

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.6 - phase: 0
(228 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
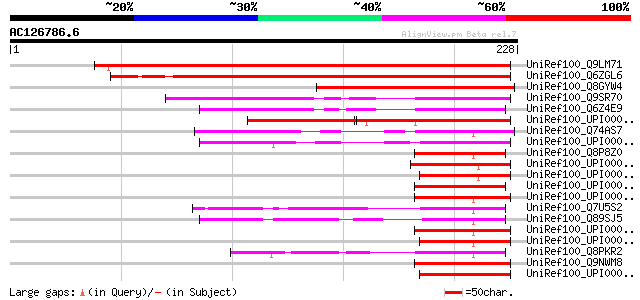
UniRef100_Q9LM71 Probable FKBP-type peptidyl-prolyl cis-trans is... 280 2e-74
UniRef100_Q6ZGL6 Putative immunophilin / FKBP-type peptidyl-prol... 259 3e-68
UniRef100_Q8GYW4 Hypothetical protein [Arabidopsis thaliana] 138 1e-31
UniRef100_Q9SR70 T22K18.11 protein [Arabidopsis thaliana] 106 5e-22
UniRef100_Q6Z4E9 Immunophilin / FKBP-type peptidyl-prolyl cis-tr... 84 4e-15
UniRef100_UPI0000295D3F UPI0000295D3F UniRef100 entry 71 2e-11
UniRef100_Q74AS7 FKBP-type peptidyl-prolyl cis-trans isomerase [... 57 4e-07
UniRef100_UPI000032020B UPI000032020B UniRef100 entry 56 6e-07
UniRef100_Q8P8Z0 FKBP-type peptidyl-prolyl cis-trans isomerase [... 54 2e-06
UniRef100_UPI00002E26BF UPI00002E26BF UniRef100 entry 54 3e-06
UniRef100_UPI0000311939 UPI0000311939 UniRef100 entry 54 4e-06
UniRef100_UPI00002AD005 UPI00002AD005 UniRef100 entry 53 5e-06
UniRef100_UPI00002A4102 UPI00002A4102 UniRef100 entry 53 7e-06
UniRef100_Q7U5S2 FKBP-type peptidyl-prolyl cis-trans isomerase (... 53 7e-06
UniRef100_Q89SJ5 Peptidyl-prolyl cis-trans isomerase [Bradyrhizo... 52 9e-06
UniRef100_UPI000034D8C4 UPI000034D8C4 UniRef100 entry 52 1e-05
UniRef100_UPI0000326439 UPI0000326439 UniRef100 entry 52 1e-05
UniRef100_Q8PKR2 FKBP-type peptidyl-prolyl cis-trans isomerase [... 52 1e-05
UniRef100_Q9NWM8 FK506 binding protein 14 precursor [Homo sapiens] 51 3e-05
UniRef100_UPI00002FF524 UPI00002FF524 UniRef100 entry 50 3e-05
>UniRef100_Q9LM71 Probable FKBP-type peptidyl-prolyl cis-trans isomerase 1,
chloroplast precursor [Arabidopsis thaliana]
Length = 232
Score = 280 bits (717), Expect = 2e-74
Identities = 138/190 (72%), Positives = 159/190 (83%), Gaps = 3/190 (1%)
Query: 39 VVALS---QSRRSTAILISSLPFTFVFLSPPPPAEARERRKKKNIPIDDYITSPDGLKYY 95
VVA S R ++ IL+SS+P T F+ P +EARERR +K IP+++Y T P+GLK+Y
Sbjct: 36 VVAFSVPISRRDASIILLSSIPLTSFFVLTPSSSEARERRSRKVIPLEEYSTGPEGLKFY 95
Query: 96 DFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKER 155
D EGKGP+A +GST QVHFDC YR ITA+S+RESKLLAGNR IAQPYEFKVG+ PGKER
Sbjct: 96 DIEEGKGPVATEGSTAQVHFDCRYRSITAISTRESKLLAGNRSIAQPYEFKVGSTPGKER 155
Query: 156 KREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGAT 215
KREFVDNPNGLFSAQAAPKPP AMY I EGM+VGGKRTVIVPPE GYG+KGMNEIPPGAT
Sbjct: 156 KREFVDNPNGLFSAQAAPKPPPAMYFITEGMKVGGKRTVIVPPEAGYGQKGMNEIPPGAT 215
Query: 216 FELNIELLQL 225
FELNIELL++
Sbjct: 216 FELNIELLRV 225
>UniRef100_Q6ZGL6 Putative immunophilin / FKBP-type peptidyl-prolyl cis-trans
isomerase [Oryza sativa]
Length = 242
Score = 259 bits (663), Expect = 3e-68
Identities = 130/180 (72%), Positives = 151/180 (83%), Gaps = 4/180 (2%)
Query: 46 RRSTAILISSLPFTFVFLSPPPPAEARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIA 105
RR+ A L+S+ F + +SPP A RR + + ++DY+TSPDGLKYYD +EGKGP A
Sbjct: 61 RRAAAQLLSAAGF-LIAVSPPSLAA---RRGRMVVSLEDYVTSPDGLKYYDLVEGKGPTA 116
Query: 106 EKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNG 165
EKGSTVQVHFDC+YRGITAVSSRE+KLLAGNR IAQPYEF VG+ PGKERKREFVD+ NG
Sbjct: 117 EKGSTVQVHFDCIYRGITAVSSREAKLLAGNRSIAQPYEFSVGSLPGKERKREFVDSANG 176
Query: 166 LFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
L+SAQA+PKPP AMYTI EGM+VGGKR VIVPPE GYGKKGMNEIPP A FEL+IELL++
Sbjct: 177 LYSAQASPKPPAAMYTITEGMKVGGKRRVIVPPELGYGKKGMNEIPPDAPFELDIELLEV 236
>UniRef100_Q8GYW4 Hypothetical protein [Arabidopsis thaliana]
Length = 95
Score = 138 bits (348), Expect = 1e-31
Identities = 67/89 (75%), Positives = 74/89 (82%)
Query: 139 IAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPP 198
+ QPYEFKVG+ PGKERKREFVDNPNGLFSAQAAPKPP AMY I EGM+VGGKRTVIVPP
Sbjct: 7 LEQPYEFKVGSTPGKERKREFVDNPNGLFSAQAAPKPPPAMYFITEGMKVGGKRTVIVPP 66
Query: 199 EKGYGKKGMNEIPPGATFELNIELLQLAS 227
E GYG+KGMNEIP ++L + L L S
Sbjct: 67 EAGYGQKGMNEIPVRTYYKLYMSLELLLS 95
>UniRef100_Q9SR70 T22K18.11 protein [Arabidopsis thaliana]
Length = 230
Score = 106 bits (264), Expect = 5e-22
Identities = 66/155 (42%), Positives = 82/155 (52%), Gaps = 24/155 (15%)
Query: 71 ARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRES 130
+R + +P D+ T P+GLKYYD G G A KGS V VH+ ++GIT ++SR+
Sbjct: 86 SRRALRASKLPESDFTTLPNGLKYYDIKVGNGAEAVKGSRVAVHYVAKWKGITFMTSRQG 145
Query: 131 KLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGG 190
+ G PY F VG ER G VEGMRVGG
Sbjct: 146 LGVGGGT----PYGFDVGQ---SERGNVLKGLDLG-----------------VEGMRVGG 181
Query: 191 KRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
+R VIVPPE YGKKG+ EIPP AT EL+IELL +
Sbjct: 182 QRLVIVPPELAYGKKGVQEIPPNATIELDIELLSI 216
>UniRef100_Q6Z4E9 Immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase- like
[Oryza sativa]
Length = 236
Score = 83.6 bits (205), Expect = 4e-15
Identities = 54/138 (39%), Positives = 70/138 (50%), Gaps = 24/138 (17%)
Query: 86 ITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEF 145
+TS +YYD G G A KGS V VH+ ++GIT ++SR+ + G PY F
Sbjct: 118 VTSEALSRYYDITVGSGLKAVKGSRVAVHYVAKWKGITFMTSRQGLGVGGGT----PYGF 173
Query: 146 KVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKK 205
+G ER G VEGM+VGG+R +IVPPE YGKK
Sbjct: 174 DIG---NSERGNVLKGLDLG-----------------VEGMKVGGQRLIIVPPELAYGKK 213
Query: 206 GMNEIPPGATFELNIELL 223
G+ EIPP AT E++ LL
Sbjct: 214 GVQEIPPNATIEVDCHLL 231
>UniRef100_UPI0000295D3F UPI0000295D3F UniRef100 entry
Length = 233
Score = 70.9 bits (172), Expect = 2e-11
Identities = 37/73 (50%), Positives = 49/73 (66%), Gaps = 3/73 (4%)
Query: 156 KREF-VDNPNGLFSAQAAPKPPQAMYT--IVEGMRVGGKRTVIVPPEKGYGKKGMNEIPP 212
KRE V+ GLF+ + PKPP +Y ++GM+VGG+RT IVP + G+ G EIPP
Sbjct: 158 KREIQVEGGGGLFTGGSGPKPPPLIYVPEALKGMKVGGRRTFIVPSDVGFEDVGEGEIPP 217
Query: 213 GATFELNIELLQL 225
GATFEL +ELL +
Sbjct: 218 GATFELVVELLDV 230
Score = 53.1 bits (126), Expect = 5e-06
Identities = 21/49 (42%), Positives = 33/49 (66%)
Query: 108 GSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERK 156
G+ HF+C +RG+T S+RE++ L GNR I++P++FK G P + K
Sbjct: 3 GTLATTHFECKFRGLTVSSTREARTLGGNRTISEPFQFKYGTLPNEFTK 51
>UniRef100_Q74AS7 FKBP-type peptidyl-prolyl cis-trans isomerase [Geobacter
sulfurreducens]
Length = 138
Score = 57.0 bits (136), Expect = 4e-07
Identities = 43/145 (29%), Positives = 59/145 (40%), Gaps = 32/145 (22%)
Query: 84 DYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPY 143
D +T+ GL Y D G G G V+VH+ T S + +P+
Sbjct: 25 DAVTTASGLSYVDLAAGSGAAPVAGKPVKVHYTGWLENGTKFDSSVDR--------GEPF 76
Query: 144 EFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYG 203
F +GA + P + V M+VGGKR +IVPP+ GYG
Sbjct: 77 VFTIGA-------------------GEVIPGWDEG----VMSMKVGGKRRLIVPPQLGYG 113
Query: 204 KKGM-NEIPPGATFELNIELLQLAS 227
G IPP AT +ELL +A+
Sbjct: 114 AAGAGGVIPPNATLIFEVELLDVAT 138
>UniRef100_UPI000032020B UPI000032020B UniRef100 entry
Length = 134
Score = 56.2 bits (134), Expect = 6e-07
Identities = 44/141 (31%), Positives = 65/141 (45%), Gaps = 32/141 (22%)
Query: 86 ITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDC-LYRGITAVSSRESKLLAGNRVIAQPYE 144
I +GL D L G+G +AEK S VH+ L +G SS++ + +P+
Sbjct: 24 IIMENGLIIEDKLIGEGVVAEKYSIATVHYTGKLEKGKVFDSSKQ--------IGREPFR 75
Query: 145 FKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGK 204
F +GA Q Q + GM+VGG+R +I+PP GYG
Sbjct: 76 FTLGA-------------------GQVIEGWDQGII----GMKVGGQRKLIIPPHLGYGD 112
Query: 205 KGMNEIPPGATFELNIELLQL 225
+ M IPP +T NIEL+++
Sbjct: 113 QDMGVIPPNSTLIFNIELIEI 133
>UniRef100_Q8P8Z0 FKBP-type peptidyl-prolyl cis-trans isomerase [Xanthomonas
campestris]
Length = 132
Score = 54.3 bits (129), Expect = 2e-06
Identities = 26/42 (61%), Positives = 32/42 (75%), Gaps = 1/42 (2%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELL 223
V GMRVGGKRT+++PPE GYG KG IPPGA+ ++ELL
Sbjct: 86 VAGMRVGGKRTLMIPPEFGYGDKGAGGVIPPGASLVFDVELL 127
>UniRef100_UPI00002E26BF UPI00002E26BF UniRef100 entry
Length = 225
Score = 53.9 bits (128), Expect = 3e-06
Identities = 24/46 (52%), Positives = 35/46 (75%), Gaps = 1/46 (2%)
Query: 181 TIVEGMRVGGKRTVIVPPEKGYGKKGMNE-IPPGATFELNIELLQL 225
T + GM+VGGKRT+I+PPE GYG++G E IPP +T +IE++ +
Sbjct: 63 TGIIGMKVGGKRTLIIPPELGYGERGAGELIPPNSTLVFDIEIIDI 108
>UniRef100_UPI0000311939 UPI0000311939 UniRef100 entry
Length = 231
Score = 53.5 bits (127), Expect = 4e-06
Identities = 23/42 (54%), Positives = 33/42 (77%), Gaps = 1/42 (2%)
Query: 185 GMRVGGKRTVIVPPEKGYGKKGMNE-IPPGATFELNIELLQL 225
GM+VGGKRT+I+PPE GYG++G E IPP +T +IE++ +
Sbjct: 73 GMKVGGKRTLIIPPELGYGERGAGELIPPNSTLVFDIEIIDI 114
>UniRef100_UPI00002AD005 UPI00002AD005 UniRef100 entry
Length = 50
Score = 53.1 bits (126), Expect = 5e-06
Identities = 21/41 (51%), Positives = 29/41 (70%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELL 223
V GM+VGGKR +++PPE GYG +G+ IPP + +ELL
Sbjct: 7 VAGMKVGGKRKLVIPPELGYGSRGIGPIPPNSVLTFEVELL 47
>UniRef100_UPI00002A4102 UPI00002A4102 UniRef100 entry
Length = 131
Score = 52.8 bits (125), Expect = 7e-06
Identities = 25/44 (56%), Positives = 32/44 (71%), Gaps = 1/44 (2%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELLQL 225
V+ M+VGGKR + +PPE GYG KG N IPPGAT +ELL++
Sbjct: 85 VKDMKVGGKRKLTIPPELGYGDKGAGNVIPPGATLIFEVELLEV 128
>UniRef100_Q7U5S2 FKBP-type peptidyl-prolyl cis-trans isomerase (PPIase) precursor
[Synechococcus sp.]
Length = 208
Score = 52.8 bits (125), Expect = 7e-06
Identities = 45/142 (31%), Positives = 60/142 (41%), Gaps = 33/142 (23%)
Query: 83 DDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQP 142
D IT+ GLK + G+G A G TV VH YRG + K + P
Sbjct: 96 DTQITA-SGLKIIELQVGEGAEAASGQTVSVH----YRG----TLENGKQFDASYDRGTP 146
Query: 143 YEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGY 202
+ F +GA + E VD GM+VGGKR +++PP+ Y
Sbjct: 147 FTFPLGAGRVIKGWDEGVD-----------------------GMKVGGKRKLVIPPDLAY 183
Query: 203 GKKGM-NEIPPGATFELNIELL 223
G +G IPP AT +ELL
Sbjct: 184 GSRGAGGVIPPNATLVFEVELL 205
>UniRef100_Q89SJ5 Peptidyl-prolyl cis-trans isomerase [Bradyrhizobium japonicum]
Length = 154
Score = 52.4 bits (124), Expect = 9e-06
Identities = 40/139 (28%), Positives = 61/139 (43%), Gaps = 28/139 (20%)
Query: 86 ITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEF 145
+T+ GL+ D G G + G +H Y G + ++ K + +P+EF
Sbjct: 40 MTTASGLQIIDTAVGTGASPQPGQICVMH----YTGWLYENGQKGKKFDSSVDRKEPFEF 95
Query: 146 KVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKK 205
+G + R G+ S M+VGGKRT+I+PP+ GYG +
Sbjct: 96 PIG------KGRVIAGWDEGVAS-----------------MKVGGKRTLIIPPQLGYGAR 132
Query: 206 GM-NEIPPGATFELNIELL 223
G IPP AT ++ELL
Sbjct: 133 GAGGVIPPNATLMFDVELL 151
>UniRef100_UPI000034D8C4 UPI000034D8C4 UniRef100 entry
Length = 71
Score = 52.0 bits (123), Expect = 1e-05
Identities = 24/44 (54%), Positives = 32/44 (72%), Gaps = 1/44 (2%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELLQL 225
V+ M+VGGKR + +PPE GYG +G N IPPGAT +ELL++
Sbjct: 25 VKDMKVGGKRKLTIPPELGYGDRGAGNVIPPGATLIFEVELLEI 68
>UniRef100_UPI0000326439 UPI0000326439 UniRef100 entry
Length = 239
Score = 52.0 bits (123), Expect = 1e-05
Identities = 24/42 (57%), Positives = 31/42 (73%), Gaps = 1/42 (2%)
Query: 185 GMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELLQL 225
GM++GGKRT+IVPPE GYGKKG N IPP + IE++ +
Sbjct: 87 GMKIGGKRTLIVPPELGYGKKGAGNIIPPNSKLIFEIEIIDV 128
>UniRef100_Q8PKR2 FKBP-type peptidyl-prolyl cis-trans isomerase [Xanthomonas
axonopodis]
Length = 132
Score = 52.0 bits (123), Expect = 1e-05
Identities = 42/126 (33%), Positives = 59/126 (46%), Gaps = 27/126 (21%)
Query: 100 GKGPIAEKGSTVQVHFD-CLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKRE 158
G G A G+ V VH+ LY A + K + A+P++F +G G + R
Sbjct: 27 GTGAEATPGAMVTVHYTGWLYDEKAA--DKHGKKFDSSLDRAEPFQFVLG---GHQVIRG 81
Query: 159 FVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFE 217
+ D V GMRVGGKRT+++PP+ GYG G IPPGA+
Sbjct: 82 WDDG--------------------VAGMRVGGKRTLMIPPDYGYGDNGAGGVIPPGASLV 121
Query: 218 LNIELL 223
++ELL
Sbjct: 122 FDVELL 127
>UniRef100_Q9NWM8 FK506 binding protein 14 precursor [Homo sapiens]
Length = 211
Score = 50.8 bits (120), Expect = 3e-05
Identities = 22/43 (51%), Positives = 32/43 (74%)
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
++GM VG KR +I+PP GYGK+G +IPP +T NI+LL++
Sbjct: 92 LKGMCVGEKRKLIIPPALGYGKEGKGKIPPESTLIFNIDLLEI 134
>UniRef100_UPI00002FF524 UPI00002FF524 UniRef100 entry
Length = 130
Score = 50.4 bits (119), Expect = 3e-05
Identities = 21/41 (51%), Positives = 29/41 (70%)
Query: 185 GMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
GM+VGGKRT+++P E GYG G IPP A+ ++ELL +
Sbjct: 87 GMKVGGKRTLMIPAEYGYGASGAGPIPPNASLVFDVELLDV 127
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 408,228,492
Number of Sequences: 2790947
Number of extensions: 17517319
Number of successful extensions: 42952
Number of sequences better than 10.0: 379
Number of HSP's better than 10.0 without gapping: 250
Number of HSP's successfully gapped in prelim test: 129
Number of HSP's that attempted gapping in prelim test: 42419
Number of HSP's gapped (non-prelim): 578
length of query: 228
length of database: 848,049,833
effective HSP length: 123
effective length of query: 105
effective length of database: 504,763,352
effective search space: 53000151960
effective search space used: 53000151960
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 72 (32.3 bits)
Medicago: description of AC126786.6


