

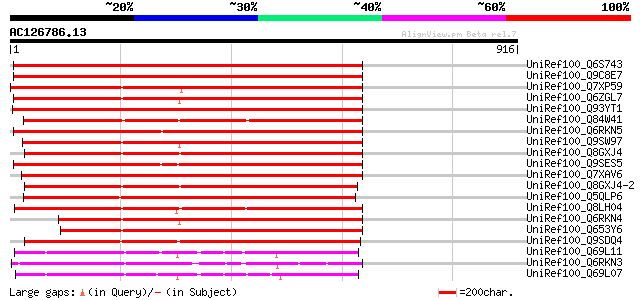
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.13 - phase: 0
(916 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6S743 GLR3.3 [Arabidopsis thaliana] 822 0.0
UniRef100_Q9C8E7 Glutamate receptor 3.3 precursor [Arabidopsis t... 822 0.0
UniRef100_Q7XP59 OSJNBa0013K16.8 protein [Oryza sativa] 743 0.0
UniRef100_Q6ZGL7 Putative glutamate receptor [Oryza sativa] 705 0.0
UniRef100_Q93YT1 Glutamate receptor 3.2 precursor [Arabidopsis t... 696 0.0
UniRef100_Q84W41 Glutamate receptor 3.6 precursor [Arabidopsis t... 693 0.0
UniRef100_Q6RKN5 Glutamate receptor 3.1 precursor [Arabidopsis t... 692 0.0
UniRef100_Q9SW97 Glutamate receptor 3.5 precursor [Arabidopsis t... 687 0.0
UniRef100_Q8GXJ4 Glutamate receptor 3.4 precursor [Arabidopsis t... 686 0.0
UniRef100_Q9SES5 Ligand gated channel-like protein [Brassica napus] 682 0.0
UniRef100_Q7XAV6 Glutamate receptor [Raphanus sativus var. sativus] 670 0.0
UniRef100_Q8GXJ4-2 Splice isoform 2 of Q8GXJ4 [Arabidopsis thali... 669 0.0
UniRef100_Q5QLP6 Putative ionotropic glutamate receptor homolog ... 660 0.0
UniRef100_Q8LH04 Glutamate receptor, ionotropic kainate 5-like p... 638 0.0
UniRef100_Q6RKN4 Putative glutamate receptor ion channel [Arabid... 620 e-176
UniRef100_Q653Y6 Putative ionotropic glutamate receptor ortholog... 619 e-175
UniRef100_Q9SDQ4 Glutamate receptor 3.7 precursor [Arabidopsis t... 540 e-151
UniRef100_Q69L11 Putative Avr9/Cf-9 rapidly elicited protein 141... 372 e-101
UniRef100_Q6RKN3 Glutamate receptor 2.7 precursor [Arabidopsis t... 371 e-101
UniRef100_Q69L07 Putative Avr9/Cf-9 rapidly elicited protein 141... 360 1e-97
>UniRef100_Q6S743 GLR3.3 [Arabidopsis thaliana]
Length = 933
Score = 822 bits (2124), Expect = 0.0
Identities = 389/634 (61%), Positives = 495/634 (77%), Gaps = 3/634 (0%)
Query: 7 WLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILH 66
W + F+ L + + +P V IG+IF+FDS IGKVAK+A+++AVKDVNSN IL
Sbjct: 5 WTFFFLSFLCSGLFRRTHSEKPKVVKIGSIFSFDSVIGKVAKIAIDEAVKDVNSNPDILS 64
Query: 67 STQLVLHMQTSNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFA 126
T+ + MQ SNCSGF GM++ALRFME D++ I+GPQ SVV+H+++H+ANELRVP+LSFA
Sbjct: 65 GTKFSVSMQNSNCSGFMGMVEALRFMEKDIVGIIGPQCSVVAHMISHMANELRVPLLSFA 124
Query: 127 ATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDA 186
TDP +S LQFP+F+RTT SDLYQM A+A I+DFYGWKEVI ++VDDD+GRNGV+AL+D
Sbjct: 125 VTDPVMSPLQFPYFIRTTQSDLYQMDAIASIVDFYGWKEVIAVFVDDDFGRNGVAALNDK 184
Query: 187 LAERRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLG 246
LA RR RI+YK G+ V++ EI N+L+ + ++Q RI+V+H +S GF +FK A YLG
Sbjct: 185 LASRRLRITYKAGLHPDTAVNKNEIMNMLIKIMLLQPRIVVIHVYSELGFAVFKEAKYLG 244
Query: 247 MMQEGYVWIATDWLSTVLDSTS-LPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTG 305
MM GYVWIATDWLST LDS+S LP E ++T+QG LVLR HTPD+D K+ F +W ++G
Sbjct: 245 MMGNGYVWIATDWLSTNLDSSSPLPAERLETIQGVLVLRPHTPDSDFKREFFKRWRKMSG 304
Query: 306 GSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHS-DKAGGLNLDAMSIFDN 364
SL LN+YGL+AYD+V L+A+ +D FF GG +S +N++ L++ K+G LNL+AM++FD
Sbjct: 305 ASLALNTYGLYAYDSVMLLARGLDKFFKDGGNISFSNHSMLNTLGKSGNLNLEAMTVFDG 364
Query: 365 GTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSP 424
G LL +IL + VGL+G ++ +RS RPAYDIINV G GVR++GYWSN+SGLS V P
Sbjct: 365 GEALLKDILGTRMVGLTGQLQFTPDRSRTRPAYDIINVAGTGVRQIGYWSNHSGLSTVLP 424
Query: 425 ETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVK 484
E LY S + L VIWPGET ++PRGWVF NNGK+L+IGVP+R SY+EFVS ++
Sbjct: 425 ELLYTKEKPNMSTSPKLRHVIWPGETFTKPRGWVFSNNGKELKIGVPLRVSYKEFVSQIR 484
Query: 485 GTD-LFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDI 543
GT+ +FKGFC+DVF AAVNLLPYAVP +F+P+G+G +NPSYT V ITTG FDG VGD+
Sbjct: 485 GTENMFKGFCIDVFTAAVNLLPYAVPVKFIPYGNGKENPSYTHMVEMITTGNFDGVVGDV 544
Query: 544 AIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVV 603
AIVTNRT+IVDFTQPYAASGLVVVAPFKK+NSG W+FL+PF MW VT C F FVGIVV
Sbjct: 545 AIVTNRTKIVDFTQPYAASGLVVVAPFKKLNSGAWAFLRPFNRLMWAVTGCCFLFVGIVV 604
Query: 604 WILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
WILEHR NDEFRG PK+Q VTILWFS ST+FF+H
Sbjct: 605 WILEHRTNDEFRGPPKRQCVTILWFSFSTMFFAH 638
>UniRef100_Q9C8E7 Glutamate receptor 3.3 precursor [Arabidopsis thaliana]
Length = 933
Score = 822 bits (2123), Expect = 0.0
Identities = 389/634 (61%), Positives = 495/634 (77%), Gaps = 3/634 (0%)
Query: 7 WLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILH 66
W + F+ L + + +P V IG+IF+FDS IGKVAK+A+++AVKDVNSN IL
Sbjct: 5 WTFFFLSFLCSGLFRRTHSEKPKVVKIGSIFSFDSVIGKVAKIAIDEAVKDVNSNPDILS 64
Query: 67 STQLVLHMQTSNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFA 126
T+ + MQ SNCSGF GM++ALRFME D++ I+GPQ SVV+H+++H+ANELRVP+LSFA
Sbjct: 65 GTKFSVSMQNSNCSGFMGMVEALRFMEKDIVGIIGPQCSVVAHMISHMANELRVPLLSFA 124
Query: 127 ATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDA 186
TDP +S LQFP+F+RTT SDLYQM A+A I+DFYGWKEVI ++VDDD+GRNGV+AL+D
Sbjct: 125 VTDPVMSPLQFPYFIRTTQSDLYQMDAIASIVDFYGWKEVIAVFVDDDFGRNGVAALNDK 184
Query: 187 LAERRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLG 246
LA RR RI+YK G+ V++ EI N+L+ + ++Q RI+V+H +S GF +FK A YLG
Sbjct: 185 LASRRLRITYKAGLHPDTAVNKNEIMNMLIKIMLLQPRIVVIHVYSELGFAVFKEAKYLG 244
Query: 247 MMQEGYVWIATDWLSTVLDSTS-LPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTG 305
MM GYVWIATDWLST LDS+S LP E ++T+QG LVLR HTPD+D K+ F +W ++G
Sbjct: 245 MMGNGYVWIATDWLSTNLDSSSPLPAERLETIQGVLVLRPHTPDSDFKREFFKRWRKMSG 304
Query: 306 GSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHS-DKAGGLNLDAMSIFDN 364
SL LN+YGL+AYD+V L+A+ +D FF GG +S +N++ L++ K+G LNL+AM++FD
Sbjct: 305 ASLALNTYGLYAYDSVMLLARGLDKFFKDGGNISFSNHSMLNTLGKSGNLNLEAMTVFDG 364
Query: 365 GTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSP 424
G LL +IL + VGL+G ++ +RS RPAYDIINV G GVR++GYWSN+SGLS V P
Sbjct: 365 GEALLKDILGTRMVGLTGQLQFTPDRSRTRPAYDIINVAGTGVRQIGYWSNHSGLSTVLP 424
Query: 425 ETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVK 484
E LY S + L VIWPGET ++PRGWVF NNGK+L+IGVP+R SY+EFVS ++
Sbjct: 425 ELLYTKEKPNMSTSPKLKHVIWPGETFTKPRGWVFSNNGKELKIGVPLRVSYKEFVSQIR 484
Query: 485 GTD-LFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDI 543
GT+ +FKGFC+DVF AAVNLLPYAVP +F+P+G+G +NPSYT V ITTG FDG VGD+
Sbjct: 485 GTENMFKGFCIDVFTAAVNLLPYAVPVKFIPYGNGKENPSYTHMVEMITTGNFDGVVGDV 544
Query: 544 AIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVV 603
AIVTNRT+IVDFTQPYAASGLVVVAPFKK+NSG W+FL+PF MW VT C F FVGIVV
Sbjct: 545 AIVTNRTKIVDFTQPYAASGLVVVAPFKKLNSGAWAFLRPFNRLMWAVTGCCFLFVGIVV 604
Query: 604 WILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
WILEHR NDEFRG PK+Q VTILWFS ST+FF+H
Sbjct: 605 WILEHRTNDEFRGPPKRQCVTILWFSFSTMFFAH 638
>UniRef100_Q7XP59 OSJNBa0013K16.8 protein [Oryza sativa]
Length = 938
Score = 743 bits (1917), Expect = 0.0
Identities = 350/643 (54%), Positives = 478/643 (73%), Gaps = 9/643 (1%)
Query: 1 MNFYYYWLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNS 60
M F +Y L+ +F + + + RP V IGA F +S+IG+VA +A+ AV D+N+
Sbjct: 1 MKFIFY-LFSIFCCLCSCAQSQNISGRPDAVRIGAQFARNSTIGRVAAVAVLAAVNDINN 59
Query: 61 NSSILHSTQLVLHMQTSNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRV 120
+S+IL T+L LHM S+C+ F G++QAL+FME D +AI+GP SS +H+++H+ANEL V
Sbjct: 60 DSNILPGTKLDLHMHDSSCNRFLGIVQALQFMEKDTVAIIGPLSSTTAHVLSHLANELHV 119
Query: 121 PMLSFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGV 180
P++SF+ATDPTLSSL++PFFVRTT+SD +QMTAVA+++++YGWK+V TI+VD+DYGRN +
Sbjct: 120 PLMSFSATDPTLSSLEYPFFVRTTVSDQFQMTAVADLVEYYGWKQVTTIFVDNDYGRNAI 179
Query: 181 SALDDALAERRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFK 240
S+L D L++RR +I YK + P EI ++L+ VAMM+SR+I++HA+ +SG ++F+
Sbjct: 180 SSLGDELSKRRSKILYKAPFR--PGASNNEIADVLIKVAMMESRVIILHANPDSGLVVFQ 237
Query: 241 VAHYLGMMQEGYVWIATDWLSTVLD-STSLPLETMDTLQGALVLRQHTPDTDRKKMFTSK 299
A LGM+ GY WIATDWL++ LD S L + + T+QG L LR HT +T RK M +SK
Sbjct: 238 QALKLGMVSNGYAWIATDWLTSYLDPSVHLDIGLLSTMQGVLTLRHHTENTRRKSMLSSK 297
Query: 300 WNNLTGGSLG-----LNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGL 354
W+ L G L++YGL+AYDTVW++A A+D FF+ GG +S + L+ GL
Sbjct: 298 WSELLKEDSGHSRFLLSTYGLYAYDTVWMLAHALDAFFNSGGNISFSPDPKLNEISGRGL 357
Query: 355 NLDAMSIFDNGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWS 414
NL+A+S+FD G LLL I + +F+G +GP+K DS +L +PAYDI++++G+G+R VGYWS
Sbjct: 358 NLEALSVFDGGQLLLEKIHQVDFLGATGPVKFDSGGNLIQPAYDIVSIIGSGLRTVGYWS 417
Query: 415 NYSGLSIVSPETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRA 474
NYSGLS++SPETLY P NR+ Q LH VIWPGET ++PRGWVFPNNG +++IGVP R
Sbjct: 418 NYSGLSVISPETLYKKPANRTRETQKLHDVIWPGETINKPRGWVFPNNGNEIKIGVPDRV 477
Query: 475 SYREFVSPVKGTDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTG 534
SYR+FVS T + +G C+DVFVAA+NLL Y VPYRFVPFG+ +NPSY+E +NKI T
Sbjct: 478 SYRQFVSVDSETGMVRGLCIDVFVAAINLLAYPVPYRFVPFGNNRENPSYSELINKIITD 537
Query: 535 YFDGAVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTAC 594
FD VGD+ I+TNRT++VDFTQPY +SGLVV+ K+ NSGGW+FLQPFT MW VT
Sbjct: 538 DFDAVVGDVTIITNRTKVVDFTQPYVSSGLVVLTSVKRQNSGGWAFLQPFTIKMWTVTGL 597
Query: 595 FFFFVGIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
FF +G VVW+LEHR+NDEFRG P +Q +T+ WFS STLFF+H
Sbjct: 598 FFLIIGTVVWMLEHRINDEFRGPPAKQLITVFWFSFSTLFFAH 640
>UniRef100_Q6ZGL7 Putative glutamate receptor [Oryza sativa]
Length = 944
Score = 705 bits (1820), Expect = 0.0
Identities = 348/637 (54%), Positives = 459/637 (71%), Gaps = 10/637 (1%)
Query: 8 LWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHS 67
L ++FL M P + +RPS VNIGAI F+S+IG V+ +A++ A++D+NS+S+IL+
Sbjct: 8 LLVLFLVMSPDGIRRSLAARPSIVNIGAILRFNSTIGGVSMIAIQAALEDINSDSTILNG 67
Query: 68 TQLVLHMQTSNCS-GFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFA 126
T L + M+ +NC GF GM++AL+FMETDVIAI+GPQ S ++HIV++VANELRVP++SFA
Sbjct: 68 TTLKVDMRDTNCDDGFLGMVEALQFMETDVIAIIGPQCSTIAHIVSYVANELRVPLMSFA 127
Query: 127 ATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDA 186
+ D TLSS+QFPFFVRT SDLYQM AVA I+D+Y WK V IY+DDDYGRNG++ LDDA
Sbjct: 128 S-DATLSSIQFPFFVRTAPSDLYQMDAVAAIVDYYRWKIVTAIYIDDDYGRNGIATLDDA 186
Query: 187 LAERRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLG 246
L +RRC+ISYK+ + + + ++ NLLV+V+ M+SR+I++H + G IF +A+ L
Sbjct: 187 LTQRRCKISYKIAFPA--NARKSDLINLLVSVSYMESRVIILHTGAGPGLKIFSLANQLS 244
Query: 247 MMQEGYVWIATDWLSTVLDS-TSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTG 305
MM GYVWIATDWLS LD+ +S+P ETM +QG L LR H P++ K SKW+ L+
Sbjct: 245 MMGNGYVWIATDWLSAYLDANSSVPDETMYGMQGVLTLRPHIPESKMKSNLISKWSRLSK 304
Query: 306 ----GSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSI 361
L +SY + YD+VW VA+A+D FF GG +S +N + L + G L+L+AMSI
Sbjct: 305 KYSYSYLRTSSYAFYVYDSVWAVARALDAFFDDGGKISFSNDSRLRDETGGTLHLEAMSI 364
Query: 362 FDNGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGL-S 420
FD G LL I ++NF G+SG ++ D+ L PAYD+IN++GNG+R VGYWSNYS L S
Sbjct: 365 FDMGNNLLEKIRKANFTGVSGQVQFDATGDLIHPAYDVINIIGNGMRTVGYWSNYSSLLS 424
Query: 421 IVSPETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFV 480
V PE LY+ PPN S ANQHL+ VIWPG+T PRGWVFP+N K+L+IGVP R S+REFV
Sbjct: 425 TVLPEVLYSEPPNNSLANQHLYDVIWPGQTAQTPRGWVFPSNAKELKIGVPNRFSFREFV 484
Query: 481 SPVKGTDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAV 540
+ T KG+C+DVF A+ LLPY V Y+F+PFG G++NP Y + V + FD A+
Sbjct: 485 TKDNVTGSMKGYCIDVFTQALALLPYPVTYKFIPFGGGNENPHYDKLVQMVEDNEFDAAI 544
Query: 541 GDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVG 600
GDIAI +RT DFTQP+ SGLV++AP KK W+FLQPFT MW VT FF VG
Sbjct: 545 GDIAITMSRTVTTDFTQPFIESGLVILAPVKKHIVNSWAFLQPFTLQMWCVTGLFFLVVG 604
Query: 601 IVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
VVW+LEHR+NDEFRGSP++Q +TI WFS STLFF+H
Sbjct: 605 AVVWVLEHRINDEFRGSPREQIITIFWFSFSTLFFAH 641
>UniRef100_Q93YT1 Glutamate receptor 3.2 precursor [Arabidopsis thaliana]
Length = 912
Score = 696 bits (1796), Expect = 0.0
Identities = 341/638 (53%), Positives = 467/638 (72%), Gaps = 6/638 (0%)
Query: 6 YWLWLVFLFMLPYLEQVYSNS---RPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNS 62
+W+ ++ F++ + + S RP +V++GAIF+ + G+V +AM+ A +DVNS+
Sbjct: 2 FWVLVLLSFIVLIGDGMISEGAGLRPRYVDVGAIFSLGTLQGEVTNIAMKAAEEDVNSDP 61
Query: 63 SILHSTQLVLHMQTSNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPM 122
S L ++L + + +GF ++ AL+FMETD +AI+GPQ+S+++H+++H+ANEL VPM
Sbjct: 62 SFLGGSKLRITTYDAKRNGFLTIMGALQFMETDAVAIIGPQTSIMAHVLSHLANELSVPM 121
Query: 123 LSFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSA 182
LSF A DP+LS+LQFPFFV+T SDL+ M A+AE+I +YGW EVI +Y DDD RNG++A
Sbjct: 122 LSFTALDPSLSALQFPFFVQTAPSDLFLMRAIAEMISYYGWSEVIALYNDDDNSRNGITA 181
Query: 183 LDDALAERRCRISYKVGIKSGPDVDRG-EITNLLVNVAMMQSRIIVVHAHSNSGFMIFKV 241
L D L RRC+ISYK + + EI N LV + M+SR+I+V+ +G IF+
Sbjct: 182 LGDELEGRRCKISYKAVLPLDVVITSPREIINELVKIQGMESRVIIVNTFPKTGKKIFEE 241
Query: 242 AHYLGMMQEGYVWIATDWLSTVLDSTS-LPLETMDTLQGALVLRQHTPDTDRKKMFTSKW 300
A LGMM++GYVWIAT WL+++LDS + LP +T ++L+G L LR HTP++ +KK F ++W
Sbjct: 242 AQKLGMMEKGYVWIATTWLTSLLDSVNPLPAKTAESLRGVLTLRIHTPNSKKKKDFVARW 301
Query: 301 NNLTGGSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGG-LNLDAM 359
N L+ G++GLN YGL+AYDTVW++A+A+ +S ++ L S K GG LNL A+
Sbjct: 302 NKLSNGTVGLNVYGLYAYDTVWIIARAVKRLLDSRANISFSSDPKLTSMKGGGSLNLGAL 361
Query: 360 SIFDNGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGL 419
SIFD G+ L+ I+ +N G++G I+ +RS+ +P+YDIINVV +G R++GYWSN+SGL
Sbjct: 362 SIFDQGSQFLDYIVNTNMTGVTGQIQFLPDRSMIQPSYDIINVVDDGFRQIGYWSNHSGL 421
Query: 420 SIVSPETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREF 479
SI+ PE+LY NRSS+NQHL+ V WPG T+ PRGWVFPNNG++LRIGVP RAS++EF
Sbjct: 422 SIIPPESLYKKLSNRSSSNQHLNNVTWPGGTSETPRGWVFPNNGRRLRIGVPDRASFKEF 481
Query: 480 VSPVKGTDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGA 539
VS + G++ +G+ +DVF AAV L+ Y VP+ FV FGDG KNP++ EFVN +T G FD
Sbjct: 482 VSRLDGSNKVQGYAIDVFEAAVKLISYPVPHEFVLFGDGLKNPNFNEFVNNVTIGVFDAV 541
Query: 540 VGDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFV 599
VGDIAIVT RTRIVDFTQPY SGLVVVAP K+N W+FL+PFTP MW VTA FF V
Sbjct: 542 VGDIAIVTKRTRIVDFTQPYIESGLVVVAPVTKLNDTPWAFLRPFTPPMWAVTAAFFLIV 601
Query: 600 GIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
G V+WILEHR+NDEFRG P++Q VTILWFS ST+FFSH
Sbjct: 602 GSVIWILEHRINDEFRGPPRKQIVTILWFSFSTMFFSH 639
>UniRef100_Q84W41 Glutamate receptor 3.6 precursor [Arabidopsis thaliana]
Length = 903
Score = 693 bits (1788), Expect = 0.0
Identities = 332/614 (54%), Positives = 452/614 (73%), Gaps = 10/614 (1%)
Query: 25 NSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFDG 84
++RP VNIG++FTF+S IGKV K+AM+ AV+DVN++ SIL++T L + M + +GF
Sbjct: 24 SARPQVVNIGSVFTFNSLIGKVIKVAMDAAVEDVNASPSILNTTTLRIIMHDTKYNGFMS 83
Query: 85 MIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTT 144
+++ L+FME++ +AI+GPQ S + +VAHVA EL++P+LSF+ATDPT+S LQFPFF+RT+
Sbjct: 84 IMEPLQFMESETVAIIGPQRSTTARVVAHVATELKIPILSFSATDPTMSPLQFPFFIRTS 143
Query: 145 LSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGP 204
+DL+QM A+A+I+ FYGW+EV+ IY DDDYGRNGV+AL D L+E+RCRISYK + P
Sbjct: 144 QNDLFQMAAIADIVQFYGWREVVAIYGDDDYGRNGVAALGDRLSEKRCRISYKAALPPAP 203
Query: 205 DVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTVL 264
R IT+LL+ VA+ +SRIIVVHA G +F VA LGMM GYVWIAT+WLST++
Sbjct: 204 --TRENITDLLIKVALSESRIIVVHASFIWGLELFNVARNLGMMSTGYVWIATNWLSTII 261
Query: 265 DSTS-LPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHAYDTVWL 323
D+ S LPL+T++ +QG + LR HTP++ K+ F +W+NLT +GL++Y L+AYDTVWL
Sbjct: 262 DTDSPLPLDTINNIQGVITLRLHTPNSIMKQNFVQRWHNLT--HVGLSTYALYAYDTVWL 319
Query: 324 VAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFVGLSGP 383
+AQAID+FF +GG VS + + G L+LDA+ +FD G + L +IL+ + +GL+G
Sbjct: 320 LAQAIDDFFKKGGNVSFSKNPIISELGGGNLHLDALKVFDGGKIFLESILQVDRIGLTGR 379
Query: 384 IKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQHLHT 443
+K S+R+L PA+D++NV+G G +GYW N+SGLS++ + + N S + Q LH+
Sbjct: 380 MKFTSDRNLVNPAFDVLNVIGTGYTTIGYWFNHSGLSVMPADEM----ENTSFSGQKLHS 435
Query: 444 VIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDLFKGFCVDVFVAAVNL 503
V+WPG + PRGWVF NNG+ LRIGVP R + E VS VK + GFCVDVF+AA+NL
Sbjct: 436 VVWPGHSIKIPRGWVFSNNGRHLRIGVPNRYRFEEVVS-VKSNGMITGFCVDVFIAAINL 494
Query: 504 LPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFTQPYAASG 563
LPYAVP+ V FG+GH NPS +E V ITTG +D VGDI I+T RT++ DFTQPY SG
Sbjct: 495 LPYAVPFELVAFGNGHDNPSNSELVRLITTGVYDAGVGDITIITERTKMADFTQPYVESG 554
Query: 564 LVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRGSPKQQFV 623
LVVVAP +K+ S +FL+PFTP MW++ A F VG V+W LEH+ NDEFRG P++Q +
Sbjct: 555 LVVVAPVRKLGSSAMAFLRPFTPQMWLIAAASFLIVGAVIWCLEHKHNDEFRGPPRRQVI 614
Query: 624 TILWFSLSTLFFSH 637
T WFS STLFFSH
Sbjct: 615 TTFWFSFSTLFFSH 628
>UniRef100_Q6RKN5 Glutamate receptor 3.1 precursor [Arabidopsis thaliana]
Length = 925
Score = 692 bits (1786), Expect = 0.0
Identities = 346/639 (54%), Positives = 463/639 (72%), Gaps = 10/639 (1%)
Query: 7 WLWLVFLFMLP---YLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSS 63
W+ L F+ +L L + S+SRP + +GAIF ++ G+ A +A + A +DVNS+ S
Sbjct: 7 WVLLSFIIVLGGGLLLSEGASSSRPPVIKVGAIFGLNTMYGETANIAFKAAEEDVNSDPS 66
Query: 64 ILHSTQLVLHMQTSNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPML 123
L ++L + M + SGF ++ AL+FMETDV+AI+GPQ+S+++H+++H+ANEL VPML
Sbjct: 67 FLGGSKLRILMNDAKRSGFLSIMGALQFMETDVVAIIGPQTSIMAHVLSHLANELTVPML 126
Query: 124 SFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSAL 183
SF A DPTLS LQFPFFV+T SDL+ M A+AE+I +YGW +V+ +Y DDD RNGV+AL
Sbjct: 127 SFTALDPTLSPLQFPFFVQTAPSDLFLMRAIAEMITYYGWSDVVALYNDDDNSRNGVTAL 186
Query: 184 DDALAERRCRISYKVGIKSGPDVDRG-EITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVA 242
D L ERRC+ISYK + + EI L+ + M+SR+IVV+ N+G MIFK A
Sbjct: 187 GDELEERRCKISYKAVLPLDVVITSPVEIIEELIKIRGMESRVIVVNTFPNTGKMIFKEA 246
Query: 243 HYLGMMQEGYVWIATDWLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNN 302
LGMM++GYVWIAT WLS+VLDS +LPL+T + G L LR HTPD+ +K+ F ++W N
Sbjct: 247 ERLGMMEKGYVWIATTWLSSVLDS-NLPLDTK-LVNGVLTLRLHTPDSRKKRDFAARWKN 304
Query: 303 LTGGS--LGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMS 360
+ +GLN YGL+AYDTVW++A+A+ GG +S +N L S K LNL A+S
Sbjct: 305 KLSNNKTIGLNVYGLYAYDTVWIIARAVKTLLEAGGNLSFSNDAKLGSLKGEALNLSALS 364
Query: 361 IFDNGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLS 420
FD G+ LL+ I+ + GL+GP++ +RS+ +P+YDIIN+V + V ++GYWSNYSGLS
Sbjct: 365 RFDQGSQLLDYIVHTKMSGLTGPVQFHPDRSMLQPSYDIINLVDDRVHQIGYWSNYSGLS 424
Query: 421 IVSPETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFV 480
IV PE+ Y+ PPNRSS+NQHL++V WPG T+ PRGW+F NNG++LRIGVP RAS+++FV
Sbjct: 425 IVPPESFYSKPPNRSSSNQHLNSVTWPGGTSVTPRGWIFRNNGRRLRIGVPDRASFKDFV 484
Query: 481 SPVKG-TDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGY-FDG 538
S V G ++ +G+C+DVF AAV LL Y VP+ F+ FGDG NP+Y E VNK+TTG FD
Sbjct: 485 SRVNGSSNKVQGYCIDVFEAAVKLLSYPVPHEFIFFGDGLTNPNYNELVNKVTTGVDFDA 544
Query: 539 AVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFF 598
VGDIAIVT RTRIVDFTQPY SGLVVVAP ++N W+FL+PFT MW VTA FF
Sbjct: 545 VVGDIAIVTKRTRIVDFTQPYIESGLVVVAPVTRLNENPWAFLRPFTLPMWAVTASFFVI 604
Query: 599 VGIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
VG +WILEHR+NDEFRG P++Q +TILWF+ ST+FFSH
Sbjct: 605 VGAAIWILEHRINDEFRGPPRRQIITILWFTFSTMFFSH 643
>UniRef100_Q9SW97 Glutamate receptor 3.5 precursor [Arabidopsis thaliana]
Length = 953
Score = 687 bits (1772), Expect = 0.0
Identities = 329/621 (52%), Positives = 433/621 (68%), Gaps = 10/621 (1%)
Query: 24 SNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFD 83
S+S PS VN+GA+FT+DS IG+ AKLA A++D+N++ SIL T+L + Q +NCSGF
Sbjct: 41 SSSLPSSVNVGALFTYDSFIGRAAKLAFVAAIEDINADQSILRGTKLNIVFQDTNCSGFV 100
Query: 84 GMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRT 143
G + AL+ ME V+A +GPQSS + HI++HVANEL VP LSFAATDPTLSSLQ+P+F+RT
Sbjct: 101 GTMGALQLMENKVVAAIGPQSSGIGHIISHVANELHVPFLSFAATDPTLSSLQYPYFLRT 160
Query: 144 TLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSG 203
T +D +QM A+ + + ++ W+EV+ I+VDD+YGRNG+S L DALA++R +ISYK
Sbjct: 161 TQNDYFQMNAITDFVSYFRWREVVAIFVDDEYGRNGISVLGDALAKKRAKISYKAAFP-- 218
Query: 204 PDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTV 263
P D I++LL +V +M+SRI VVH + +SG IF VA LGMM GYVWI TDWL T
Sbjct: 219 PGADNSSISDLLASVNLMESRIFVVHVNPDSGLNIFSVAKSLGMMGSGYVWITTDWLLTA 278
Query: 264 LDSTSLPLE--TMDTLQGALVLRQHTPDTDRKKMFTSKWNNLT-----GGSLGLNSYGLH 316
LDS PL+ +D LQG + R +TP++D K+ F +W NL G NSY L+
Sbjct: 279 LDSME-PLDPRALDLLQGVVAFRHYTPESDNKRQFKGRWKNLRFKESLKSDDGFNSYALY 337
Query: 317 AYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSN 376
AYD+VWLVA+A+D FFSQG V+ +N SL + G+ L + IF+ G L IL N
Sbjct: 338 AYDSVWLVARALDVFFSQGNTVTFSNDPSLRNTNDSGIKLSKLHIFNEGERFLQVILEMN 397
Query: 377 FVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSS 436
+ GL+G I+ +SE++ PAYDI+N+ G RVGYWSN++G S+ PETLY+ P N S+
Sbjct: 398 YTGLTGQIEFNSEKNRINPAYDILNIKSTGPLRVGYWSNHTGFSVAPPETLYSKPSNTSA 457
Query: 437 ANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDLFKGFCVDV 496
+Q L+ +IWPGE PRGWVFP NGK L+IGVP R SY+ + S K KGFC+D+
Sbjct: 458 KDQRLNEIIWPGEVIKPPRGWVFPENGKPLKIGVPNRVSYKNYASKDKNPLGVKGFCIDI 517
Query: 497 FVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFT 556
F AA+ LLPY VP ++ +GDG KNPSY ++++ FD AVGD+ I+TNRT+ VDFT
Sbjct: 518 FEAAIQLLPYPVPRTYILYGDGKKNPSYDNLISEVAANIFDVAVGDVTIITNRTKFVDFT 577
Query: 557 QPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRG 616
QP+ SGLVVVAP K S WSFL+PFT MW VT F FVG V+WILEHR N+EFRG
Sbjct: 578 QPFIESGLVVVAPVKGAKSSPWSFLKPFTIEMWAVTGALFLFVGAVIWILEHRFNEEFRG 637
Query: 617 SPKQQFVTILWFSLSTLFFSH 637
P++Q +T+ WFS ST+FFSH
Sbjct: 638 PPRRQIITVFWFSFSTMFFSH 658
>UniRef100_Q8GXJ4 Glutamate receptor 3.4 precursor [Arabidopsis thaliana]
Length = 959
Score = 686 bits (1771), Expect = 0.0
Identities = 331/612 (54%), Positives = 437/612 (71%), Gaps = 4/612 (0%)
Query: 27 RPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFDGMI 86
RPS VN+GA+FT+DS IG+ AK A++ A+ DVN++ S+L +L + Q SNCSGF G +
Sbjct: 57 RPSSVNVGALFTYDSFIGRAAKPAVKAAMDDVNADQSVLKGIKLNIIFQDSNCSGFIGTM 116
Query: 87 QALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLS 146
AL+ ME V+A +GPQSS ++H++++VANEL VP+LSF ATDPTLSSLQFP+F+RTT +
Sbjct: 117 GALQLMENKVVAAIGPQSSGIAHMISYVANELHVPLLSFGATDPTLSSLQFPYFLRTTQN 176
Query: 147 DLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGPDV 206
D +QM A+A+ + + GW++VI I+VDD+ GRNG+S L D LA++R RISYK I P
Sbjct: 177 DYFQMHAIADFLSYSGWRQVIAIFVDDECGRNGISVLGDVLAKKRSRISYKAAIT--PGA 234
Query: 207 DRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTVLDS 266
D I +LLV+V +M+SR+ VVH + +SG +F VA LGMM GYVWIATDWL T +DS
Sbjct: 235 DSSSIRDLLVSVNLMESRVFVVHVNPDSGLNVFSVAKSLGMMASGYVWIATDWLPTAMDS 294
Query: 267 TS-LPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHAYDTVWLVA 325
+ +TMD LQG + R +T ++ K+ F ++W NL G NSY ++AYD+VWLVA
Sbjct: 295 MEHVDSDTMDLLQGVVAFRHYTIESSVKRQFMARWKNLRPND-GFNSYAMYAYDSVWLVA 353
Query: 326 QAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFVGLSGPIK 385
+A+D FF + ++ +N +LH + L A+S+F+ G + IL N G++GPI+
Sbjct: 354 RALDVFFRENNNITFSNDPNLHKTNGSTIQLSALSVFNEGEKFMKIILGMNHTGVTGPIQ 413
Query: 386 LDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQHLHTVI 445
DS+R+ PAY+++N+ G R VGYWSN+SGLS+V PETLY+ PPN S+ANQ L +I
Sbjct: 414 FDSDRNRVNPAYEVLNLEGTAPRTVGYWSNHSGLSVVHPETLYSRPPNTSTANQRLKGII 473
Query: 446 WPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDLFKGFCVDVFVAAVNLLP 505
+PGE T PRGWVFPNNGK LRIGVP R SY ++VS K +G+C+DVF AA+ LLP
Sbjct: 474 YPGEVTKPPRGWVFPNNGKPLRIGVPNRVSYTDYVSKDKNPPGVRGYCIDVFEAAIELLP 533
Query: 506 YAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFTQPYAASGLV 565
Y VP ++ +GDG +NPSY VN++ FD AVGDI IVTNRTR VDFTQP+ SGLV
Sbjct: 534 YPVPRTYILYGDGKRNPSYDNLVNEVVADNFDVAVGDITIVTNRTRYVDFTQPFIESGLV 593
Query: 566 VVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRGSPKQQFVTI 625
VVAP K+ S WSFL+PFT MW VT FF FVG +VWILEHR N EFRG P++Q +TI
Sbjct: 594 VVAPVKEAKSSPWSFLKPFTIEMWAVTGGFFLFVGAMVWILEHRFNQEFRGPPRRQLITI 653
Query: 626 LWFSLSTLFFSH 637
WFS ST+FFSH
Sbjct: 654 FWFSFSTMFFSH 665
>UniRef100_Q9SES5 Ligand gated channel-like protein [Brassica napus]
Length = 912
Score = 682 bits (1761), Expect = 0.0
Identities = 342/635 (53%), Positives = 454/635 (70%), Gaps = 8/635 (1%)
Query: 7 WLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILH 66
W ++V L L + S SRP + +GAIF ++ G A LA + A +DVNS+ S L
Sbjct: 6 WGFIVLGAFLGLLSEGASTSRPRVIKVGAIFGLNTMYGHTASLAFKAAEEDVNSDPSFLG 65
Query: 67 STQLVLHMQTSNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFA 126
++L + + + SGF ++ AL+FMETDV+AI+G Q+S+++H+++H+ANEL VPMLSF
Sbjct: 66 GSKLRIMISDAQRSGFLSIMGALQFMETDVVAIIGLQTSIMAHVLSHLANELTVPMLSFT 125
Query: 127 ATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDA 186
A DPTLS LQFPFFV+T +DL+ M A+AE+I +YGW +V+ +Y DDD RNGV+AL D
Sbjct: 126 ALDPTLSPLQFPFFVQTAPNDLFLMRAIAEMITYYGWSDVVVLYNDDDNSRNGVTALGDE 185
Query: 187 LAERRCRISYKVGIKSGPDVDR-GEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYL 245
L ERRC+ISYK + + EI L + M+SRIIVV+ N+G MIF+ A L
Sbjct: 186 LEERRCKISYKAVLPLDVVITSPAEIIEELTKIRGMESRIIVVNTFPNTGKMIFEEAKKL 245
Query: 246 GMMQEGYVWIATDWLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTG 305
GMM++GYVWIAT WLS+++DS PL+ + +L G L LR HTPD +K+ F ++W
Sbjct: 246 GMMEKGYVWIATTWLSSLVDS-DFPLD-LKSLNGVLTLRLHTPDPRKKRDFAARWKK--N 301
Query: 306 GSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNG 365
++GLN YGL+AYDTVW++AQA+ +F GG ++ ++ L + K LNL A+S FD G
Sbjct: 302 KTIGLNVYGLYAYDTVWIIAQAVKSFLEAGGNLTFSHDAKLSNLKGEALNLSALSRFDEG 361
Query: 366 TLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPE 425
LL+ I+R+ GL+GP++ +RS+ +P+YDIINVV G R++GYWSN+SGLS+V PE
Sbjct: 362 PQLLDYIMRTKMSGLTGPVQFHRDRSMVQPSYDIINVVDGGFRQIGYWSNHSGLSVVPPE 421
Query: 426 TLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKG 485
+ Y P NRSS+NQHL++V WPG T+ PRGWVFPNNGK LRIGVP RAS+++FVS V G
Sbjct: 422 SFYNKPSNRSSSNQHLNSVTWPGGTSVTPRGWVFPNNGKLLRIGVPNRASFKDFVSRVNG 481
Query: 486 TDLFK--GFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGY-FDGAVGD 542
+ K G+C+DVF AAV LL Y VP+ F+ FGDG +NP+Y + VNK+ TG FD AVGD
Sbjct: 482 SSSHKVQGYCIDVFEAAVKLLSYPVPHEFIFFGDGLQNPNYNDLVNKVATGVDFDAAVGD 541
Query: 543 IAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIV 602
IAIVT RTRIVD+TQPY SGLVVVAP +N W+FL+PFTP MW VTA FF VG V
Sbjct: 542 IAIVTKRTRIVDYTQPYIESGLVVVAPVTALNENPWAFLRPFTPPMWAVTASFFMVVGAV 601
Query: 603 VWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
+WILEHR NDEFRG P++Q +TILWF+ ST+FFSH
Sbjct: 602 IWILEHRTNDEFRGPPRRQIITILWFTFSTMFFSH 636
>UniRef100_Q7XAV6 Glutamate receptor [Raphanus sativus var. sativus]
Length = 915
Score = 670 bits (1728), Expect = 0.0
Identities = 326/622 (52%), Positives = 446/622 (71%), Gaps = 7/622 (1%)
Query: 22 VYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSG 81
V RP +N+GAIF+ + G+VA +A++ A DVNS+ + L ++L + M + +G
Sbjct: 22 VSEGRRPHDINVGAIFSLSTLYGQVADIALKAAEDDVNSDPTFLPGSKLRILMYDAKRNG 81
Query: 82 FDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFV 141
F +++AL+FMETD +AI+GPQ+S+++H+++++ANEL VPM SF A DP+LS LQFPFFV
Sbjct: 82 FLSIMKALQFMETDSVAIIGPQTSIMAHVLSYLANELNVPMCSFTALDPSLSPLQFPFFV 141
Query: 142 RTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIK 201
+T SDL+ M A+AE+I +YGW +VI +Y DDD RNGV++L D L RRC+ISYK +
Sbjct: 142 QTAPSDLFLMRAIAEMITYYGWSDVIALYNDDDNSRNGVTSLGDELEGRRCKISYKAVLP 201
Query: 202 SGPDVDRG-EITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWL 260
+ EI LV + M+SR+I+V+ +G M+F+ A LGM GYVWIAT W+
Sbjct: 202 LDVVIKTPREIVRELVKIQKMESRVIIVNTFPKTGKMVFEEARRLGMTGRGYVWIATTWM 261
Query: 261 STVLDST---SLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHA 317
+++LDS SLP + ++L+G L LR HTP + +K+ F ++WN L+ GS+GLN YGL+A
Sbjct: 262 TSLLDSADPLSLP-KVAESLRGVLTLRIHTPVSRKKRDFAARWNKLSNGSVGLNVYGLYA 320
Query: 318 YDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNF 377
YDTVW++A+A+ N + + + L K G LNL A+S+FD G L+ I+++
Sbjct: 321 YDTVWIIARAVKNLLDSRANIPFSGDSKLDHLKGGSLNLGALSMFDQGQQFLDYIVKTKM 380
Query: 378 VGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSA 437
G++GP++ +RS+ +PAYDIINVVG G+R++GYWSN+SGLS++ PE L++ P NRSS+
Sbjct: 381 SGVTGPVQFLPDRSMVQPAYDIINVVGGGLRQIGYWSNHSGLSVIPPELLFSKPSNRSSS 440
Query: 438 NQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDL--FKGFCVD 495
NQHL V WPG + PRGWVFPNNG++LRIGVP RAS+++FVS V G+ G+ ++
Sbjct: 441 NQHLENVTWPGGGSVTPRGWVFPNNGRRLRIGVPNRASFKDFVSRVNGSSSSHIDGYSIN 500
Query: 496 VFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDF 555
VF AA+ LL Y VP+ F+ FGD KNP+Y + VN +TTG FD VGDIAIVT RTRIVDF
Sbjct: 501 VFEAAIKLLSYPVPHEFILFGDSLKNPNYNDLVNNVTTGVFDAVVGDIAIVTKRTRIVDF 560
Query: 556 TQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFR 615
TQPY SGLVVVAP K+N W+FL+PFTP MW VTA FF VG V+WILEHR+NDEFR
Sbjct: 561 TQPYIESGLVVVAPVTKLNDTPWAFLRPFTPPMWAVTAAFFLIVGSVIWILEHRINDEFR 620
Query: 616 GSPKQQFVTILWFSLSTLFFSH 637
G P++Q VTILWFS ST+FFSH
Sbjct: 621 GPPRRQIVTILWFSFSTMFFSH 642
>UniRef100_Q8GXJ4-2 Splice isoform 2 of Q8GXJ4 [Arabidopsis thaliana]
Length = 669
Score = 669 bits (1726), Expect = 0.0
Identities = 323/602 (53%), Positives = 428/602 (70%), Gaps = 4/602 (0%)
Query: 27 RPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFDGMI 86
RPS VN+GA+FT+DS IG+ AK A++ A+ DVN++ S+L +L + Q SNCSGF G +
Sbjct: 57 RPSSVNVGALFTYDSFIGRAAKPAVKAAMDDVNADQSVLKGIKLNIIFQDSNCSGFIGTM 116
Query: 87 QALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLS 146
AL+ ME V+A +GPQSS ++H++++VANEL VP+LSF ATDPTLSSLQFP+F+RTT +
Sbjct: 117 GALQLMENKVVAAIGPQSSGIAHMISYVANELHVPLLSFGATDPTLSSLQFPYFLRTTQN 176
Query: 147 DLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGPDV 206
D +QM A+A+ + + GW++VI I+VDD+ GRNG+S L D LA++R RISYK I P
Sbjct: 177 DYFQMHAIADFLSYSGWRQVIAIFVDDECGRNGISVLGDVLAKKRSRISYKAAIT--PGA 234
Query: 207 DRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTVLDS 266
D I +LLV+V +M+SR+ VVH + +SG +F VA LGMM GYVWIATDWL T +DS
Sbjct: 235 DSSSIRDLLVSVNLMESRVFVVHVNPDSGLNVFSVAKSLGMMASGYVWIATDWLPTAMDS 294
Query: 267 TS-LPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHAYDTVWLVA 325
+ +TMD LQG + R +T ++ K+ F ++W NL G NSY ++AYD+VWLVA
Sbjct: 295 MEHVDSDTMDLLQGVVAFRHYTIESSVKRQFMARWKNLRPND-GFNSYAMYAYDSVWLVA 353
Query: 326 QAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFVGLSGPIK 385
+A+D FF + ++ +N +LH + L A+S+F+ G + IL N G++GPI+
Sbjct: 354 RALDVFFRENNNITFSNDPNLHKTNGSTIQLSALSVFNEGEKFMKIILGMNHTGVTGPIQ 413
Query: 386 LDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQHLHTVI 445
DS+R+ PAY+++N+ G R VGYWSN+SGLS+V PETLY+ PPN S+ANQ L +I
Sbjct: 414 FDSDRNRVNPAYEVLNLEGTAPRTVGYWSNHSGLSVVHPETLYSRPPNTSTANQRLKGII 473
Query: 446 WPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDLFKGFCVDVFVAAVNLLP 505
+PGE T PRGWVFPNNGK LRIGVP R SY ++VS K +G+C+DVF AA+ LLP
Sbjct: 474 YPGEVTKPPRGWVFPNNGKPLRIGVPNRVSYTDYVSKDKNPPGVRGYCIDVFEAAIELLP 533
Query: 506 YAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFTQPYAASGLV 565
Y VP ++ +GDG +NPSY VN++ FD AVGDI IVTNRTR VDFTQP+ SGLV
Sbjct: 534 YPVPRTYILYGDGKRNPSYDNLVNEVVADNFDVAVGDITIVTNRTRYVDFTQPFIESGLV 593
Query: 566 VVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRGSPKQQFVTI 625
VVAP K+ S WSFL+PFT MW VT FF FVG +VWILEHR N EFRG P++Q +TI
Sbjct: 594 VVAPVKEAKSSPWSFLKPFTIEMWAVTGGFFLFVGAMVWILEHRFNQEFRGPPRRQLITI 653
Query: 626 LW 627
W
Sbjct: 654 FW 655
>UniRef100_Q5QLP6 Putative ionotropic glutamate receptor homolog GLR4 [Oryza sativa]
Length = 637
Score = 660 bits (1702), Expect = 0.0
Identities = 317/603 (52%), Positives = 428/603 (70%), Gaps = 5/603 (0%)
Query: 26 SRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFDGM 85
SRP+ + IGA+FTFDS IG+ A+E AV DVN++ +L T+L + Q +NCSGF G
Sbjct: 36 SRPAELRIGALFTFDSVIGRAVMPAIELAVADVNADPGVLPGTKLSVITQDTNCSGFLGT 95
Query: 86 IQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTL 145
++AL + DV+A+LGPQSS ++H+++H NE VP++SFAA+DPTLSSL++P+FVR T
Sbjct: 96 MEALELLAKDVVAVLGPQSSSIAHVISHAVNEFHVPLVSFAASDPTLSSLEYPYFVRATT 155
Query: 146 SDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGPD 205
SD +QM+A+A II+ Y W+EVI IYVDDDYGR G++AL DALA+++ +I+YK K P
Sbjct: 156 SDYFQMSAIASIINQYRWREVIAIYVDDDYGRGGITALGDALAKKKSKIAYKA--KLPPG 213
Query: 206 VDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTVLD 265
R I ++L++V MQSR+ VVH + +SG +F A LGMM GY WIATDWLS VLD
Sbjct: 214 ASRTTIEDMLMHVNEMQSRVYVVHVNPDSGLAVFAAAKSLGMMSTGYAWIATDWLSAVLD 273
Query: 266 STS-LPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLT--GGSLGLNSYGLHAYDTVW 322
S+ + + M+ QG ++LRQH D+ + S+WNNLT GG +SY + YD+VW
Sbjct: 274 SSDHISTDRMELTQGVIMLRQHVSDSGIQHSLVSRWNNLTRNGGHSSFSSYSMRTYDSVW 333
Query: 323 LVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFVGLSG 382
LVA+A+++F S+G VS + +L K L LD++ +NG LL + +NF G+SG
Sbjct: 334 LVARAVEDFLSEGNAVSFSADPNLQDIKGSNLQLDSLRSLNNGERLLEKVWHTNFTGVSG 393
Query: 383 PIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQHLH 442
++ +ER L PA+DI+N+ G G R +GYWSN S LS+V+PE L++ P + S+ N LH
Sbjct: 394 LVQFTAERDLIHPAFDILNIGGTGFRTIGYWSNISDLSVVAPEKLHSEPLDSSTNNIELH 453
Query: 443 TVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDLFKGFCVDVFVAAVN 502
VIWPG+T+ +PRGWVFP +GK LRIGVP+R SY+EFV P KG D KGF VDVF AAV
Sbjct: 454 GVIWPGQTSEKPRGWVFPYHGKPLRIGVPLRTSYKEFVMPDKGPDGVKGFSVDVFKAAVG 513
Query: 503 LLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFTQPYAAS 562
LLPY V + F+ FGDG KNPSY + + K++ +FD A+GDIAIVTNRTR+VDFTQPY S
Sbjct: 514 LLPYPVSFDFILFGDGLKNPSYNDLIEKVSDNHFDAAIGDIAIVTNRTRLVDFTQPYTES 573
Query: 563 GLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRGSPKQQF 622
GL+++AP ++I S W+FL+PFT MW V F FVG VVW+LEHR N EFRG P+QQ
Sbjct: 574 GLIILAPAREIESNAWAFLKPFTFQMWSVLGVLFLFVGAVVWVLEHRTNTEFRGPPRQQI 633
Query: 623 VTI 625
+T+
Sbjct: 634 MTV 636
>UniRef100_Q8LH04 Glutamate receptor, ionotropic kainate 5-like protein [Oryza
sativa]
Length = 955
Score = 638 bits (1646), Expect = 0.0
Identities = 328/640 (51%), Positives = 435/640 (67%), Gaps = 19/640 (2%)
Query: 10 LVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQ 69
L+FL + L +RP+ V+IGA+FTFDS IG+ AK+A+E AV DVN + +L+ T
Sbjct: 10 LLFLLWVVALAVPGLAARPANVSIGALFTFDSVIGRAAKVAIELAVADVNRDDGVLNGTY 69
Query: 70 LVLHMQTSNCSGFDGMIQALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATD 129
L + Q + CSGF G+IQ L+ ME V+A++GPQSS + H+V+HVA+ELR+P++SFAATD
Sbjct: 70 LSVVEQDTKCSGFIGIIQ-LQVMEKKVVAVVGPQSSGIGHVVSHVADELRIPLVSFAATD 128
Query: 130 PTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAE 189
PTL S Q+P+F+R T SD +QM AVA+II Y W+E IYVD+DYGR + AL D L
Sbjct: 129 PTLGSSQYPYFLRATHSDFFQMAAVADIISHYAWREATLIYVDNDYGRAALDALGDHLQS 188
Query: 190 RRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQ 249
R ++SY+ + P DR IT+LL+ V+MM+SR+IVVHA+ +SG IF A LGMM
Sbjct: 189 MRSKVSYRAPLP--PAADRAAITDLLLRVSMMESRVIVVHANPDSGLDIFAAAQSLGMMS 246
Query: 250 EGYVWIATDWLSTVLDSTSLP---LETMDTLQGALVLRQHTPDTDRKKMFTSKW------ 300
GYVWIAT+WL+ +LDS S P + LQG + LRQ+TPD+D K+ S++
Sbjct: 247 SGYVWIATEWLAALLDSDSSPPRKTTALALLQGVVTLRQYTPDSDAKRSLMSRFAARLQA 306
Query: 301 NNLTGGSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGG--LNLDA 358
+N TGG +N+Y L AYD VW+ A+A+D G VS ++ L ++ G L L A
Sbjct: 307 HNTTGG---INAYVLFAYDAVWMAARAVDQLLVDGSNVSFSDDARLRAENETGSALRLGA 363
Query: 359 MSIFDNGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSG 418
+ +FD G LL+ + NF G++G ++ +R+L PAY+++NV G GVRRVGYWSN +
Sbjct: 364 LKVFDQGEQLLSKMKTLNFTGVTGQVRFGDDRNLADPAYEVLNVGGTGVRRVGYWSNRTR 423
Query: 419 LSIVSPETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYRE 478
LS+ +PE N + + L++VIWPGET S PRGWVFPNNGK LRIGVP R +Y++
Sbjct: 424 LSVTAPEQ-EQNGKKKKQQGEELYSVIWPGETASTPRGWVFPNNGKALRIGVPYRTTYKQ 482
Query: 479 FVSP-VKGTDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFD 537
FVS G D G+C+DVF AAV LL Y VP +V GDG KNPSY E V ++ G D
Sbjct: 483 FVSKDAGGPDGASGYCIDVFKAAVALLAYPVPVSYVVVGDGVKNPSYGELVQRVAEGELD 542
Query: 538 GAVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFF 597
AVGDI+IVTNRTR+VDFTQPY SGLV+V ++ S W+FL+PFT MW VT FF
Sbjct: 543 AAVGDISIVTNRTRVVDFTQPYVESGLVIVTAVRERASSAWAFLKPFTREMWAVTGGFFL 602
Query: 598 FVGIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
FVG VVW+LEHR N +FRGSP++Q VT+ WFS ST+FF+H
Sbjct: 603 FVGAVVWVLEHRSNTDFRGSPRKQLVTVFWFSFSTMFFAH 642
>UniRef100_Q6RKN4 Putative glutamate receptor ion channel [Arabidopsis thaliana]
Length = 851
Score = 620 bits (1600), Expect = e-176
Identities = 297/557 (53%), Positives = 386/557 (68%), Gaps = 10/557 (1%)
Query: 88 ALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLSD 147
AL+ ME V+A +GPQSS + HI++HVANEL VP LSFAATDPTLSSLQ+P+F+ TT +D
Sbjct: 3 ALQLMENKVVAAIGPQSSGIGHIISHVANELHVPFLSFAATDPTLSSLQYPYFLCTTQND 62
Query: 148 LYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGPDVD 207
+QM A+ + + ++ W+EV+ I+VDD+YGRNG+S L DALA++R +ISYK P D
Sbjct: 63 YFQMNAITDFVSYFRWREVVAIFVDDEYGRNGISVLGDALAKKRAKISYKAAFP--PGAD 120
Query: 208 RGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTVLDST 267
I++LL +V +M+SRI VVH + +SG IF VA LGMM GYVWI TDWL T LDS
Sbjct: 121 NSSISDLLASVNLMESRIFVVHVNPDSGLNIFSVAKSLGMMGSGYVWITTDWLLTALDSM 180
Query: 268 SLPLE--TMDTLQGALVLRQHTPDTDRKKMFTSKWNNLT-----GGSLGLNSYGLHAYDT 320
PL+ +D LQG + R +TP++D K+ F +W NL G NSY L+AYD+
Sbjct: 181 E-PLDPRALDLLQGVVAFRHYTPESDNKRQFKGRWKNLRFKESLKSDDGFNSYALYAYDS 239
Query: 321 VWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFVGL 380
VWLVA+A+D FFSQG V+ +N SL + G+ L + IF+ G L IL N+ GL
Sbjct: 240 VWLVARALDVFFSQGNTVTFSNDPSLRNTNDSGIKLSKLHIFNEGERFLQVILEMNYTGL 299
Query: 381 SGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQH 440
+G I+ +SE++ PAYDI+N+ G RVGYWSN++G S+V PETLY+ P N S+ NQ
Sbjct: 300 TGQIEFNSEKNRINPAYDILNIKITGPLRVGYWSNHTGFSVVPPETLYSKPSNTSAKNQR 359
Query: 441 LHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDLFKGFCVDVFVAA 500
L+ +IWPGE PRGWVFP NGK L+IGVP R SY+ + S K KGFC+D+F AA
Sbjct: 360 LNEIIWPGEVIKPPRGWVFPENGKPLKIGVPNRVSYKNYASKDKNPLGVKGFCIDIFEAA 419
Query: 501 VNLLPYAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFTQPYA 560
+ LLPY VP ++ +GDG KNPSY ++++ FD AVGD+ I+TNRT+ VDFTQP+
Sbjct: 420 IQLLPYPVPRTYILYGDGKKNPSYDNLISEVAANIFDVAVGDVTIITNRTKFVDFTQPFI 479
Query: 561 ASGLVVVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRGSPKQ 620
SGLVVVAP K S WSFL+PFT MW VT F FVG V+WILEHR N+EFRG P++
Sbjct: 480 ESGLVVVAPVKGAKSSPWSFLKPFTIEMWAVTGALFLFVGAVIWILEHRFNEEFRGPPRR 539
Query: 621 QFVTILWFSLSTLFFSH 637
Q +T+ WFS ST+FFSH
Sbjct: 540 QIITVFWFSFSTMFFSH 556
>UniRef100_Q653Y6 Putative ionotropic glutamate receptor ortholog GLR6 [Oryza sativa]
Length = 845
Score = 619 bits (1595), Expect = e-175
Identities = 289/550 (52%), Positives = 393/550 (70%), Gaps = 6/550 (1%)
Query: 92 METDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLSDLYQM 151
ME +V+A++GPQSS + H+++HV NEL VP+LSFAATDPTLS+ ++P+F+R+T+SD +QM
Sbjct: 1 MEKNVVAVIGPQSSGIGHVISHVVNELHVPLLSFAATDPTLSASEYPYFLRSTMSDYFQM 60
Query: 152 TAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGPDVDRGEI 211
AVA I+D+Y WKEV I+VDDDYGR V+AL DALA R RISYK + P+ + I
Sbjct: 61 HAVASIVDYYQWKEVTAIFVDDDYGRGAVAALSDALALSRARISYKAAVP--PNSNAATI 118
Query: 212 TNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTVLDST-SLP 270
++L MM+SR+ VVH + ++G IF +A+ L MM GYVWI TDWL+ V+DS+ S
Sbjct: 119 NDVLFRANMMESRVFVVHVNPDAGMRIFSIANKLRMMDSGYVWIVTDWLAAVMDSSMSGD 178
Query: 271 LETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGG---SLGLNSYGLHAYDTVWLVAQA 327
L+TM +QG +VLRQH PD++ K+ F SKWNN+ + GLNSYG +AYD+VW+VA+A
Sbjct: 179 LKTMSYMQGLIVLRQHFPDSETKREFISKWNNVARNRSIASGLNSYGFYAYDSVWIVARA 238
Query: 328 IDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFVGLSGPIKLD 387
ID G ++ + LH L L A+ +FD+G LL +L +NF GL+G ++ D
Sbjct: 239 IDQLLDNGEEINFSADPRLHDSMNSTLRLSALKLFDSGEQLLQQLLLTNFTGLTGQLQFD 298
Query: 388 SERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQHLHTVIWP 447
S+R+L RPAYDI+N+ G+ +GYWSNYSGLS+ +PE LY PN S++ Q L V+WP
Sbjct: 299 SDRNLVRPAYDILNIGGSVPHLIGYWSNYSGLSVAAPEILYEKQPNTSTSAQRLKNVVWP 358
Query: 448 GETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTDLFKGFCVDVFVAAVNLLPYA 507
G + S+P+GWVFPNNG+ LR+GVP + S++E +S G D G+C+++F AA+ LLPY
Sbjct: 359 GHSASKPKGWVFPNNGQPLRVGVPNKPSFKELMSRDTGPDNVTGYCIEIFNAAIKLLPYP 418
Query: 508 VPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFTQPYAASGLVVV 567
VP +F+ GDG KNP+Y + +N + D AVGD AIV NRT+I +F+QPY SGLV+V
Sbjct: 419 VPCQFIVIGDGLKNPNYDDIINMVAANSLDAAVGDFAIVRNRTKIAEFSQPYIESGLVIV 478
Query: 568 APFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRGSPKQQFVTILW 627
P K+ +S W+FL+PFT MW VT F FVGIVVWILEHR N+EFRGSP++Q +TI W
Sbjct: 479 VPVKEASSSAWAFLKPFTLEMWCVTGVLFIFVGIVVWILEHRTNEEFRGSPRRQMITIFW 538
Query: 628 FSLSTLFFSH 637
FS ST+FF+H
Sbjct: 539 FSFSTMFFAH 548
>UniRef100_Q9SDQ4 Glutamate receptor 3.7 precursor [Arabidopsis thaliana]
Length = 921
Score = 540 bits (1390), Expect = e-151
Identities = 272/609 (44%), Positives = 397/609 (64%), Gaps = 9/609 (1%)
Query: 27 RPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQLVLHMQTSNCSGFDGMI 86
RP VNIGA+F FDS IG+ AK+A+E AV DVN++ S L T+L L M+ S C+ F G
Sbjct: 27 RPQLVNIGAVFAFDSVIGRAAKVALEAAVSDVNNDKSFLKETELRLLMEDSACNVFRGSF 86
Query: 87 QALRFMETDVIAILGPQSSVVSHIVAHVANELRVPMLSFAATDPTLSSLQFPFFVRTTLS 146
A +E +V+A++GP SS V+H ++ +A L P++SFAATDPTLS+LQFPFF+RTT +
Sbjct: 87 GAFELLEKEVVAMIGPISSSVAHTISDIAKGLHFPLVSFAATDPTLSALQFPFFLRTTPN 146
Query: 147 DLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAERRCRISYKVGIKSGPDV 206
D +QM+A+ ++I+FYGWKEVI++Y DD+ GRNGVSALDD L ++R RISYKV +
Sbjct: 147 DAHQMSALVDLINFYGWKEVISVYSDDELGRNGVSALDDELYKKRSRISYKVPL--SVHS 204
Query: 207 DRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQEGYVWIATDWLSTVLDS 266
D +TN L + R+ ++H + IF +A L MM YVW+ATDWLS LDS
Sbjct: 205 DEKFLTNALNKSKSIGPRVYILHFGPDPLLRIFDIAQKLQMMTHEYVWLATDWLSVTLDS 264
Query: 267 TSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWNNLTGGSLGLNSYGLHAYDTVWLVAQ 326
S T+ L+G + LRQH P++ + + FT K + + +N+Y LHAYDTVW++A
Sbjct: 265 LS-DKGTLKRLEGVVGLRQHIPESVKMEHFTHKLQS----NRSMNAYALHAYDTVWMIAH 319
Query: 327 AIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFDNGTLLLNNILRSNFVGLSGPIKL 386
I+ ++G ++ + L + L+L+ + F++G LLL +L+ NF G++G ++
Sbjct: 320 GIEELLNEGINITFSYSEKLLHARGTKLHLEKIKFFNSGELLLEKLLKVNFTGIAGQVQF 379
Query: 387 DSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVSPETLYANPPNRSSANQHLHTVIW 446
S R++ Y+IINV V VG+WS G S+V+P+T ++ +++ L + W
Sbjct: 380 GSGRNVIGCDYEIINVNKTDVHTVGFWSKNGGFSVVAPKTRHSQKKTSFVSDEKLGDITW 439
Query: 447 PGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPVKGTD-LFKGFCVDVFVAAVNLLP 505
PG +PRGWV ++ L+I VP R S+ EFV+ K + +GFC+DVF+ A+ +P
Sbjct: 440 PGGGREKPRGWVIADSADPLKIVVPRRVSFVEFVTEEKNSSHRIQGFCIDVFIEALKFVP 499
Query: 506 YAVPYRFVPFGDGHKNPSYTEFVNKITTGYFDGAVGDIAIVTNRTRIVDFTQPYAASGLV 565
Y+VPY F PFG+GH +P+Y + +T G +D AVGDIAIV +R+++VDF+QPYA++GLV
Sbjct: 500 YSVPYIFEPFGNGHSSPNYNHLIQMVTDGVYDAAVGDIAIVPSRSKLVDFSQPYASTGLV 559
Query: 566 VVAPFKKINSGGWSFLQPFTPFMWIVTACFFFFVGIVVWILEHRVNDEFRGSPKQQFVTI 625
VV P N+ W FL+PFT +W V F + +V+WILEHR+N++FRG P++Q T+
Sbjct: 560 VVIPANDDNA-TWIFLRPFTSRLWCVVLVSFLVIAVVIWILEHRINEDFRGPPRRQLSTM 618
Query: 626 LWFSLSTLF 634
L FS STLF
Sbjct: 619 LLFSFSTLF 627
>UniRef100_Q69L11 Putative Avr9/Cf-9 rapidly elicited protein 141 [Oryza sativa]
Length = 933
Score = 372 bits (954), Expect = e-101
Identities = 230/637 (36%), Positives = 342/637 (53%), Gaps = 34/637 (5%)
Query: 10 LVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQ 69
+ FLF+ + Q + S +N+G I S +GK+A+ ++ AV+D T+
Sbjct: 12 IFFLFLSLTVAQNITGSGEDTLNVGVILHLKSLVGKMARTSILMAVEDFYKAHRNF-KTK 70
Query: 70 LVLHMQTSNCSGFDGMIQALRFMET-DVIAILGPQSSVVSHIVAHVANELRVPMLSFAAT 128
LVLH++ SN +A+ +E +V AI+GPQ S + V+ + N+ +VP++SF AT
Sbjct: 71 LVLHIRDSNGDDIQAASEAIDLLENYNVRAIVGPQKSSEATFVSDLGNKSQVPVISFTAT 130
Query: 129 DPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALA 188
+PTLSS+ P+F+R TLSD+ Q+ +A + YGW+EV+ IY D DYGR + L DAL
Sbjct: 131 NPTLSSINVPYFLRGTLSDVAQVNTLAALAKAYGWREVVPIYEDTDYGRGIIPYLADALQ 190
Query: 189 ERRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMM 248
E + Y+ I + D +I L + MQ+R+ VVH +N G ++FK A LGMM
Sbjct: 191 EFGASMPYRSAISESANTD--QIERELYKLMTMQTRVYVVHMSTNIGSILFKKAKDLGMM 248
Query: 249 QEGYVWIATDWLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWN------N 302
E Y WI TD +S + +S S + ++ + GA+ +R + P + FT++WN N
Sbjct: 249 SEDYAWILTDGISNIANSLSPSI--LEEMSGAIGVRFYVPASKELDDFTTRWNKRFKEDN 306
Query: 303 LTGGSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIF 362
L+ +GL YDT+W +AQA + + D L + I
Sbjct: 307 PNDPPSQLSIFGLWGYDTIWALAQAAEKVRMADAIF------QKQKDTKNTTCLGTLRIS 360
Query: 363 DNGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIV 422
G LL++IL S F GLSG L R L + IINVVG+ ++ +G+W+ G+
Sbjct: 361 TIGPKLLDSILLSKFRGLSGEFDL-RNRQLELSTFQIINVVGSQLKEIGFWTAKHGIF-- 417
Query: 423 SPETLYANPPNRSSANQ--HLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFV 480
L N ++ N L+ V+WPGE + P+GW P NGK+LRIGV A Y EF+
Sbjct: 418 --RQLNKNKSKTTNMNSMPDLNPVVWPGEVYTVPKGWQIPTNGKKLRIGVRTNA-YPEFM 474
Query: 481 ----SPVKGTDLFKGFCVDVFVAAVNLLPYAVPYRFVPF--GDGHKNPSYTEFVNKITTG 534
+PV G+ +DVF + LPYA+PY +V F G G + SY +FV ++ G
Sbjct: 475 KVESNPVTNEITASGYAIDVFEEVLKRLPYAIPYEYVSFDNGQGINSGSYNDFVYQVYLG 534
Query: 535 YFDGAVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFK-KINSGGWSFLQPFTPFMWIVTA 593
+D A+GDI I NRT VDFT PY SG+ ++ P + N W FL+P T +W +
Sbjct: 535 VYDAAIGDITIRYNRTSYVDFTLPYTESGVAMIVPVRDDRNKNTWVFLKPLTTDLWFGSI 594
Query: 594 CFFFFVGIVVWILEHRVND-EFRGSPKQQFVTILWFS 629
FF + IV+W+LE R N+ E GS +Q ++FS
Sbjct: 595 AFFVYTAIVIWLLERRSNNAELTGSFLRQLGIAIYFS 631
>UniRef100_Q6RKN3 Glutamate receptor 2.7 precursor [Arabidopsis thaliana]
Length = 952
Score = 371 bits (953), Expect = e-101
Identities = 216/649 (33%), Positives = 354/649 (54%), Gaps = 41/649 (6%)
Query: 3 FYYYWLWLVFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNS 62
F YY++ V F+L +E ++ + + +G + +S K+ ++ ++ D
Sbjct: 13 FMYYFVLFVCGFVL--MEGCLGQNQTTEIKVGVVLDLHTSFSKLCLTSINISLSDFYKYH 70
Query: 63 SILHSTQLVLHMQTSNCSGFDGMIQALRFMETD-VIAILGPQSSVVSHIVAHVANELRVP 121
S ++T+L +H++ S AL ++ + V AI+GP++S+ + + +A++ +VP
Sbjct: 71 SD-YTTRLAIHIRDSMEDVVQASSAALDLIKNEQVSAIIGPRTSMQAEFMIRLADKSQVP 129
Query: 122 MLSFAATDPTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVS 181
++F+AT P L+S+ P+FVR TL D Q+ A+A I+ +GW+ V+ IYVD+++G +
Sbjct: 130 TITFSATCPLLTSINSPYFVRATLDDSSQVKAIAAIVKSFGWRNVVAIYVDNEFGEGILP 189
Query: 182 ALDDALAERRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKV 241
L DAL + + + + I + D +I L + MQ+R+ VVH GF F+
Sbjct: 190 LLTDALQDVQAFVVNRCLIPQEANDD--QILKELYKLMTMQTRVFVVHMPPTLGFRFFQK 247
Query: 242 AHYLGMMQEGYVWIATDWLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWN 301
A +GMM+EGYVW+ TD + +L S +++ +QG L +R H P + + K F +W
Sbjct: 248 AREIGMMEEGYVWLLTDGVMNLLKSNERG-SSLENMQGVLGVRSHIPKSKKLKNFRLRWE 306
Query: 302 NL---TGGSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDK--AGG--- 353
+ G +N + L AYD++ +A A++ TN SL D A G
Sbjct: 307 KMFPKKGNDEEMNIFALRAYDSITALAMAVEK----------TNIKSLRYDHPIASGNNK 356
Query: 354 LNLDAMSIFDNGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYW 413
NL + + G LL + F GL+G +L + + L +D+IN++G+ R +G W
Sbjct: 357 TNLGTLGVSRYGPSLLKALSNVRFNGLAGEFELINGQ-LESSVFDVINIIGSEERIIGLW 415
Query: 414 SNYSGLSIVSPETLYANPPNRSSA-NQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPI 472
+G+ + A N +S + L VIWPG++ P+GW P NGK LR+G+P+
Sbjct: 416 RPSNGI-------VNAKSKNTTSVLGERLGPVIWPGKSKDVPKGWQIPTNGKMLRVGIPV 468
Query: 473 RASYREFVS----PVKGTDLFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGHKNPSYTEFV 528
+ + EFV P+ G+C+++F A + LPY+V +++ F +N Y E V
Sbjct: 469 KKGFLEFVDAKIDPISNAMTPTGYCIEIFEAVLKKLPYSVIPKYIAFLSPDEN--YDEMV 526
Query: 529 NKITTGYFDGAVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFKKINSGGWSFLQPFTPFM 588
++ TG +D VGD+ IV NR+ VDFT PY SG+ ++ P K N W FL+P++ +
Sbjct: 527 YQVYTGAYDAVVGDVTIVANRSLYVDFTLPYTESGVSMMVPLKD-NKNTWVFLRPWSLDL 585
Query: 589 WIVTACFFFFVGIVVWILEHRVNDEFRGSPKQQFVTILWFSLSTLFFSH 637
W+ TACFF F+G +VWILEHRVN +FRG P Q T WF+ ST+ F+H
Sbjct: 586 WVTTACFFVFIGFIVWILEHRVNTDFRGPPHHQIGTSFWFAFSTMNFAH 634
>UniRef100_Q69L07 Putative Avr9/Cf-9 rapidly elicited protein 141 [Oryza sativa]
Length = 934
Score = 360 bits (923), Expect = 1e-97
Identities = 220/634 (34%), Positives = 338/634 (52%), Gaps = 30/634 (4%)
Query: 11 VFLFMLPYLEQVYSNSRPSFVNIGAIFTFDSSIGKVAKLAMEQAVKDVNSNSSILHSTQL 70
+FLF+ + Q + + +++G I S +GK+A+ ++ AV+D S T+L
Sbjct: 11 LFLFLSLTVAQNITKNGAGTLDVGVILHLKSLVGKIARTSVLMAVEDFYSVHRNF-KTKL 69
Query: 71 VLHMQTSNCSGFDGMIQALRFMET-DVIAILGPQSSVVSHIVAHVANELRVPMLSFAATD 129
VLH++ SN +A+ +E +V AI+GPQ S V+++ N+ +VP++SF AT+
Sbjct: 70 VLHIRDSNGDDVQAASEAIDLLENYNVRAIVGPQKSSEVTFVSNLGNKSQVPVISFTATN 129
Query: 130 PTLSSLQFPFFVRTTLSDLYQMTAVAEIIDFYGWKEVITIYVDDDYGRNGVSALDDALAE 189
P LSS+ P+F+R TLSD+ Q+ +A +I Y W+EV+ IY D DYGR + L DAL E
Sbjct: 130 PALSSINVPYFLRGTLSDVAQVNTIAALIKAYDWREVVPIYEDTDYGRGIIPYLADALQE 189
Query: 190 RRCRISYKVGIKSGPDVDRGEITNLLVNVAMMQSRIIVVHAHSNSGFMIFKVAHYLGMMQ 249
+ Y+ I D ++ L + MQ+R+ VVH N ++F A LGMM
Sbjct: 190 FGAFMPYRSAISESATTD--QLERELYKLMTMQTRVYVVHMSLNIASILFAKAKDLGMMS 247
Query: 250 EGYVWIATDWLSTVLDSTSLPLETMDTLQGALVLRQHTPDTDRKKMFTSKWN------NL 303
E Y WI TD +S +++ SL ++ + GA+ +R + P + FT++WN N
Sbjct: 248 EDYAWILTDGISNIVN--SLNTSILEKMNGAIGVRFYVPASKELDDFTTRWNKRFKEDNP 305
Query: 304 TGGSLGLNSYGLHAYDTVWLVAQAIDNFFSQGGVVSCTNYTSLHSDKAGGLNLDAMSIFD 363
L+++GL YDT+W +AQA + + D +L + I
Sbjct: 306 NDPPSQLSTFGLWGYDTIWALAQAAEKVRMADAIF------RKQKDGKNSTSLGTLGIST 359
Query: 364 NGTLLLNNILRSNFVGLSGPIKLDSERSLFRPAYDIINVVGNGVRRVGYWSNYSGLSIVS 423
G LL++IL S F GLSG L R L + IINVVG + +G+W G+
Sbjct: 360 IGPELLDSILHSKFQGLSGEFDL-GNRQLEFSTFQIINVVGGRSKEIGFWITKHGIFRQI 418
Query: 424 PETLYANPPNRSSANQHLHTVIWPGETTSRPRGWVFPNNGKQLRIGVPIRASYREFVSPV 483
E + + ++ L+ V+WPGE + P+GW P NGK+LR+GV + Y EF+
Sbjct: 419 NENI--SKTTNVNSMPGLNRVMWPGEVYTVPKGWQIPTNGKKLRVGVR-TSGYPEFMKVE 475
Query: 484 KGTD----LFKGFCVDVFVAAVNLLPYAVPYRFVPFGDGH--KNPSYTEFVNKITTGYFD 537
+ T G+ +DVF A+ LPYA+PY +V F DG + SY +FV ++ G +D
Sbjct: 476 RNTATNEITASGYAIDVFEEALKRLPYAIPYEYVAFDDGQGVNSGSYNDFVYQVHLGVYD 535
Query: 538 GAVGDIAIVTNRTRIVDFTQPYAASGLVVVAPFK-KINSGGWSFLQPFTPFMWIVTACFF 596
A+GDI I NRT VDFT PY SG+ ++ P K + W FL+P T +W + FF
Sbjct: 536 AAIGDITIRYNRTSYVDFTLPYTESGVAMIVPVKDDRDKNTWVFLKPLTTGLWFGSIAFF 595
Query: 597 FFVGIVVWILEHRVND-EFRGSPKQQFVTILWFS 629
+ +V+W+LE R+N+ E GS +Q ++FS
Sbjct: 596 IYTAVVIWLLERRINNAELTGSFFRQLGIAIYFS 629
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.338 0.147 0.498
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,499,582,607
Number of Sequences: 2790947
Number of extensions: 61901468
Number of successful extensions: 202322
Number of sequences better than 10.0: 1384
Number of HSP's better than 10.0 without gapping: 486
Number of HSP's successfully gapped in prelim test: 900
Number of HSP's that attempted gapping in prelim test: 199548
Number of HSP's gapped (non-prelim): 2132
length of query: 916
length of database: 848,049,833
effective HSP length: 137
effective length of query: 779
effective length of database: 465,690,094
effective search space: 362772583226
effective search space used: 362772583226
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC126786.13


