDLIQQMGREIVRQESPEHPGKRSRLWSTEDIVHVLEDNTGT 531
Query: 542 SKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCL 601
KI+ I ++ E V+ G AF KM L+TLII +FS G K L++L+W GC
Sbjct: 532 CKIQSIILDFSKSEKVVQWDGMAFVKMISLRTLIIRK-MFSKGPKNF-QILKMLEWWGCP 589
Query: 602 SKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSGLS 661
SK L S + I + + + F +M+VL D CE+LT PD+SG
Sbjct: 590 SKSLPSDFKPEKLAILKLPYSGFMSLE-----LPNFLHMRVLNFDRCEFLTRTPDLSGFP 644
Query: 662 NLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHFRPLGLASLKKLILYECECLD 721
L++L F C+NL+ IH+S+G L+KLE ++ GC KLE F P+ L SL+ + L C L
Sbjct: 645 ILKELFFVFCENLVEIHDSVGFLDKLEIMNFEGCSKLETFPPIKLTSLESINLSHCSSLV 704
Query: 722 NFPELLCKMAHIKEIDISNTSIGELPFSFQNLS-----ELH---------------ELTV 761
+FPE+L KM +I + + T+I +LP S + L ELH EL V
Sbjct: 705 SFPEILGKMENITHLSLEYTAISKLPNSIRELVRLQSLELHNCGMVQLPSSIVTLRELEV 764
Query: 762 TS-----GMKFPK-----------IVFSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDL 805
S G++F K + S + +++L ++SDE + L W N+ LDL
Sbjct: 765 LSICQCEGLRFSKQDEDVKNKSLLMPSSYLKQVNLWSCSISDEFIDTGLAWFANVKSLDL 824
Query: 806 SFSNFKILPECLRECH-----------HLVEIN-----------VMC------------- 830
S +NF ILP C++EC HL EI + C
Sbjct: 825 SANNFTILPSCIQECRLLRKLYLDYCTHLHEIRGIPPNLETLSAIRCTSLKDLDLAVPLE 884
Query: 831 ---------------CESLEEIRGIPPNLKELCARYCKSLSSSSRRMLMSQ--------- 866
CE+L+EIRGIPP+++ L A C+SL++S RRML+ Q
Sbjct: 885 STKEGCCLRQLILDDCENLQEIRGIPPSIEFLSATNCRSLTASCRRMLLKQELHEAGNKR 944
Query: 867 -----------------------------PSIT-CIFILPKGNEYATSVNVFVNGYEIEI 896
P I+ C+ L + + V +NG +++
Sbjct: 945 YSLPGTRIPEWFEHCSRGQSISFWFRNKFPVISLCLAGLMHKHPFGLKPIVSINGNKMKT 1004
Query: 897 GCYWSLFFF-----TDHTTLFHTSKLNELIKTQCEYNIEKGLLKNEW------IYVEFKL 945
F+F TDH +F ++ + E N+++ + +N+W + V+FK
Sbjct: 1005 EFQRRWFYFEFPVLTDHILIFGERQI------KFEDNVDEVVSENDWNHVVVSVDVDFKW 1058
Query: 946 KDHENSVYAQRGIHVWNEKSNTEEENVVFTDP 977
E V + G+HV KS+ E+ + F DP
Sbjct: 1059 NPTEPLV-VRTGLHVIKPKSSVED--IRFIDP 1087
>UniRef100_Q8H6S7 Resistance protein KR3 [Glycine max]
Length = 636
Score = 579 bits (1492), Expect = e-163
Identities = 308/603 (51%), Positives = 421/603 (69%), Gaps = 23/603 (3%)
Query: 15 KYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIFI 74
+Y VF++FRG DTR+ FTG+L+KAL +KGI F+D+N ++RGDEI +L +AI+ SRI I
Sbjct: 34 RYDVFINFRGEDTRFAFTGHLHKALCNKGIRAFMDENDIKRGDEIRATLEEAIKGSRIAI 93
Query: 75 PVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEKR 134
VFS +YASSSFCLDEL I+ CY+ K LV+PVF+ V+P+ VR +GSY E LA E+R
Sbjct: 94 TVFSKDYASSSFCLDELATILGCYREKTLLVIPVFYKVDPSDVRRLQGSYAEGLARLEER 153
Query: 135 FQNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISRQP--LHV 191
F + +N WKKAL + A L+G+H GYE+K I KIV + +KI++ ++V
Sbjct: 154 FHPNMEN------WKKALQKVAELAGHHFKDGAGYEFKFIRKIVDDVFDKINKAEASIYV 207
Query: 192 ATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCFL 251
A +PVGL V++++ LL+ GS D + M+GI+G+GG+GKSTLAR +YN D F+ SCFL
Sbjct: 208 ADHPVGLHLEVEKIRKLLEAGSSDAISMIGIHGMGGVGKSTLARAVYNLHTDHFDDSCFL 267
Query: 252 HDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILLILDDVDNLK 311
+VRE S ++ LK LQ LL + EI L +G +IK +L KK+LL+LDDVD K
Sbjct: 268 QNVREESNRHGLKRLQSILLSQILKKEINLASEQQGTSMIKNKLKGKKVLLVLDDVDEHK 327
Query: 312 QLHALAGGLDW----FGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAF 367
QL A+ G W FG +IITTR+K LL+S+G++ TH V+ L++ +A++LL+ AF
Sbjct: 328 QLQAIVGKSVWSESEFGTRLVLIITTRDKQLLTSYGVKRTHEVKELSKKDAIQLLKRKAF 387
Query: 368 KS-DKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQK 426
K+ D+V Y +LN V + GLPL LEV+GSNLFGKSI++W+ + Y RIPNKEI K
Sbjct: 388 KTYDEVDQSYNQVLNDVVTWTSGLPLALEVIGSNLFGKSIKEWESAIKQYQRIPNKEILK 447
Query: 427 ILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKIS 486
ILKVS+DALEEEE+SVFLDI CC KGY+ +E EDIL + YD+C+ +H+GVL KSL++IS
Sbjct: 448 ILKVSFDALEEEEKSVFLDITCCLKGYKCREIEDILHSLYDNCMKYHIGVLVDKSLIQIS 507
Query: 487 TYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEM 546
+ V LHDLI++MGKE+ RQ+SPKE G+R RLW +DII VLK+N+GTS++++
Sbjct: 508 D-------DRVTLHDLIENMGKEIDRQKSPKETGKRRRLWLLKDIIQVLKDNSGTSEVKI 560
Query: 547 IYMN--LHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKC 604
I ++ + + I+ G AFK+M LK LII NG+ S G YLP SLR+L+W S C
Sbjct: 561 ICLDFPISDKQETIEWNGNAFKEMKNLKALIIRNGILSQGPNYLPESLRILEWHRHPSHC 620
Query: 605 LSS 607
L S
Sbjct: 621 LPS 623
>UniRef100_Q9FPK8 Putative resistance protein [Glycine max]
Length = 522
Score = 505 bits (1300), Expect = e-141
Identities = 273/540 (50%), Positives = 377/540 (69%), Gaps = 23/540 (4%)
Query: 1 MQSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEI- 59
M S+S + Y++ VFLSFRG DTR GF GNLYKALT+KG +TF + L RG+EI
Sbjct: 1 MAGSERSSSRVWHYEFDVFLSFRGEDTRLGFVGNLYKALTEKGFHTFF-REKLVRGEEIA 59
Query: 60 -TPSLL-KAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVV 117
+PS++ KAI+ SR+F+ VFS NYASS+ CL+EL+ I+ + R VLPVF+ V+P+ V
Sbjct: 60 ASPSVVEKAIQHSRVFVVVFSQNYASSTRCLEELLSILRFSQDNRRPVLPVFYYVDPSDV 119
Query: 118 RHRKGSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGY-HDSPPGYEYKLIGKI 176
+ G YGEALA HEKRF ++ +++ W+KAL +AA LSG+ GYEY+LI KI
Sbjct: 120 GLQTGMYGEALAMHEKRFNSES---DKVMKWRKALCEAAALSGWPFKHGDGYEYELIEKI 176
Query: 177 VKYISNKISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQ 236
V+ +S KI+R PVGLQ R+ ++ LLD S GVH++GIYG+GG+GK+TLAR
Sbjct: 177 VEGVSKKINR--------PVGLQYRMLELNGLLDAASLSGVHLIGIYGVGGIGKTTLARA 228
Query: 237 IYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERL 295
+Y+ VA QF+ CFL +VREN+ ++ L +LQ+ +L +T G +I+L V +GI ++K+RL
Sbjct: 229 LYDSVAVQFDALCFLDEVRENAMKHGLVHLQQTILAETVGEKDIRLPSVKQGITLLKQRL 288
Query: 296 CRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNE 355
K++LL+LDD++ +QL AL G WFG GSRVIITTR++ LL SHG+E + VE L +
Sbjct: 289 QEKRVLLVLDDINESEQLKALVGSPGWFGPGSRVIITTRDRQLLESHGVEKIYEVENLAD 348
Query: 356 TEALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDG 415
EALELL W AFK+DKV + + + RA+ YA GLPL LEV+GSNLFG+ I +W++TLD
Sbjct: 349 GEALELLCWKAFKTDKVYPDFINKIYRALTYASGLPLALEVIGSNLFGREIVEWQYTLDL 408
Query: 416 YDRIPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLG 475
Y++I +K+IQKILK+S+DAL+E E+ +FLDIAC FKG + + E I+ Y + +
Sbjct: 409 YEKIHDKDIQKILKISFDALDEHEKDLFLDIACFFKGCKLAQVESIVSGRYGDSLKAIID 468
Query: 476 VLAGKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVL 535
VL K+L+KI + V++HDLI+ MG+E+VRQESPK PG SRLW ED+ VL
Sbjct: 469 VLLEKTLIKIDEH------GRVKMHDLIQQMGREIVRQESPKHPGNCSRLWSPEDVADVL 522
>UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 498 bits (1283), Expect = e-139
Identities = 321/856 (37%), Positives = 495/856 (57%), Gaps = 54/856 (6%)
Query: 14 YKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIF 73
+ Y VFLSFRG D R F +LY AL K INTF D L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELVSSIEESRIA 75
Query: 74 IPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL I+ C KG++V+PVF+ V+P+ VR +K +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 134 RFQNDPKNMERLQGWKKALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SRQPLH 190
RFQ D ++Q W+ AL +AAN+SG+ ++ G+E +++ KI + I ++ S++
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCF 250
A VG++S + QV +L GS GVH +GI G+ G+GK+TLAR IY+ + QF+G+CF
Sbjct: 191 NARNLVGMESHMHQVYKMLGIGSG-GVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 251 LHDVRENSAQNNLKYLQEKLLLKTTGLE-IKLDHVSEGIPVIKERLCRKKILLILDDVDN 309
LH+VR+ SA+ L+ LQE LL + ++ ++++ EG + K+RL KK+LL+LDDVD+
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKS 369
+ QL+ALAG +WFG GSR+IITT++K LL + E + ++ LN E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 370 DKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILK 429
++ +ED+ + + + GLPL L+V+GS L+G+ +++W ++ +IP EI K L+
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 430 VSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYY 489
S+ L EQ +FLDIAC F G + K+ + + C + VL K L+
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKK-KDSVTRILESFHFCPVIGIKVLMEKCLITTLQ-- 486
Query: 490 PSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYM 549
+ +H LI+DMG +VR+E+ +P SRLW++EDI VL+ N GT KIE + +
Sbjct: 487 -----GRITIHQLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKIEGMSL 541
Query: 550 NLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSSI 609
+L + E V + GKAF +MT+L+ L +N G ++LP LR L W G SK L +S
Sbjct: 542 HLTNEEEV-NFGGKAFMQMTRLRFLKFQNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSF 600
Query: 610 LNKASEITSQ*FCFSQQKKSFLVCMQK----FQNMKVLTLDYCEYLTHIPDVSGLSNLEK 665
K ++ S + KKS ++ + K +K + L + + L +PD S NLE+
Sbjct: 601 --KGDQLVS-----LKLKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLER 653
Query: 666 LSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHF-RPLGLASLKKLILYECECLDNFP 724
L C +L+ I+ SI +L KL L+ CR L+ + + L L+ L+L C L FP
Sbjct: 654 LVLEECTSLVEINFSIENLGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTFP 713
Query: 725 ELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGMKFPKIVFSNMTKLSLSFFN 784
E+ KM + E+ + TS+ ELP S +NLS + + ++ ++ L S F
Sbjct: 714 EIEEKMNCLAELYLDATSLSELPASVENLSGVGVINLS--------YCKHLESLPSSIFR 765
Query: 785 LSDECLPIVLKWCVNMTHLDLS-FSNFKILPECLRECHHLVEINV--MCCESLEEIRGIP 841
L +CL LD+S S K LP+ L L +++ +++ +
Sbjct: 766 L--KCLKT----------LDVSGCSKLKNLPDDLGLLVGLEQLHCTHTAIQTIPSSMSLL 813
Query: 842 PNLKELCARYCKSLSS 857
NLK L C +LSS
Sbjct: 814 KNLKRLSLSGCNALSS 829
>UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum]
Length = 1136
Score = 493 bits (1270), Expect = e-138
Identities = 319/856 (37%), Positives = 492/856 (57%), Gaps = 54/856 (6%)
Query: 14 YKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIF 73
+ Y VFLSFRG D R F +LY AL K INTF D L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELVSSIEESRIA 75
Query: 74 IPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL I+ C KG++V+PVF+ V+P+ VR +K +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 134 RFQNDPKNMERLQGWKKALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SRQPLH 190
RFQ D ++Q W+ AL +AAN+SG+ ++ G+E +++ KI + I ++ S++
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTANGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCF 250
A VG++S + +V +L GS GVH +GI G+ G+GK+TLAR IY+ + QF+G+CF
Sbjct: 191 NARNLVGMESHMHKVYKMLGIGSG-GVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 251 LHDVRENSAQNNLKYLQEKLLLKTTGLE-IKLDHVSEGIPVIKERLCRKKILLILDDVDN 309
LH+VR+ SA+ L+ LQE LL + ++ ++++ EG + K+RL KK+LL+LDDVD+
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKS 369
+ QL+ALAG +WFG GSR+IITT++K LL + E + ++ LN E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 370 DKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILK 429
++ +ED+ + + + GLPL L+V+GS L+G+ +++W ++ +IP EI K L+
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 430 VSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYY 489
S+ L EQ +FLDIAC F G + K+ + + C + VL K L+ I
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKK-KDSVTRILESFHFCPVIGIKVLMEKCLITILQ-- 486
Query: 490 PSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYM 549
+ +H LI+DMG +VR+E+ +P SR+W++EDI VL+ N GT K E + +
Sbjct: 487 -----GRITIHQLIQDMGWHIVRREATDDPRMCSRMWKREDICPVLERNLGTDKNEGMSL 541
Query: 550 NLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSSI 609
+L + E V + GKAF +MT+L+ L N G ++LP LR L W G SK L +S
Sbjct: 542 HLTNEEEV-NFGGKAFMQMTRLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSF 600
Query: 610 LNKASEITSQ*FCFSQQKKSFLVCMQK----FQNMKVLTLDYCEYLTHIPDVSGLSNLEK 665
K ++ + KKS ++ + K +K + L + + L PD S NLE+
Sbjct: 601 --KGDQLVG-----LKLKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRTPDFSVTPNLER 653
Query: 666 LSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHF-RPLGLASLKKLILYECECLDNFP 724
L C +L+ I+ SI +L KL L+ CR L+ + + L L+ L+L C L FP
Sbjct: 654 LVLEECTSLVEINFSIENLGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTFP 713
Query: 725 ELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGMKFPKIVFSNMTKLSLSFFN 784
E+ KM + E+ + TS+ ELP S +NLS + + ++ ++ L S F
Sbjct: 714 EIEEKMNCLAELYLGATSLSELPASVENLSGVGVINLS--------YCKHLESLPSSIFR 765
Query: 785 LSDECLPIVLKWCVNMTHLDLS-FSNFKILPECLRECHHLVEINV--MCCESLEEIRGIP 841
L +CL LD+S S K LP+ L L E++ +++ +
Sbjct: 766 L--KCLKT----------LDVSGCSKLKNLPDDLGLLVGLEELHCTHTAIQTIPSSMSLL 813
Query: 842 PNLKELCARYCKSLSS 857
NLK L C +LSS
Sbjct: 814 KNLKHLSLSGCNALSS 829
>UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 492 bits (1266), Expect = e-137
Identities = 319/856 (37%), Positives = 492/856 (57%), Gaps = 54/856 (6%)
Query: 14 YKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIF 73
+ Y VFLSFRG D R F +LY AL K INTF D L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELMSSIEESRIA 75
Query: 74 IPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL I+ C KG++V+PVF+ V+P+ VR +K +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 134 RFQNDPKNMERLQGWKKALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SRQPLH 190
RFQ D ++Q W+ AL +AAN+SG+ ++ G+E +++ KI + I ++ S++
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCF 250
A VG++S + +V +L GS GVH +GI G+ G+GK+TLAR IY+ + QF+G+CF
Sbjct: 191 NARNLVGMESHMLKVYKMLGIGSG-GVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 251 LHDVRENSAQNNLKYLQEKLLLKTTGLE-IKLDHVSEGIPVIKERLCRKKILLILDDVDN 309
LH+VR+ SA+ L+ LQE LL + ++ +++++ EG + K+RL KK+LL+LDDVD+
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINNSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKS 369
+ QL+ALAG +WFG GSR+IITT++K LL + E + ++ LN E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 370 DKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILK 429
++ +ED+ + + + GLPL L+V+GS L+G+ +++W ++ +IP EI K L+
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 430 VSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKSLVKISTYY 489
S+ L EQ +FLDIAC F G + K+ + + C + VL K L+ I
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKK-KDSVTRILESFHFCPVIGIKVLMEKCLITILQ-- 486
Query: 490 PSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIYM 549
+ +H LI+DMG +VR+E+ +P SRLW++EDI VL+ N GT K E + +
Sbjct: 487 -----GRITIHQLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKNEGMSL 541
Query: 550 NLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSSI 609
+L + E V + GKAF +MT+L+ L N G ++LP LR L W G SK L +S
Sbjct: 542 HLTNEEEV-NFGGKAFMQMTRLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSF 600
Query: 610 LNKASEITSQ*FCFSQQKKSFLVCMQK----FQNMKVLTLDYCEYLTHIPDVSGLSNLEK 665
K ++ + KKS ++ + K +K + L + + L PD S NLE+
Sbjct: 601 --KGDQLVG-----LKLKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRTPDFSVTPNLER 653
Query: 666 LSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHF-RPLGLASLKKLILYECECLDNFP 724
L C +L+ I+ SI +L KL L+ CR L+ + + L L+ L+L C L FP
Sbjct: 654 LVLEECTSLVEINFSIENLGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTFP 713
Query: 725 ELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGMKFPKIVFSNMTKLSLSFFN 784
E+ KM + E+ + TS+ LP S +NLS + + ++ ++ L S F
Sbjct: 714 EIEEKMNCLAELYLGATSLSGLPASVENLSGVGVINLS--------YCKHLESLPSSIFR 765
Query: 785 LSDECLPIVLKWCVNMTHLDLS-FSNFKILPECLRECHHLVEINV--MCCESLEEIRGIP 841
L +CL LD+S S K LP+ L L +++ ++ +
Sbjct: 766 L--KCLKT----------LDVSGCSKLKNLPDDLGLLVGLEKLHCTHTAIHTIPSSMSLL 813
Query: 842 PNLKELCARYCKSLSS 857
NLK L R C +LSS
Sbjct: 814 KNLKRLSLRGCNALSS 829
>UniRef100_Q84KB4 MRGH5 [Cucumis melo]
Length = 1092
Score = 487 bits (1254), Expect = e-136
Identities = 313/846 (36%), Positives = 494/846 (57%), Gaps = 36/846 (4%)
Query: 5 SSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLL 64
SSS+S + + Y VFLSFRG DTR FTG+LY L KG+N FID +GL+RG++I+ +L
Sbjct: 10 SSSSSPIFNWSYDVFLSFRGEDTRSNFTGHLYMFLRQKGVNVFID-DGLERGEQISETLF 68
Query: 65 KAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSY 124
K I+ S I I +FS NYASS++CLDELV I+ C K+KG+ VLP+F+ V+P+ VR + G +
Sbjct: 69 KTIQNSLISIVIFSENYASSTWCLDELVEIMECKKSKGQKVLPIFYKVDPSDVRKQNGWF 128
Query: 125 GEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKI 184
E LA+HE F ME++ W+ AL+ AANLSG+H E LI IVK + + +
Sbjct: 129 REGLAKHEANF------MEKIPIWRDALTTAANLSGWHLGARK-EAHLIQDIVKEVLSIL 181
Query: 185 SR-QPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVAD 243
+ +PL+ + VG+ S+++ + + + V+M+GIYGIGG+GK+TLA+ +Y+ +A
Sbjct: 182 NHTKPLNANEHLVGIDSKIEFLYRKEEMYKSECVNMLGIYGIGGIGKTTLAKALYDKMAS 241
Query: 244 QFEGSCFLHDVRENSAQ-NNLKYLQEKLLLKTTGLEIKLDHVSEGIPVIKERLCRKKILL 302
QFEG C+L DVRE S + L LQ+KLL + ++++ + GI +IK RL KK+L+
Sbjct: 242 QFEGCCYLRDVREASKLFDGLTQLQKKLLFQILKYDLEVVDLDWGINIIKNRLRSKKVLI 301
Query: 303 ILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELL 362
+LDDVD L+QL AL GG DWFG G+++I+TTRNK LL SHG + + V+GL++ EA+EL
Sbjct: 302 LLDDVDKLEQLQALVGGHDWFGQGTKIIVTTRNKQLLVSHGFDKMYEVQGLSKHEAIELF 361
Query: 363 RWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKS-IEDWKHTLDGYDRIPN 421
R AFK+ + S Y D+ RA Y G PL L V+GS L +S + +W LDG++
Sbjct: 362 RRHAFKNLQPSSNYLDLSERATRYCTGHPLALIVLGSFLCDRSDLAEWSGILDGFENSLR 421
Query: 422 KEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHHLGVLAGKS 481
K+I+ IL++S+D LE+E + +FLDI+C G + + +L + + + L S
Sbjct: 422 KDIKDILQLSFDGLEDEVKEIFLDISCLLVGKRVSYVKKML-SECHSILDFGITKLKDLS 480
Query: 482 LVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGT 541
L++ + V++HDLIK MG ++V ES +PG+RSRLW ++DI+ V N+G+
Sbjct: 481 LIRFED-------DRVQMHDLIKQMGHKIVHDESHDQPGKRSRLWLEKDILEVFSNNSGS 533
Query: 542 SKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIE-NGLFSGGLKYLPSSLRVLKWKGC 600
++ I + L + VID +AF+ M L+ L+++ N F +KYLP+ L+ +KW
Sbjct: 534 DAVKAIKLVLTDPKRVIDLDPEAFRSMKNLRILMVDGNVRFCKKIKYLPNGLKWIKWHRF 593
Query: 601 LSKCLSSSILNK-ASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEYLTHIPDVSG 659
L S + K + Q + K CM+ +K+L L + L I + S
Sbjct: 594 AHPSLPSCFITKDLVGLDLQHSFITNFGKGLQNCMR----LKLLDLRHSVILKKISESSA 649
Query: 660 LSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHF--RPLGLASLKKLILYEC 717
NLE+L + C NL TI S L KL L + C L+ + +L+ L L C
Sbjct: 650 APNLEELYLSNCSNLKTIPKSFLSLRKLVTLDLHHCVNLKKIPRSYISWEALEDLDLSHC 709
Query: 718 ECLDNFPELLCKMAHIKEIDISN-TSIGELPFSFQNLSELHELTV---TSGMKFPKIVFS 773
+ L+ P+ + ++++ + T++ + S +L++L L + ++ K P+ +
Sbjct: 710 KKLEKIPD-ISSASNLRSLSFEQCTNLVMIHDSIGSLTKLVTLKLQNCSNLKKLPRYISW 768
Query: 774 N-MTKLSLSFFNLSDECLPIVLKWCVNMTHLDL-SFSNFKILPECLRECHHLVEINVMCC 831
N + L+LS+ +E +P N+ HL L ++ +++ + + LV +N+ C
Sbjct: 769 NFLQDLNLSWCKKLEE-IP-DFSSTSNLKHLSLEQCTSLRVVHDSIGSLSKLVSLNLEKC 826
Query: 832 ESLEEI 837
+LE++
Sbjct: 827 SNLEKL 832
Score = 89.4 bits (220), Expect = 5e-16
Identities = 111/429 (25%), Positives = 175/429 (39%), Gaps = 100/429 (23%)
Query: 527 RQEDIIHVLKENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLK 586
R I+ + E++ +E +Y++ S I K +F + KL TL + + + LK
Sbjct: 636 RHSVILKKISESSAAPNLEELYLSNCSNLKTIPK---SFLSLRKLVTLDLHHCV---NLK 689
Query: 587 YLPSSLRVLKWKGC----LSKCLSSSIL---NKASEITSQ*F-----------CFSQQKK 628
+P S + W+ LS C + + AS + S F K
Sbjct: 690 KIPRSY--ISWEALEDLDLSHCKKLEKIPDISSASNLRSLSFEQCTNLVMIHDSIGSLTK 747
Query: 629 SFLVCMQKFQNMKVL------------TLDYCEYLTHIPDVSGLSNLEKLSFTCCDNLIT 676
+ +Q N+K L L +C+ L IPD S SNL+ LS C +L
Sbjct: 748 LVTLKLQNCSNLKKLPRYISWNFLQDLNLSWCKKLEEIPDFSSTSNLKHLSLEQCTSLRV 807
Query: 677 IHNSIGHLNKLEWLSAYGCRKLEHFRP-LGLASLKKLILYECECLDNFPELLCKMAHIKE 735
+H+SIG L+KL L+ C LE L L SL+ L L C L+ FPE+ M +
Sbjct: 808 VHDSIGSLSKLVSLNLEKCSNLEKLPSYLKLKSLQNLTLSGCCKLETFPEIDENMKSLYI 867
Query: 736 IDISNTSIGELPFSF---------------------------QNLSELH----------- 757
+ + +T+I ELP S ++L ELH
Sbjct: 868 LRLDSTAIRELPPSIGYLTHLYMFDLKGCTNLISLPCTTHLLKSLGELHLSGSSRFEMFS 927
Query: 758 -----------------ELTVTSGM---KFPK--IVFSNMTKLSLSFFNLSD-ECLPIVL 794
E ++TS + PK + F + T L L N+S+ + L I+
Sbjct: 928 YIWDPTINPVCSSSKIMETSLTSEFFHSRVPKESLCFKHFTLLDLEGCNISNVDFLEILC 987
Query: 795 KWCVNMTHLDLSFSNFKILPECLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKS 854
+++ + LS +NF LP CL + L + + C+ L+EI +P ++ + A C S
Sbjct: 988 NVASSLSSILLSENNFSSLPSCLHKFMSLRNLELRNCKFLQEIPNLPLCIQRVDATGCVS 1047
Query: 855 LSSSSRRML 863
LS S +L
Sbjct: 1048 LSRSPNNIL 1056
>UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuberosum]
Length = 1121
Score = 483 bits (1244), Expect = e-134
Identities = 321/860 (37%), Positives = 491/860 (56%), Gaps = 62/860 (7%)
Query: 14 YKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLKAIEESRIF 73
+ Y VFLSFRG + R F +LY AL K INTF D L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGENVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELMSSIEESRIA 75
Query: 74 IPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL II C KG++V+PVF+ V+P+ VR +K +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIIECKNVKGQIVVPVFYDVDPSTVRRQKNIFGEAFSKHEA 135
Query: 134 RFQNDPKNMERLQGWKKALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SRQPLH 190
RF+ D +++ W+ AL +AAN+SG+ ++ G+E ++I KI + I ++ S++
Sbjct: 136 RFEED-----KVKKWRAALEEAANISGWDLPNTSNGHEARVIEKITEDIMVRLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQFEGSCF 250
A VG++S + QV +L GS GV +GI G+ G+GK+TLAR IY+ + QFEG+CF
Sbjct: 191 NARNVVGMESHMHQVYKMLGIGSG-GVRFLGILGMSGVGKTTLARVIYDNIQSQFEGACF 249
Query: 251 LHDVRENSAQNNLKYLQEKLLLKTTGLE-IKLDHVSEGIPVIKERLCRKKILLILDDVDN 309
LH+VR+ SA+ L++LQE LL + ++ ++++ EG + K+RL KK+LL+LDDVD+
Sbjct: 250 LHEVRDRSAKQGLEHLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLRWMAFKS 369
+ QL+ALAG +WFG GSR+IITT++K LL + E + + L++ E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMGTLDKYESLQLFKQHAFKK 369
Query: 370 DKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKEIQKILK 429
+ +ED+ + + + GLPL L+V+GS L+G+ +++W ++ +IP EI K L+
Sbjct: 370 NHSTKEFEDLSAQVIEHTGGLPLALKVLGSFLYGRGLDEWISEVERLKQIPQNEILKKLE 429
Query: 430 VSYDALEEEEQSVFLDIACCFKGYQWKEFEDIL-CAHYDHCITHHLGVLAGKSLVKISTY 488
S+ L EQ +FLDIAC F G + IL H+ I + VL K L+ I
Sbjct: 430 PSFTGLNNIEQKIFLDIACFFSGKKKDSVTRILESFHFSPVI--GIKVLMEKCLITILK- 486
Query: 489 YPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIHVLKENTGTSKIEMIY 548
+ +H LI++MG +VR+E+ P SRLW++EDI VL++N T KIE +
Sbjct: 487 ------GRITIHQLIQEMGWHIVRREASYNPRICSRLWKREDICPVLEQNLCTDKIEGMS 540
Query: 549 MNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLKYLPSSLRVLKWKGCLSKCLSSS 608
++L + E V + GKA +MT L+ L N G ++LP LR L W G SK L +S
Sbjct: 541 LHLTNEEEV-NFGGKALMQMTSLRFLKFRNAYVYQGPEFLPDELRWLDWHGYPSKNLPNS 599
Query: 609 ILNKASEITSQ*FCFSQQKKSFLVCMQK----FQNMKVLTLDYCEYLTHIPDVSGLSNLE 664
K ++ S + KKS ++ + K +K + L + + L +PD S NLE
Sbjct: 600 F--KGDQLVS-----LKLKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLE 652
Query: 665 KLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKLEHF-RPLGLASLKKLILYECECLDNF 723
+L C +L+ I+ SIG L KL L+ CR L+ + + L L+ L+L C L F
Sbjct: 653 RLVLEECTSLVEINFSIGDLGKLVLLNLKNCRNLKTIPKRIRLEKLEVLVLSGCSKLRTF 712
Query: 724 PELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGMKFPKIVFSNMTKLSLSFF 783
PE+ KM + E+ + TS+ ELP S +N S + + ++ ++ L S F
Sbjct: 713 PEIEEKMNRLAELYLGATSLSELPASVENFSGVGVINLS--------YCKHLESLPSSIF 764
Query: 784 NLSDECLPIVLKWCVNMTHLDLS-FSNFKILPECLRECHHLVEINVMCC-----ESLEEI 837
L +CL LD+S S K LP+ + LV I + C +++
Sbjct: 765 RL--KCLKT----------LDVSGCSKLKNLPD---DLGLLVGIEKLHCTHTAIQTIPSS 809
Query: 838 RGIPPNLKELCARYCKSLSS 857
+ NLK L C +LSS
Sbjct: 810 MSLLKNLKHLSLSGCNALSS 829
>UniRef100_Q6URA2 TIR-NBS-LRR type R protein 7 [Malus baccata]
Length = 1095
Score = 482 bits (1240), Expect = e-134
Identities = 315/882 (35%), Positives = 490/882 (54%), Gaps = 50/882 (5%)
Query: 2 QSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITP 61
++ SSS+S+S + Y +FLSFRG DTR GFTG+L+ AL D+G ++D++ L RG+EI
Sbjct: 9 EASSSSSSMSKLWNYDLFLSFRGEDTRNGFTGHLHAALKDRGYQAYMDQDDLNRGEEIKE 68
Query: 62 SLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRK 121
L +AIE SRI I VFS YA SS+CLDELV I+ C GR VLP+F+ V+P+ VR +
Sbjct: 69 ELFRAIEGSRISIIVFSKRYADSSWCLDELVKIMECRSKLGRHVLPIFYHVDPSHVRKQD 128
Query: 122 GSYGEALAEHEKRF------QNDPKNMERLQGWKKALSQAANLSGYHD---SPPGYEYKL 172
G EA +HE+ + ER++ WKKAL++AANLSG HD + G E L
Sbjct: 129 GDLAEAFLKHEEGIGEGTDGKKREAKQERVKQWKKALTEAANLSG-HDLRITDNGREANL 187
Query: 173 IGK-IVKYISNK--ISRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLG 229
+ IV I K +S L VA + VG+ SR+Q + S L G + V MVGI+G+GGLG
Sbjct: 188 CPREIVDNIITKWLMSTNKLRVAKHQVGINSRIQDIISRLSSGGSN-VIMVGIWGMGGLG 246
Query: 230 KSTLARQIYNFVADQFEGSCFLHDVRENSAQNNLKYLQEKLLLKTTGLEIKLDHVSEGIP 289
K+T A+ IYN + +F+ FL DV ++++ L YLQ++L+ + K+ V EGI
Sbjct: 247 KTTAAKAIYNQIHHEFQFKSFLPDVGNAASKHGLVYLQKELIYDILKTKSKISSVDEGIG 306
Query: 290 VIKERLCRKKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHA 349
+I+++ +++L+I+D++D + QL A+ G DWFG GSR+IITTR++ LL ++ T+
Sbjct: 307 LIEDQFRHRRVLVIMDNIDEVGQLDAIVGNPDWFGPGSRIIITTRDEHLLKQ--VDKTYV 364
Query: 350 VEGLNETEALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDW 409
+ L+E EALEL W AF ++ Y ++ + V+Y GLPL LEV+GS LF + I +W
Sbjct: 365 AQKLDEREALELFSWHAFGNNWPNEEYLELSEKVVSYCGGLPLALEVLGSFLFKRPIAEW 424
Query: 410 KHTLDGYDRIPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHC 469
K L+ R P +I K L++S++ L++ ++++FLDI+C F G E +D + D C
Sbjct: 425 KSQLEKLKRTPEGKIIKSLRISFEGLDDAQKAIFLDISCFFIG----EDKDYVAKVLDGC 480
Query: 470 ---ITHHLGVLAGKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLW 526
T + VL + LV + N + +HDL+++M K ++ ++SP +PG+ SRLW
Sbjct: 481 GFYATIGISVLRERCLVTVEH-------NKLNMHDLLREMAKVIISEKSPGDPGKWSRLW 533
Query: 527 RQEDIIHVLKENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGGLK 586
+ ++I+VL +GT ++E + + +AF + KL+ L + +G K
Sbjct: 534 DKREVINVLTNKSGTEEVEGLALPWGYRHDTAFST-EAFANLKKLRLLQLCRVELNGEYK 592
Query: 587 YLPSSLRVLKWKGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVC--MQKFQNMKVLT 644
+LP L L W C K + N+ + + Q K V + N+K L
Sbjct: 593 HLPKELIWLHWFECPLKSIPDDFFNQDKLVVLE----MQWSKLVQVWEGSKSLHNLKTLD 648
Query: 645 LDYCEYLTHIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNKLEWLSAYGCRKL-----E 699
L L PD S + NLE+L C L IH SIGHL +L ++ C KL +
Sbjct: 649 LSESRSLQKSPDFSQVPNLEELILYNCKELSEIHPSIGHLKRLSLVNLEWCDKLISLPGD 708
Query: 700 HFRPLGLASLKKLILYECECLDNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHEL 759
++ S++ L+L C L E + +M ++ ++ T I E+P S L L L
Sbjct: 709 FYKS---KSVEALLLNGCLILRELHEDIGEMISLRTLEAEYTDIREVPPSIVRLKNLTRL 765
Query: 760 TVTS--GMKFPKIV--FSNMTKLSLSFFNLSDECLPIVLKWCVNMTHLDLSFSNFKILPE 815
+++S + P + +++ +L+LS F L+D+ +P L +++ L+L ++F LP
Sbjct: 766 SLSSVESIHLPHSLHGLNSLRELNLSSFELADDEIPKDLGSLISLQDLNLQRNDFHTLPS 825
Query: 816 CLRECHHLVEINVMCCESLEEIRGIPPNLKELCARYCKSLSS 857
L L + + CE L I +P NLK L A C +L +
Sbjct: 826 -LSGLSKLETLRLHHCEQLRTITDLPTNLKFLLANGCPALET 866
>UniRef100_Q84ZV0 R 14 protein [Glycine max]
Length = 641
Score = 478 bits (1230), Expect = e-133
Identities = 248/437 (56%), Positives = 322/437 (72%), Gaps = 6/437 (1%)
Query: 6 SSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITPSLLK 65
++T+ S + Y VFLSFRG DTRYGFTGNLY+AL +KGI+TF D+ L GDEITP+L K
Sbjct: 2 AATTRSLPFIYDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRKGSYG 125
AI+ESRI I V S NYA SSFCLDELV I+HC K++G LV+PVF+ V+P+ +RH+KGSYG
Sbjct: 62 AIQESRIAITVLSQNYAFSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDLRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
EA+ +H+KRF++ ME+LQ W+ AL Q A+LSG+H YEYK IG IV+ +S KI
Sbjct: 121 EAMIKHQKRFES---KMEKLQKWRMALKQVADLSGHHFKDGDAYEYKFIGSIVEEVSRKI 177
Query: 185 SRQPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYNFVADQ 244
+R LHV YPVGL+S+V + LLD GSDD VH++GI+G+ GLGK+TL+ +YN +A
Sbjct: 178 NRASLHVLDYPVGLESQVTDLMKLLDVGSDDVVHIIGIHGMRGLGKTTLSLAVYNLIALH 237
Query: 245 FEGSCFLHDVRENSAQNNLKYLQEKLLLKTTG-LEIKLDHVSEGIPVIKERLCRKKILLI 303
F+ SCFL +VRE S ++ LK+LQ LLLK G +I L EG +I+ RL RKK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLLKLLGEKDINLTSWQEGASMIQHRLRRKKVLLI 297
Query: 304 LDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETEALELLR 363
LDD D +QL A+ G DWFG GSRVIITTR+K LL HGIE T+ V+ LN+ AL+LL
Sbjct: 298 LDDADRHEQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHGIERTYEVKVLNDNAALQLLT 357
Query: 364 WMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIEDWKHTLDGYDRIPNKE 423
W AF+ +K+ YE +LNR VAYA GLPL LEV+GS+LF K++ +W++ ++ Y RIP E
Sbjct: 358 WNAFRREKIDPSYEHVLNRVVAYASGLPLALEVIGSHLFEKTVAEWEYAVEHYSRIPIDE 417
Query: 424 IQKILKVSYDALEEEEQ 440
I ILKVS+DA ++E Q
Sbjct: 418 IVDILKVSFDATKQETQ 434
>UniRef100_Q84KB3 MRGH63 [Cucumis melo]
Length = 943
Score = 475 bits (1222), Expect = e-132
Identities = 330/917 (35%), Positives = 492/917 (52%), Gaps = 99/917 (10%)
Query: 2 QSPSSSTSISYEYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDKNGLQRGDEITP 61
Q+ SS+S + + + VFLSFRG DTR FT +L L +GIN FIDK L RG+EI+
Sbjct: 3 QAGRSSSSSCFRWSFDVFLSFRGEDTRSNFTSHLNMTLRQRGINVFIDKK-LSRGEEISS 61
Query: 62 SLLKAIEESRIFIPVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFGVEPTVVRHRK 121
SLL+AIEES++ I V S +YASSS+CL+ELV II C K +G++VLP+F+ V+P+ V ++
Sbjct: 62 SLLEAIEESKVSIIVISESYASSSWCLNELVKIIMCNKLRGQVVLPIFYKVDPSEVGNQS 121
Query: 122 GSYGEALAEHEKRFQNDPKNMERLQGWKKALSQAANLSGYHDSPPGYEYKLIGKIVKYIS 181
G +GE A+ E RF +D +++ WK+AL +++SG+ E LI IV+ +
Sbjct: 122 GRFGEEFAKLEVRFSSD-----KMEAWKEALITVSHMSGWPVLQRDDEANLIQNIVQEVW 176
Query: 182 NKISR--QPLHVATYPVGLQSRVQQVKSLLDEGSDDGVHMVGIYGIGGLGKSTLARQIYN 239
++ R L VA YPVG+ QV++LL +G MVG+YGIGG+GK+TLA+ +YN
Sbjct: 177 KELDRATMQLDVAKYPVGIDI---QVRNLLPHVMSNGTTMVGLYGIGGMGKTTLAKALYN 233
Query: 240 FVADQFEGSCFLHDVRENSAQ-NNLKYLQEKLLLKT-TGLEIKLDHVSEGIPVIKERLCR 297
+AD FEG CFL ++RE S Q L LQ +LL + IK+ ++ G+ +I+ RL
Sbjct: 234 KIADDFEGCCFLPNIREASNQYGGLVQLQRELLREILVDDSIKVSNLPRGVTIIRNRLYS 293
Query: 298 KKILLILDDVDNLKQLHALAGGLDWFGCGSRVIITTRNKDLLSSHGIESTHAVEGLNETE 357
KKILLILDDVD +QL AL GG DWFG GS+VI TTRNK LL +HG + +V GL+ E
Sbjct: 294 KKILLILDDVDTREQLQALVGGHDWFGHGSKVIATTRNKQLLVTHGFDKMQSVVGLDYDE 353
Query: 358 ALELLRWMAFKSDKVPSGYEDILNRAVAYAFGLPLVLEVVGSNLFGKSIED---WKHTLD 414
ALEL W F++ + Y ++ RAV Y GLPL LEV+GS F SI+D +K LD
Sbjct: 354 ALELFSWHCFRNSHPLNDYLELSKRAVDYCKGLPLALEVLGS--FLHSIDDPFNFKRILD 411
Query: 415 GYDR-IPNKEIQKILKVSYDALEEEEQSVFLDIACCFKGYQWKEFEDILCAHYDHCITHH 473
Y++ +KEIQ L++SYD LE+E + +F I+CCF + + +L A C+
Sbjct: 412 EYEKYYLDKEIQDSLRISYDGLEDEVKEIFCYISCCFVREDINKVKMMLEACGCICLEKG 471
Query: 474 LGVLAGKSLVKISTYYPSGSINDVRLHDLIKDMGKEVVRQESPKEPGERSRLWRQEDIIH 533
+ L SL+ I G N V +HD+I+ MG+ + E+ K +R RL ++D ++
Sbjct: 472 ITKLMNLSLLTI------GRFNRVEMHDIIQQMGRTIHLSETSKS-HKRKRLLIKDDAMN 524
Query: 534 VLKENTGTSKIEMIYMNLHSMESVIDKKGKAFKKMTKLKTLIIENGLFSGG--LKYLPSS 591
VLK N +++I N + +D +AF+K+ L L + N S L+YLPSS
Sbjct: 525 VLKGNKEARAVKVIKFNF-PKPTELDIDSRAFEKVKNLVVLEVGNATSSKSTTLEYLPSS 583
Query: 592 LRVLKW-KGCLSKCLSSSILNKASEITSQ*FCFSQQKKSFLVCMQKFQNMKVLTLDYCEY 650
LR + W + S + + E+ + ++ C + +K + L +
Sbjct: 584 LRWMNWPQFPFSSLPPTYTMENLVELKLPYSSIKHFGQGYMSC----ERLKEINLTDSNF 639
Query: 651 LTHIPDVSGLSNLEKLSFTCCDNLITIHNSIGHLNK------------------------ 686
L IPD+S NL+ L C+NL+ +H SIG LNK
Sbjct: 640 LVEIPDLSTAINLKYLDLVGCENLVKVHESIGSLNKLVALHLSSSVKGFEQFPSHLKLKS 699
Query: 687 LEWLSAYGCR------------KLEHFRPLG--------------LASLKKLILYECECL 720
L++LS CR K + +G L SLK L LY C+ L
Sbjct: 700 LKFLSMKNCRIDEWCPQFSEEMKSIEYLSIGYSIVTHQLSPTIGYLTSLKHLTLYYCKEL 759
Query: 721 DNFPELLCKMAHIKEIDISNTSIGELPFSFQNLSELHELTVTSGMKFPKIVFSNMTKLSL 780
P + +++++ + + ++ + P L+ ++ S + + +TKL L
Sbjct: 760 TTLPSTIYRLSNLTSLIVLDSDLSTFP-------SLNHPSLPSSLFY-------LTKLRL 805
Query: 781 SFFNLSD-ECLPIVLKWCVNMTHLDLSFSNFKILPECLRECHHLVEINVMCCESLEEIRG 839
+++ + L ++ ++ LDLS +NF LP C+ L + M CE LEEI
Sbjct: 806 VGCKITNLDFLETIVYVAPSLKELDLSENNFCRLPSCIINFKSLKYLYTMDCELLEEISK 865
Query: 840 IPPNLKELCARYCKSLS 856
+P + A CKSL+
Sbjct: 866 VPEGVICTSAAGCKSLA 882
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,764,346,360
Number of Sequences: 2790947
Number of extensions: 77550562
Number of successful extensions: 239444
Number of sequences better than 10.0: 2669
Number of HSP's better than 10.0 without gapping: 1051
Number of HSP's successfully gapped in prelim test: 1639
Number of HSP's that attempted gapping in prelim test: 229845
Number of HSP's gapped (non-prelim): 6157
length of query: 1040
length of database: 848,049,833
effective HSP length: 138
effective length of query: 902
effective length of database: 462,899,147
effective search space: 417535030594
effective search space used: 417535030594
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC126790.9


