

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.10 - phase: 0 /pseudo
(1308 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
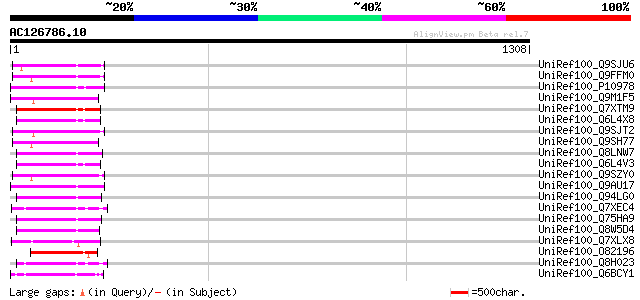
UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidop... 144 2e-32
UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidops... 144 2e-32
UniRef100_P10978 Retrovirus-related Pol polyprotein from transpo... 142 9e-32
UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana] 140 3e-31
UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa] 138 1e-30
UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa] 137 2e-30
UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidop... 136 4e-30
UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidop... 136 4e-30
UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa] 136 5e-30
UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa] 135 1e-29
UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana] 134 1e-29
UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense] 131 1e-28
UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sa... 129 5e-28
UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa] 129 8e-28
UniRef100_Q75HA9 Putative polyprotein [Oryza sativa] 125 7e-27
UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza... 125 9e-27
UniRef100_Q7XLX8 OSJNBa0042I15.12 protein [Oryza sativa] 124 2e-26
UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabid... 119 8e-25
UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Or... 118 1e-24
UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas] 113 3e-23
>UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 838
Score = 144 bits (363), Expect = 2e-32
Identities = 95/246 (38%), Positives = 143/246 (57%), Gaps = 19/246 (7%)
Query: 7 DIEKFTGGNDFGLWKVKMRAI---------LIQQKCVEALKGEAQMAAHLTPAEKTELND 57
++EK G D+ LWK K+ A L + + +E + A+ + LT E L +
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 58 K---AVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVE 114
K A S +I+ LG+ VLR+V +E TA M LD L+M KSL +R LKQ+LY Y+M +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 115 SKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLE 174
S I E + +F K+I +L N+ V++ DED+A+ LL +LP+ F+ KDT+ YGK T+ L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKT-TLALD 185
Query: 175 EVQAALRTKELTKFKELK-VDDSGEGLNV-SRGRSQNRGKGKGKNSRSKSRSKGDGNKTQ 232
E+ A+R+K L K + +S + L V RGRS+ R K +N +S+SRSK K
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSERN-KSQSRSKSREKKV- 243
Query: 233 YKCFIC 238
C++C
Sbjct: 244 --CWVC 247
>UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 144 bits (363), Expect = 2e-32
Identities = 97/254 (38%), Positives = 143/254 (56%), Gaps = 28/254 (11%)
Query: 7 DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKT------------- 53
++EKF G D+ LWK K+ A + +E L E + + E +
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 54 -------ELNDKAVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQ 106
E KA S II+ LG+ VLR+V ++ TA M LD L+M KSL +R LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 107 LYFYRMVESKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYG 166
LY Y+M E+ + E + +F K+I +L N+ V + DED+A+ LL +LPR F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 167 KEGTITLEEVQAALRTKELTKFKELK-VDDSGEGLNV-SRGRSQNRGKGKGKNSRSKSRS 224
K T+ LEE+ +A+R+K L K + ++ +GL V RGRS+ RGKG KN +S+S+S
Sbjct: 187 KT-TLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKN-KSRSKS 244
Query: 225 KGDGNKTQYKCFIC 238
KG G C+IC
Sbjct: 245 KGAGK----TCWIC 254
>UniRef100_P10978 Retrovirus-related Pol polyprotein from transposon TNT 1-94
[Contains: Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Endonuclease] [Nicotiana tabacum]
Length = 1328
Score = 142 bits (357), Expect = 9e-32
Identities = 84/238 (35%), Positives = 133/238 (55%), Gaps = 5/238 (2%)
Query: 3 GSK*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSA 62
G K ++ KF G N F W+ +MR +LIQQ + L +++ + + +L+++A SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQL 122
I + L D V+ + E TA +W +L+SLYM+K+L ++ LK+QLY M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 TEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRT 182
FN +I LAN+ V +E+EDKA+ LL +LP S++N T+L+GK TI L++V +AL
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 183 KELTKFKELKVDDSGEGL-NVSRGRSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICH 239
E + K ++ G+ L RGRS R S ++ +SK C+ C+
Sbjct: 182 NEKMR---KKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCN 236
>UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1363
Score = 140 bits (352), Expect = 3e-31
Identities = 87/244 (35%), Positives = 143/244 (57%), Gaps = 24/244 (9%)
Query: 3 GSK*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDK---- 58
G++ ++EKF G D+ +WK K+ A + L+ +EK++ ++K
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEERE 62
Query: 59 -----------AVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQL 107
A S I++ + D+VLR++ +ET+A +M LD LYM+K+L +R LKQ+L
Sbjct: 63 KMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKL 122
Query: 108 YFYRMVESKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGK 167
Y ++M E+ I + EF I+ +L N++V + DED+A+ LL +LP+ F+ KDT+ Y
Sbjct: 123 YSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSS 182
Query: 168 EGTI-TLEEVQAALRTKELTKFKELK--VDDSGEGLNV-----SRGRSQNRGKGKGKNSR 219
T+ +L+EV AA+ ++EL +F +K + EGL V +RGRS+ + KGKGK S+
Sbjct: 183 GKTVLSLDEVAAAIYSREL-EFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGKRSK 241
Query: 220 SKSR 223
SKS+
Sbjct: 242 SKSK 245
>UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa]
Length = 1181
Score = 138 bits (348), Expect = 1e-30
Identities = 80/213 (37%), Positives = 129/213 (60%), Gaps = 7/213 (3%)
Query: 17 FGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREVS 76
F LW+VKMRA+L QQ+ +AL G + + EK + KA+S I + L + +L+EV
Sbjct: 17 FSLWQVKMRAVLAQQELDDALSGFDKRTQDWSNDEKKR-DRKAMSYIHLHLSNNILQEVL 75
Query: 77 RETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANID 136
+E TA +W KL+ + MTK L + LKQ+L+ +++ + +M+ L+ F +I+ +L +++
Sbjct: 76 KEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSAFKEIVADLESME 135
Query: 137 VNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDDS 196
V +++D AL LLC+LP S+ NF+DT+LY ++ T+TL+EV AL KE ++K
Sbjct: 136 VKYDEKDLALILLCSLPSSYANFRDTILYSRD-TLTLKEVYDALHAKE-----KMKKMVP 189
Query: 197 GEGLNVSRGRSQNRGKGKGKNSRSKSRSKGDGN 229
EG N RG + KN+ +KSR K +
Sbjct: 190 SEGSNSQAEGLVVRGSQQEKNTNNKSRDKSSSS 222
>UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa]
Length = 1211
Score = 137 bits (346), Expect = 2e-30
Identities = 81/213 (38%), Positives = 126/213 (59%), Gaps = 7/213 (3%)
Query: 17 FGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREVS 76
F LW+VKMRA+L QQ +AL G + + EK + K +S I + L + +L+EV
Sbjct: 17 FSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKR-DRKTMSYIHLHLSNNILQEVL 75
Query: 77 RETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANID 136
+E TA +W KL+ + MTK L + LKQ+L+ +++ + +M+ L+ F KI+ +L +++
Sbjct: 76 KEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSAFKKIVADLESME 135
Query: 137 VNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDDS 196
V ++ED L LLC+LP S+ NF+DT+LY + T+TL+EV AL KE ++K
Sbjct: 136 VKYDEEDLCLILLCSLPSSYANFRDTILYSCD-TLTLKEVYDALHAKE-----KIKKMVP 189
Query: 197 GEGLNVSRGRSQNRGKGKGKNSRSKSRSKGDGN 229
EG N RG+ + KN+ SKSR K +
Sbjct: 190 SEGSNSQAEGLVVRGRQQEKNTNSKSRDKSSSS 222
>UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 136 bits (343), Expect = 4e-30
Identities = 94/255 (36%), Positives = 141/255 (54%), Gaps = 30/255 (11%)
Query: 7 DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDK-------- 58
++EKF G D+ +WK K+ A L ALK E + + + TE +K
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 59 -------AVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYR 111
A SAI++ + D+VLR++ +E +A +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 112 MVESKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYG-KEGT 170
M E+ I + EF +II +L N +V + DED+A+ LL +LP+ F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 171 ITLEEVQAALRTKELTKFKELK-VDDSGEGLNV-----SRGRSQNRG-KGKGKNSRSKSR 223
++L+EV AA+ +KEL K + EGL V +RGR++ RG K SRSKSR
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEKTETRGRTEQRGNNNNNKKSRSKSR 246
Query: 224 SKGDGNKTQYKCFIC 238
SK C+IC
Sbjct: 247 SKKG-------CWIC 254
>UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1356
Score = 136 bits (343), Expect = 4e-30
Identities = 84/239 (35%), Positives = 141/239 (58%), Gaps = 22/239 (9%)
Query: 7 DIEKFTGGNDFGLWKVKMRA---ILIQQKCVEALKGEAQMAAHLTPAEKT---------- 53
++EKF G D+ +WK K+ A IL ++ + + + L +++
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 54 --ELNDKAVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYR 111
E KA SAI++ + D+VLR++ +E+TA +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 112 MVESKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGK-EGT 170
M E+ + + EF +II +L N++V + DED+A+ LL ALP++F+ KDT+ Y +
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 171 ITLEEVQAALRTKEL---TKFKELKVDDSG---EGLNVSRGRSQNRGKGKGKNSRSKSR 223
+TL+EV AA+ +KEL + K +KV G + N ++G+ + +GKGKGK +SK +
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENKGKGEQKGKGKGKKGKSKKK 245
>UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa]
Length = 1280
Score = 136 bits (342), Expect = 5e-30
Identities = 86/227 (37%), Positives = 136/227 (59%), Gaps = 15/227 (6%)
Query: 17 FGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREVS 76
F LW+VKMRA+L QQ +AL G + + EK + + KA+S I + L + +L+EV
Sbjct: 52 FSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKK-DRKAMSYIHLHLSNNILQEVL 110
Query: 77 RETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANID 136
+E TA +W KL+ + MTK L + LKQ+L+ +++ + +M+ L+ F +I+ +L +I+
Sbjct: 111 KEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSTFKEIVADLESIE 170
Query: 137 VNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDD- 195
V ++ED L LLC+LP S+ NF+DT+LY + T+ L+EV AL KE K K++ +
Sbjct: 171 VKYDEEDLGLILLCSLPSSYANFRDTILYSHD-TLILKEVYDALHAKE--KMKKMVPSEG 227
Query: 196 ---SGEGLNVSRGRSQ-----NRGKGKGKNS-RSKSRSKGDGNKTQY 233
EGL V RGR Q N+ + K +S R +S+S+G +Y
Sbjct: 228 SNSQAEGL-VVRGRQQEKNTKNQSRDKSSSSYRGRSKSRGRYKSCKY 273
>UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa]
Length = 1243
Score = 135 bits (339), Expect = 1e-29
Identities = 76/213 (35%), Positives = 129/213 (59%), Gaps = 7/213 (3%)
Query: 17 FGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREVS 76
F LW+VKMRA+L QQ +AL G + + EK + KA+S I + L + +L+EV
Sbjct: 17 FSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKR-DRKAISYIHLHLSNNILQEVL 75
Query: 77 RETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANID 136
+E TA +W KL+ + MTK L + LKQ+L+ +++ + + +M+ L+ F +I+ +L +++
Sbjct: 76 KEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLSAFKEIVADLESME 135
Query: 137 VNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDDS 196
V +++D L LLC+LP S+ NF+ T+LY ++ T+TL+EV A KE ++K +
Sbjct: 136 VKYDEDDLGLILLCSLPSSYANFRGTILYSRD-TLTLKEVYDAFHAKE-----KMKKMVT 189
Query: 197 GEGLNVSRGRSQNRGKGKGKNSRSKSRSKGDGN 229
EG N RG+ + KN++++SR K +
Sbjct: 190 SEGSNSQAEGLVVRGRQQKKNTKNQSRDKSSSS 222
>UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana]
Length = 1230
Score = 134 bits (338), Expect = 1e-29
Identities = 91/250 (36%), Positives = 135/250 (53%), Gaps = 23/250 (9%)
Query: 7 DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEK-------------T 53
++EKF G D+ LWK K+ A + AL+ ++ L E+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 54 ELNDKAVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMV 113
E KA S I++ + D+VLR+ +E TA SM LD LYM+K+L +R LKQ+LY Y+M
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 114 ESKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGK-EGTIT 172
E+ + + EF ++I +L N +V + DED+A+ LL +LP+ F+ KDT+ YG T++
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 173 LEEVQAALRTKELTKFKELK-VDDSGEGLNVS---RGRSQNRGKGKGKNSRSKSRSKGDG 228
++EV AA+ +KEL K + EGL V R + K KG RS+SRSKG
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKGNKGRSRSRSKG-- 244
Query: 229 NKTQYKCFIC 238
C+IC
Sbjct: 245 ---WKGCWIC 251
>UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 131 bits (330), Expect = 1e-28
Identities = 80/237 (33%), Positives = 131/237 (54%), Gaps = 4/237 (1%)
Query: 3 GSK*DIEKFTGGND-FGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVS 61
G K ++ KF G F +W+ +M+ +LIQQ +AL G+++ + + EL++KA S
Sbjct: 3 GVKYEVAKFNGDKPVFSMWQRRMKDLLIQQGLHKALGGKSKKPESMKLEDWEELDEKAAS 62
Query: 62 AIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQ 121
AI + L D V+ + E +A +W KL++LYM+K+L ++ LK+QLY M E +
Sbjct: 63 AIRLHLTDDVVNNIVDEESACGIWTKLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSH 122
Query: 122 LTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALR 181
L N +I LAN+ V +E+EDK + LL +LP S++ T+L+GK+ +I L++V +AL
Sbjct: 123 LNVLNGLITQLANLGVKIEEEDKRIVLLNSLPSSYDTLSTTILHGKD-SIQLKDVTSALL 181
Query: 182 TKELTKFKELKVDDSGEGLNVSRGRSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFIC 238
E K ++ + + SRGRS R S ++ +SK C+ C
Sbjct: 182 LNE--KMRKKPENHGQVFITESRGRSYQRSSSNYGRSGARGKSKVRSKSKARNCYNC 236
>UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1326
Score = 129 bits (325), Expect = 5e-28
Identities = 83/222 (37%), Positives = 132/222 (59%), Gaps = 12/222 (5%)
Query: 17 FGLWKVKMRAILIQQKCV-EALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREV 75
F LW+VKMRAIL Q + EAL+ + + AE+ + KA+ I + L + +L+EV
Sbjct: 17 FSLWQVKMRAILAQTSDLDEALESFGKKKSTEWTAEEKRKDRKALLLIQLHLSNDILQEV 76
Query: 76 SRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANI 135
+E TA +W KL+S+ M+K L + +K +L+ +++ ES ++ ++ F +I+ +L +I
Sbjct: 77 LQEKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHISVFKEIVVDLVSI 136
Query: 136 DVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDD 195
+V +DED L LLC+LP S+ NF+DT+L ++ +TL EV AL+ +E K K + D
Sbjct: 137 EVQFDDEDLGLLLLCSLPSSYANFRDTILLSRD-ELTLAEVYEALQNRE--KMKGMVQSD 193
Query: 196 S----GEGLNVSRGRSQNRGKGKGKN---SRSKSRSKGDGNK 230
+ GE L V RGRS+ R + S+S+ RSK G K
Sbjct: 194 ASSSKGEALQV-RGRSEQRTYNDSSDRDKSQSRGRSKSRGKK 234
>UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa]
Length = 382
Score = 129 bits (323), Expect = 8e-28
Identities = 80/244 (32%), Positives = 139/244 (56%), Gaps = 16/244 (6%)
Query: 4 SK*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSAI 63
SK ++ KF G +F LW+++++ +L QQ +AL + M + + E+ +A + I
Sbjct: 7 SKFEVVKFDGTGNFVLWQMRLKDLLAQQGISKAL--QETMPEKIDADKWNEMKAQAAATI 64
Query: 64 IMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLT 123
+ L D V+ +V E + +W+KL SL+M+KSL + LKQQLY ++ E + + +
Sbjct: 65 RLSLSDSVMYQVMDEKSPKEIWDKLASLHMSKSLTSKLYLKQQLYGLQVQEESDLRKHVD 124
Query: 124 EFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTK 183
FN+++ +L+ +DV L+DEDKA+ LLC+LP S+E+ T+ +GK+ T+ EE+ ++L +
Sbjct: 125 VFNQLVVDLSKLDVKLDDEDKAIILLCSLPLSYEHVVTTLTHGKD-TVKTEEIISSLLAR 183
Query: 184 ELTKFKELKVDDSGEGLNVSRGRS-QNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNLV 242
+L + K+ E S+G+S + K + SKS+ KG +C+ CH
Sbjct: 184 DLRRSKK------NEATKASQGKSLLVKDKHDHEAGVSKSKEKG------ARCYKCHEFG 231
Query: 243 TSRR 246
RR
Sbjct: 232 HIRR 235
>UniRef100_Q75HA9 Putative polyprotein [Oryza sativa]
Length = 1322
Score = 125 bits (315), Expect = 7e-27
Identities = 78/220 (35%), Positives = 128/220 (57%), Gaps = 8/220 (3%)
Query: 17 FGLWKVKMRAILIQQKCV-EALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREV 75
F LW+VKMRA+L Q + EAL+ + AE+ + KA+S I + L + +L+EV
Sbjct: 17 FSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLIQLHLSNDILQEV 76
Query: 76 SRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANI 135
++ TA +W KL+S+ M+K L + +K +L+ +++ ES ++ ++ F +I+ +L ++
Sbjct: 77 LQKKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLHESGSVLNHISVFKEIVADLVSM 136
Query: 136 DVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTK--FKELKV 193
+V +DED L LLC+LP S+ NF+ T+L ++ +TL EV AL+ +E K +
Sbjct: 137 EVQFDDEDLGLLLLCSLPSSYANFRHTILLSRD-ELTLAEVYEALQNREKMKGMVQSYAS 195
Query: 194 DDSGEGLNVSRGRSQNRGKGKGKN---SRSKSRSKGDGNK 230
GE L V RGRS+ R + S+S+ RSK G K
Sbjct: 196 SSKGEALQV-RGRSEQRTYNDSNDHDKSQSRGRSKSRGKK 234
>UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 125 bits (314), Expect = 9e-27
Identities = 79/215 (36%), Positives = 129/215 (59%), Gaps = 10/215 (4%)
Query: 17 FGLWKVKMRAILIQQKCV-EALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREV 75
F LW+VKMRA+L Q + EAL+ + AE+ + KA+S I + L + +L++V
Sbjct: 14 FSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLIQLHLSNDILQKV 73
Query: 76 SRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANI 135
+E TA +W KL+S+ M+K L + +K +L+ +++ ES ++ ++ F +II +L ++
Sbjct: 74 LQEKTAAELWFKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHISVFKEIIADLVSM 133
Query: 136 DVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDD 195
+V +DED L LLC+LP + NF+DT+L ++ +TL EV AL+ +E K K + D
Sbjct: 134 EVQFDDEDLGLLLLCSLPSLYANFRDTILLSRD-ELTLAEVYEALQNRE--KMKGMVQSD 190
Query: 196 S----GEGLNVSRGRSQNRGKGKGKNSRSKSRSKG 226
+ G+ L V RGRS+ R N R KS+S+G
Sbjct: 191 ASSSKGKALQV-RGRSEQR-TYNDSNDRDKSQSRG 223
>UniRef100_Q7XLX8 OSJNBa0042I15.12 protein [Oryza sativa]
Length = 432
Score = 124 bits (311), Expect = 2e-26
Identities = 75/230 (32%), Positives = 127/230 (54%), Gaps = 11/230 (4%)
Query: 5 K*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSAII 64
K D+ KF G +FGLW+ +++ +L QQ+ +ALKG E+ +L +A + I
Sbjct: 43 KFDLVKFDGFGNFGLWQTRVKDLLAQQEVSKALKGVKPAKMEDDDCEEMQL--QAAATIR 100
Query: 65 MCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTE 124
CL D+++ V ET+ +W KL++ +M+K+L +++ LKQ+LY +M E + +
Sbjct: 101 RCLSDQIMYHVMEETSPKIIWEKLEAQFMSKTLTNKRYLKQKLYGLKMQEGADLTAHVNV 160
Query: 125 FNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGT------ITLEEVQA 178
FN+++ +L +DV ++DEDKA+ LLC+LP S+E+ M KEG + ++E A
Sbjct: 161 FNQLVTDLVKMDVEVDDEDKAIVLLCSLPESYEH---VMRKNKEGETSQAEGLLVKEDHA 217
Query: 179 ALRTKELTKFKELKVDDSGEGLNVSRGRSQNRGKGKGKNSRSKSRSKGDG 228
+ K+ K K +K E ++ R +GK K + S + S DG
Sbjct: 218 GHKVKKNKKKKMVKCFRCDEWGHIRRECPTLKGKAKANVATSSNDSDSDG 267
>UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1137
Score = 119 bits (297), Expect = 8e-25
Identities = 72/176 (40%), Positives = 110/176 (61%), Gaps = 13/176 (7%)
Query: 52 KTELNDKAVSAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYR 111
+ + ++KA+ I + +GDKVLR + TA W LD LY+ KSL +R L+ ++Y YR
Sbjct: 37 RIDQDEKAMDMIFINVGDKVLRNIENSKTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYR 96
Query: 112 MVESKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFENFKDTMLYGKEGTI 171
M +SK + E + EF K+I +L N+ + + DE +A+ +L ALP S++ K+T+ YG+EG I
Sbjct: 97 MQDSKTLEENVDEFQKMISDLNNLQIQVPDEVQAILILSALPDSYDMLKETLKYGREG-I 155
Query: 172 TLEEVQAALRTKELTKFKELKVDDS------GEGLNVSRGRSQNRGKGKGKNSRSK 221
L++V +A ++KEL EL+ D S GEGL V RG+SQ RG K++ K
Sbjct: 156 KLDDVISAAKSKEL----ELR-DSSGGSRPVGEGLYV-RGKSQARGSDGPKSTEGK 205
>UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Oryza sativa]
Length = 556
Score = 118 bits (295), Expect = 1e-24
Identities = 77/230 (33%), Positives = 128/230 (55%), Gaps = 16/230 (6%)
Query: 18 GLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSAIIMCLGDKVLREVSR 77
G +++++ +L QQ +AL E M + + E+ +A + I + L D V+ +V
Sbjct: 152 GSSQMRLKDLLAQQGISKAL--EETMPEKMDVGKWVEMKAQATAIIRLSLSDFVMYQVMD 209
Query: 78 ETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANIDV 137
E T +W+KL SLYM+KSL + LKQQLY +M E + + + FN+++ +L+ +DV
Sbjct: 210 EKTPKEIWDKLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRKHVDVFNQLVVDLSKLDV 269
Query: 138 NLEDEDKALHLLCALPRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDDSG 197
L+DEDKA+ LLC+LP SFE+ T+ +GK+ T+ EE+ ++L ++L + K+
Sbjct: 270 KLDDEDKAIILLCSLPPSFEHVVTTLTHGKD-TVKTEEIISSLLARDLRRSKK------N 322
Query: 198 EGLNVSRGRS-QNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNLVTSRR 246
E + S+ S + K + SKS+ KG +C+ CH RR
Sbjct: 323 EAMEASQAESLLVKAKHDHEAGVSKSKEKG------ARCYKCHEFGHIRR 366
>UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 113 bits (283), Expect = 3e-23
Identities = 80/242 (33%), Positives = 133/242 (54%), Gaps = 19/242 (7%)
Query: 2 MGSK*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEK-TELNDKAV 60
M +K +IEKF G N F LWK+K++AIL + C+ A+ ++ T +K +E+N+ A+
Sbjct: 1 MAAKFEIEKFNGKN-FSLWKLKVKAILRKDNCLAAI---SERPVDFTDDKKWSEMNEDAM 56
Query: 61 SAIIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIME 120
+ + + + D VL + + TA +W+ L+ LY KSL ++ LK++LY RM ES + E
Sbjct: 57 ADLYLSIADGVLSSIEEKKTANEIWDHLNRLYEAKSLHNKIFLKRKLYTLRMSESTSVTE 116
Query: 121 QLTEFNKIIDNLANIDVNLEDEDKALHLLCALPRSFE----NFKDTMLYGKEGTITLEEV 176
L N + L ++ +E +++A LL +LP S++ N + +L + ++V
Sbjct: 117 HLNTLNTLFSQLTSLSCKIEPQERAELLLQSLPDSYDQLIINLTNNIL---TDYLVFDDV 173
Query: 177 QAALRTKELTK--FKELKVD-DSGEGLNVSRGRSQNRGKGKGKNSRSKSRSKGDGNKTQY 233
AA+ +E + ++ +V+ E L V RGRS RG+ G+ RSKS K N T Y
Sbjct: 174 AAAVLEEESRRKNKEDRQVNLQQAEALTVMRGRSTERGQSSGR-GRSKSSKK---NLTCY 229
Query: 234 KC 235
C
Sbjct: 230 NC 231
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.360 0.161 0.601
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,862,340,229
Number of Sequences: 2790947
Number of extensions: 69064688
Number of successful extensions: 295524
Number of sequences better than 10.0: 325
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 170
Number of HSP's that attempted gapping in prelim test: 294859
Number of HSP's gapped (non-prelim): 537
length of query: 1308
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1169
effective length of database: 460,108,200
effective search space: 537866485800
effective search space used: 537866485800
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 81 (35.8 bits)
Medicago: description of AC126786.10


