

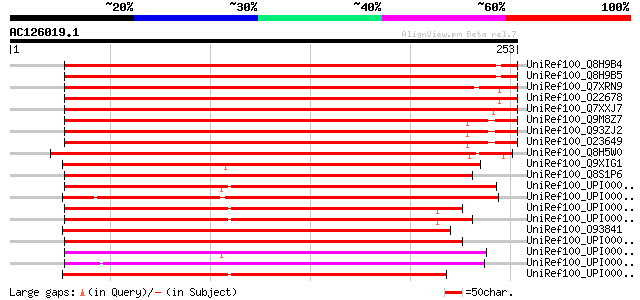
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126019.1 + phase: 0
(253 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8H9B4 UDP-glucose:sterol 3-O-glucosyltransferase [Pan... 373 e-102
UniRef100_Q8H9B5 UDP-glucose:sterol 3-O-glucosyltransferase [Pan... 372 e-102
UniRef100_Q7XRN9 OSJNBb0080H08.21 protein [Oryza sativa] 363 2e-99
UniRef100_O22678 UDP-glucose:sterol glucosyltransferase [Avena s... 362 7e-99
UniRef100_Q7XXJ7 OSJNBb0080H08.20 protein [Oryza sativa] 360 2e-98
UniRef100_Q9M8Z7 UDP-glucose:sterol glucosyltransferase [Arabido... 352 4e-96
UniRef100_Q93ZJ2 AT3g07020/F17A9_17 [Arabidopsis thaliana] 352 4e-96
UniRef100_O23649 UDP-glucose:sterol glucosyltransferase [Arabido... 352 4e-96
UniRef100_Q8H5W0 Putative UDP-glucose:sterol glucosyltransferase... 338 6e-92
UniRef100_Q9XIG1 Putative UDP-glucose:sterol glucosyltransferase... 273 4e-72
UniRef100_Q8S1P6 Putative sterol glucosyltransferase [Oryza sativa] 271 9e-72
UniRef100_UPI000023CA18 UPI000023CA18 UniRef100 entry 216 4e-55
UniRef100_UPI000023594A UPI000023594A UniRef100 entry 214 1e-54
UniRef100_UPI0000219F77 UPI0000219F77 UniRef100 entry 211 1e-53
UniRef100_UPI000023ED09 UPI000023ED09 UniRef100 entry 209 5e-53
UniRef100_O93841 CHIP6 [Colletotrichum gloeosporioides] 209 5e-53
UniRef100_UPI0000234B30 UPI0000234B30 UniRef100 entry 206 5e-52
UniRef100_UPI000021A76A UPI000021A76A UniRef100 entry 204 2e-51
UniRef100_UPI000023F6DB UPI000023F6DB UniRef100 entry 203 3e-51
UniRef100_UPI000023EA4E UPI000023EA4E UniRef100 entry 201 2e-50
>UniRef100_Q8H9B4 UDP-glucose:sterol 3-O-glucosyltransferase [Panax ginseng]
Length = 602
Score = 373 bits (958), Expect = e-102
Identities = 178/226 (78%), Positives = 193/226 (84%), Gaps = 2/226 (0%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGPKIDVVGFCFLDLAS+YEPPESLV WL G KPIY+GFGSLPVQDP+ MT++IVEAL
Sbjct: 379 DWGPKIDVVGFCFLDLASSYEPPESLVNWLNGGTKPIYIGFGSLPVQDPEKMTKVIVEAL 438
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
E TGQRGII+KGWGGLG+L EPKD+IY LDNVPHDWLFL C AVVHHGGAGTTAAGLKAA
Sbjct: 439 EITGQRGIINKGWGGLGNLAEPKDTIYSLDNVPHDWLFLQCAAVVHHGGAGTTAAGLKAA 498
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQPFWGERVH RGVGP PIPID FSLPKL+ AI FML+PKVKE AI+LAK
Sbjct: 499 CPTTIVPFFGDQPFWGERVHARGVGPAPIPIDEFSLPKLVDAIKFMLEPKVKESAIQLAK 558
Query: 208 AMENEDGVTGAVKVFFKQLPKNKPEPNTEPSSSSCFSYIARCFGYS 253
AME+EDGV GAVK FFK LP K E P S F +++CFG S
Sbjct: 559 AMEDEDGVAGAVKAFFKHLPCRKTEAEPTPVPSGFF--LSKCFGCS 602
>UniRef100_Q8H9B5 UDP-glucose:sterol 3-O-glucosyltransferase [Panax ginseng]
Length = 609
Score = 372 bits (956), Expect = e-102
Identities = 176/226 (77%), Positives = 193/226 (84%), Gaps = 2/226 (0%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGPK+DVVGFCFLDLASNYEPPESLV WL+ G KPIY+GFGSLPVQ+P+ MT++IVEAL
Sbjct: 386 DWGPKVDVVGFCFLDLASNYEPPESLVNWLKAGTKPIYIGFGSLPVQEPEKMTKVIVEAL 445
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
E TGQRGII+KGWGGLG+L EPKD+IY LDNVPHDWLFL C AVVHHGGAGTTAAGLKAA
Sbjct: 446 EITGQRGIINKGWGGLGNLAEPKDAIYSLDNVPHDWLFLQCAAVVHHGGAGTTAAGLKAA 505
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQPFWGERVH RGVGP PIPID FSLPKL+ AI FMLDPKVKE A+ LAK
Sbjct: 506 CPTTIVPFFGDQPFWGERVHARGVGPVPIPIDEFSLPKLVDAIRFMLDPKVKESAVRLAK 565
Query: 208 AMENEDGVTGAVKVFFKQLPKNKPEPNTEPSSSSCFSYIARCFGYS 253
ME+EDGV GAV+ FFK LP K P EP S F +++CFG S
Sbjct: 566 DMEDEDGVAGAVRAFFKHLPFRKTAPEPEPVPSRFF--LSKCFGCS 609
>UniRef100_Q7XRN9 OSJNBb0080H08.21 protein [Oryza sativa]
Length = 609
Score = 363 bits (933), Expect = 2e-99
Identities = 173/230 (75%), Positives = 193/230 (83%), Gaps = 6/230 (2%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGPKIDVVGFCFLDLASNY PPE LVKWLE GDKPIYVGFGSLPVQDP MTE+IV+AL
Sbjct: 382 DWGPKIDVVGFCFLDLASNYVPPEPLVKWLEAGDKPIYVGFGSLPVQDPAKMTEVIVKAL 441
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
E TGQRGII+KGWGGLG L EPKD +YLLDN PHDWLFL CKAVVHHGGAGTTAAGLKAA
Sbjct: 442 EITGQRGIINKGWGGLGTLAEPKDFVYLLDNCPHDWLFLQCKAVVHHGGAGTTAAGLKAA 501
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQPFWG+RVH RGVGP PIP+D FSL KL+ AINFM++PKVK++A+ELAK
Sbjct: 502 CPTTIVPFFGDQPFWGDRVHARGVGPLPIPVDQFSLQKLVDAINFMMEPKVKDNAVELAK 561
Query: 208 AMENEDGVTGAVKVFFKQLPKNKPEPNTEPSSSSCF----SYIARCFGYS 253
AME+EDGV+GAV+ F + LP E T P +S F +++C G S
Sbjct: 562 AMESEDGVSGAVRAFLRHLPSRAEE--TAPQQTSSFLEFLGPVSKCLGCS 609
>UniRef100_O22678 UDP-glucose:sterol glucosyltransferase [Avena sativa]
Length = 608
Score = 362 bits (928), Expect = 7e-99
Identities = 170/230 (73%), Positives = 193/230 (83%), Gaps = 4/230 (1%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGPKIDVVGFCFLDLAS+YEPPE LVKWLE GDKPIYVGFGSLPVQDP MTE I++AL
Sbjct: 379 DWGPKIDVVGFCFLDLASDYEPPEELVKWLEAGDKPIYVGFGSLPVQDPTKMTETIIQAL 438
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
E TGQRGII+KGWGGLG L EPKDSIY+LDN PHDWLFL CKAVVHHGGAGTTAAGLKAA
Sbjct: 439 EMTGQRGIINKGWGGLGTLAEPKDSIYVLDNCPHDWLFLQCKAVVHHGGAGTTAAGLKAA 498
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQ FWG+RVH RGVGP PIP++ F+L KL+ A+ FML+P+VKE A+ELAK
Sbjct: 499 CPTTIVPFFGDQQFWGDRVHARGVGPVPIPVEQFNLQKLVDAMKFMLEPEVKEKAVELAK 558
Query: 208 AMENEDGVTGAVKVFFKQLPKNKPEPNTEPSSSSCF----SYIARCFGYS 253
AME+EDGVTGAV+ F K LP +K + N+ P + F +++C G S
Sbjct: 559 AMESEDGVTGAVRAFLKHLPSSKEDENSPPPTPHGFLEFLGPVSKCLGCS 608
>UniRef100_Q7XXJ7 OSJNBb0080H08.20 protein [Oryza sativa]
Length = 307
Score = 360 bits (924), Expect = 2e-98
Identities = 170/228 (74%), Positives = 190/228 (82%), Gaps = 2/228 (0%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWG KIDVVGFCFLDLASNY PPE L+KWLE GDKPIYVGFGSLPVQDP MTE+IV+AL
Sbjct: 80 DWGLKIDVVGFCFLDLASNYVPPEPLIKWLEAGDKPIYVGFGSLPVQDPAKMTEVIVKAL 139
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
E TGQRGII+KGWGGLG L EPKD +YLLDN PHDWLFL CKAVVHHGGAGTTAAGLKAA
Sbjct: 140 EITGQRGIINKGWGGLGTLAEPKDFVYLLDNCPHDWLFLQCKAVVHHGGAGTTAAGLKAA 199
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQPFWG+RVH RGVGP PIP+D FSL KL+ AINFM++PKVKE A+ELAK
Sbjct: 200 CPTTIVPFFGDQPFWGDRVHARGVGPLPIPVDQFSLRKLVDAINFMMEPKVKEKAVELAK 259
Query: 208 AMENEDGVTGAVKVFFKQLPKNKPEPNTEPSSS--SCFSYIARCFGYS 253
AME+EDGV+G V+ F + LP E +P+SS +++C G S
Sbjct: 260 AMESEDGVSGVVRAFLRHLPLRAEETTPQPTSSFLEFLGPVSKCLGCS 307
>UniRef100_Q9M8Z7 UDP-glucose:sterol glucosyltransferase [Arabidopsis thaliana]
Length = 637
Score = 352 bits (904), Expect = 4e-96
Identities = 172/229 (75%), Positives = 189/229 (82%), Gaps = 6/229 (2%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGP+IDVVGFC+LDLASNYEPP LV+WLE GDKPIY+GFGSLPVQ+P+ MTEIIVEAL
Sbjct: 412 DWGPQIDVVGFCYLDLASNYEPPAELVEWLEAGDKPIYIGFGSLPVQEPEKMTEIIVEAL 471
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
+ T QRGII+KGWGGLG+L EPKD +YLLDNVPHDWLF CKAVVHHGGAGTTAAGLKA+
Sbjct: 472 QRTKQRGIINKGWGGLGNLKEPKDFVYLLDNVPHDWLFPRCKAVVHHGGAGTTAAGLKAS 531
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQPFWGERVH RGVGP PIP+D FSL KL AINFMLD KVK A LAK
Sbjct: 532 CPTTIVPFFGDQPFWGERVHARGVGPSPIPVDEFSLHKLEDAINFMLDDKVKSSAETLAK 591
Query: 208 AMENEDGVTGAVKVFFKQLP---KNKPEPNTEPSSSSCFSYIARCFGYS 253
AM++EDGV GAVK FFK LP +N +P EPS F +CFG S
Sbjct: 592 AMKDEDGVAGAVKAFFKHLPSAKQNISDPIPEPSG---FLSFRKCFGCS 637
>UniRef100_Q93ZJ2 AT3g07020/F17A9_17 [Arabidopsis thaliana]
Length = 555
Score = 352 bits (904), Expect = 4e-96
Identities = 172/229 (75%), Positives = 189/229 (82%), Gaps = 6/229 (2%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGP+IDVVGFC+LDLASNYEPP LV+WLE GDKPIY+GFGSLPVQ+P+ MTEIIVEAL
Sbjct: 330 DWGPQIDVVGFCYLDLASNYEPPAELVEWLEAGDKPIYIGFGSLPVQEPEKMTEIIVEAL 389
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
+ T QRGII+KGWGGLG+L EPKD +YLLDNVPHDWLF CKAVVHHGGAGTTAAGLKA+
Sbjct: 390 QRTKQRGIINKGWGGLGNLKEPKDFVYLLDNVPHDWLFPRCKAVVHHGGAGTTAAGLKAS 449
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQPFWGERVH RGVGP PIP+D FSL KL AINFMLD KVK A LAK
Sbjct: 450 CPTTIVPFFGDQPFWGERVHARGVGPSPIPVDEFSLHKLEDAINFMLDDKVKSSAETLAK 509
Query: 208 AMENEDGVTGAVKVFFKQLP---KNKPEPNTEPSSSSCFSYIARCFGYS 253
AM++EDGV GAVK FFK LP +N +P EPS F +CFG S
Sbjct: 510 AMKDEDGVAGAVKAFFKHLPSAKQNISDPIPEPSG---FLSFRKCFGCS 555
>UniRef100_O23649 UDP-glucose:sterol glucosyltransferase [Arabidopsis thaliana]
Length = 637
Score = 352 bits (904), Expect = 4e-96
Identities = 172/229 (75%), Positives = 189/229 (82%), Gaps = 6/229 (2%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGP+IDVVGFC+LDLASNYEPP LV+WLE GDKPIY+GFGSLPVQ+P+ MTEIIVEAL
Sbjct: 412 DWGPQIDVVGFCYLDLASNYEPPAELVEWLEAGDKPIYIGFGSLPVQEPEKMTEIIVEAL 471
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
+ T QRGII+KGWGGLG+L EPKD +YLLDNVPHDWLF CKAVVHHGGAGTTAAGLKA+
Sbjct: 472 QRTKQRGIINKGWGGLGNLKEPKDFVYLLDNVPHDWLFPRCKAVVHHGGAGTTAAGLKAS 531
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQPFWGERVH RGVGP PIP+D FSL KL AINFMLD KVK A LAK
Sbjct: 532 CPTTIVPFFGDQPFWGERVHARGVGPSPIPVDEFSLHKLEDAINFMLDDKVKSSAETLAK 591
Query: 208 AMENEDGVTGAVKVFFKQLP---KNKPEPNTEPSSSSCFSYIARCFGYS 253
AM++EDGV GAVK FFK LP +N +P EPS F +CFG S
Sbjct: 592 AMKDEDGVAGAVKAFFKHLPSAKQNISDPIPEPSG---FLSFRKCFGCS 637
>UniRef100_Q8H5W0 Putative UDP-glucose:sterol glucosyltransferase [Oryza sativa]
Length = 617
Score = 338 bits (868), Expect = 6e-92
Identities = 160/238 (67%), Positives = 190/238 (79%), Gaps = 8/238 (3%)
Query: 21 YIIFLFADWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMT 80
Y++ DWGPKIDVVGFCFLDLASNY+PPE L+KWLE G+KPIY+GFGSLP+ +P +T
Sbjct: 378 YLVPKPKDWGPKIDVVGFCFLDLASNYKPPEPLLKWLESGEKPIYIGFGSLPIPEPDKLT 437
Query: 81 EIIVEALETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTT 140
IIVEALE TGQRGII+KGWGGLG+L EPK+ +Y++DN+PHDWLFL CKAVVHHGGAGTT
Sbjct: 438 RIIVEALEITGQRGIINKGWGGLGNLEEPKEFVYVIDNIPHDWLFLQCKAVVHHGGAGTT 497
Query: 141 AAGLKAACPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKE 200
AA LKAACPTTIVPFFGDQ FWG VH RG+G PP+P++ L L+ AI FM+DPKVKE
Sbjct: 498 AASLKAACPTTIVPFFGDQFFWGNMVHARGLGAPPVPVEQLQLHLLVDAIKFMMDPKVKE 557
Query: 201 HAIELAKAMENEDGVTGAVKVFFKQLPK----NKPEPNTEPSSSSCFSY---IARCFG 251
A+ELAKA+E+EDGV GAVK F K LP+ KP+P PSS+ + + RCFG
Sbjct: 558 RAVELAKAIESEDGVDGAVKAFLKHLPQPRSLEKPQP-APPSSTFMQPFLLPVKRCFG 614
>UniRef100_Q9XIG1 Putative UDP-glucose:sterol glucosyltransferase [Arabidopsis
thaliana]
Length = 615
Score = 273 bits (697), Expect = 4e-72
Identities = 120/210 (57%), Positives = 158/210 (75%), Gaps = 1/210 (0%)
Query: 27 ADWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEA 86
+DWGP +DVVG+CFL+L S Y+P E + W+E G P+Y+GFGS+P+ DPK +II+E
Sbjct: 374 SDWGPLVDVVGYCFLNLGSKYQPREEFLHWIERGSPPVYIGFGSMPLDDPKQTMDIILET 433
Query: 87 LETTGQRGIISKGWGGLGDL-TEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLK 145
L+ T QRGI+ +GWGGLG+L TE ++++L+++ PHDWLF C AVVHHGGAGTTA GLK
Sbjct: 434 LKDTEQRGIVDRGWGGLGNLATEVPENVFLVEDCPHDWLFPQCSAVVHHGGAGTTATGLK 493
Query: 146 AACPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIEL 205
A CPTTIVPFFGDQ FWG+R++++G+GP PIPI S+ L ++I FML P+VK +EL
Sbjct: 494 AGCPTTIVPFFGDQFFWGDRIYEKGLGPAPIPIAQLSVENLSSSIRFMLQPEVKSQVMEL 553
Query: 206 AKAMENEDGVTGAVKVFFKQLPKNKPEPNT 235
AK +ENEDGV AV F + LP P P +
Sbjct: 554 AKVLENEDGVAAAVDAFHRHLPPELPLPES 583
>UniRef100_Q8S1P6 Putative sterol glucosyltransferase [Oryza sativa]
Length = 637
Score = 271 bits (694), Expect = 9e-72
Identities = 123/204 (60%), Positives = 153/204 (74%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWGP +DVVG+CFL+L + Y+PP+ L +WL+ G KPIY+GFGS+P+ D K +T +I++AL
Sbjct: 402 DWGPLVDVVGYCFLNLGTKYQPPQELSQWLQQGPKPIYIGFGSMPLGDEKKVTSVILDAL 461
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
TGQRGIIS+GWG LG +E +++L++ PHDWLF C AVVHHGGAGTTAAGL A
Sbjct: 462 RETGQRGIISRGWGDLGSFSEVPVDVFILEDCPHDWLFPRCAAVVHHGGAGTTAAGLVAG 521
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
CPTTIVPFFGDQ FWGER+H +GVGP PIPI ++ L AI FMLDP+VK +ELA
Sbjct: 522 CPTTIVPFFGDQFFWGERIHAQGVGPAPIPIAELTVEALSNAIRFMLDPEVKSRTMELAI 581
Query: 208 AMENEDGVTGAVKVFFKQLPKNKP 231
A+ NEDGV AV F + LP P
Sbjct: 582 AIGNEDGVAAAVDSFHRHLPAELP 605
>UniRef100_UPI000023CA18 UPI000023CA18 UniRef100 entry
Length = 1260
Score = 216 bits (550), Expect = 4e-55
Identities = 107/218 (49%), Positives = 136/218 (62%), Gaps = 3/218 (1%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWG ID+ GF FL+LAS+Y P L +L DG P+Y+GFGS+ V DP MTE+I EA+
Sbjct: 377 DWGRHIDISGFYFLNLASSYTPDPELAAFLRDGPPPVYIGFGSIVVDDPNAMTEMIFEAV 436
Query: 88 ETTGQRGIISKGWGGLG--DLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLK 145
+ +G R ++SKGWGGLG DL +P D +Y+L NVPHDWLF H VVHHGGAGTTAAG+K
Sbjct: 437 KLSGVRALVSKGWGGLGADDLGKP-DGVYMLGNVPHDWLFEHVSCVVHHGGAGTTAAGIK 495
Query: 146 AACPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIEL 205
A PT +VPFFGDQPFWG + GP PIP + KL AI F + P+ E A +
Sbjct: 496 AGKPTLVVPFFGDQPFWGAMIARAKAGPDPIPYKQLTAEKLAEAIKFCVKPETLEQAKAM 555
Query: 206 AKAMENEDGVTGAVKVFFKQLPKNKPEPNTEPSSSSCF 243
+ + E G K F L +K PS ++ +
Sbjct: 556 GQKIREEKGTDVGGKSFHDHLEVDKLRCTLAPSRAAAW 593
>UniRef100_UPI000023594A UPI000023594A UniRef100 entry
Length = 749
Score = 214 bits (546), Expect = 1e-54
Identities = 106/218 (48%), Positives = 142/218 (64%), Gaps = 3/218 (1%)
Query: 27 ADWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEA 86
ADW +DV GF F D A +Y+PP+SL +L++G P+Y+GFGS+ ++DPK T II++A
Sbjct: 297 ADWPAHVDVCGFFFRD-APDYKPPQSLDAFLQNGPPPVYIGFGSIVIEDPKKFTAIILDA 355
Query: 87 LETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKA 146
+ + G R IIS+GW LG EP +SIY +D+ PH+WLF H AVVHHGGAGTTA GL+
Sbjct: 356 VRSLGVRAIISRGWSKLGG--EPSESIYYIDDCPHEWLFQHVCAVVHHGGAGTTACGLRN 413
Query: 147 ACPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELA 206
PT I+PFFGDQPFWG V G GP PIP + KL I F L P+++ A +A
Sbjct: 414 GRPTAIIPFFGDQPFWGNLVAVSGAGPKPIPYRDVTTTKLAEVIEFCLQPQIQHAAQTIA 473
Query: 207 KAMENEDGVTGAVKVFFKQLPKNKPEPNTEPSSSSCFS 244
M+ EDGV AV F + LP +K + P+ S+ ++
Sbjct: 474 SRMQYEDGVKTAVDSFHRNLPLDKMRCDLLPNRSAAWT 511
>UniRef100_UPI0000219F77 UPI0000219F77 UniRef100 entry
Length = 1323
Score = 211 bits (538), Expect = 1e-53
Identities = 102/200 (51%), Positives = 132/200 (66%), Gaps = 2/200 (1%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWG +ID+ GF FLDLAS+++PP+ LVK+L+DGD PIY+GFGS+ V DP TE+I EA+
Sbjct: 525 DWGDEIDISGFVFLDLASSFKPPDDLVKFLDDGDPPIYIGFGSIVVDDPDHFTEMIFEAV 584
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
+ G R ++SKGWG LG P D++Y+L+N PHDWLF A V HGGAGTTA LK
Sbjct: 585 KQAGVRALVSKGWGKLGGENVP-DNVYMLENTPHDWLFPRVSACVIHGGAGTTAISLKCG 643
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
PT +VPFFGDQ FWG + G GP P+P + KL I + L + K+ A E+A+
Sbjct: 644 KPTMVVPFFGDQHFWGSMLERCGAGPEPVPYKRLTAEKLAEGIKYCLSDEAKKAATEIAR 703
Query: 208 AMENE-DGVTGAVKVFFKQL 226
+E E DG A + F K L
Sbjct: 704 DIEQEGDGAENACRSFHKHL 723
>UniRef100_UPI000023ED09 UPI000023ED09 UniRef100 entry
Length = 1156
Score = 209 bits (532), Expect = 5e-53
Identities = 105/205 (51%), Positives = 130/205 (63%), Gaps = 2/205 (0%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWG +IDV GF FLDLAS +EPP+ L ++ GD P+Y+GFGS+ V D TE+I +A+
Sbjct: 522 DWGSEIDVAGFVFLDLASTFEPPKELEDFIAAGDTPVYIGFGSIVVDDADKFTEMIFKAV 581
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
E G R ++SKGWGGLG P +++++LDN PHDWLF KA V HGGAGTTA LK
Sbjct: 582 EMAGVRALVSKGWGGLGGDDVP-ENVFMLDNTPHDWLFPRVKACVIHGGAGTTAIALKCG 640
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
PT IVPFFGDQ FWG+ V VGP PIP KL I + L K +E A ++AK
Sbjct: 641 KPTMIVPFFGDQNFWGKMVSSADVGPEPIPYKHLDAEKLAEGIEYCLSDKAREAAEKIAK 700
Query: 208 AMENE-DGVTGAVKVFFKQLPKNKP 231
+ E DG AVK F +QL P
Sbjct: 701 RIAEEGDGAENAVKEFHRQLNLESP 725
>UniRef100_O93841 CHIP6 [Colletotrichum gloeosporioides]
Length = 914
Score = 209 bits (532), Expect = 5e-53
Identities = 96/194 (49%), Positives = 127/194 (64%)
Query: 27 ADWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEA 86
ADW I++ GF FL LA Y PP L K+LE G PIY+GFGS+ V DPK +T++I +A
Sbjct: 336 ADWDDHINITGFSFLSLADKYTPPPDLTKFLEAGPPPIYIGFGSIVVDDPKALTQLIFDA 395
Query: 87 LETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKA 146
++ G R I+SKGWGG+G ++IYL+ N PHDWLF AVVHHGGAGT+AAG+ A
Sbjct: 396 VKIAGVRAIVSKGWGGVGGGDNVPENIYLIGNCPHDWLFKRVSAVVHHGGAGTSAAGIAA 455
Query: 147 ACPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELA 206
PT +VPFFGDQPFWG+ + G GP P+P + L A+I+F L P+V+ ++A
Sbjct: 456 GRPTVVVPFFGDQPFWGQMIARAGAGPVPVPFKEMTAETLAASISFALKPEVQVAVQQMA 515
Query: 207 KAMENEDGVTGAVK 220
+ + EDG K
Sbjct: 516 ERIGAEDGAGDTAK 529
>UniRef100_UPI0000234B30 UPI0000234B30 UniRef100 entry
Length = 1139
Score = 206 bits (524), Expect = 5e-52
Identities = 98/199 (49%), Positives = 123/199 (61%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DWG IDVVGF L A +Y+PP+ L +L+ G P+Y+GFGS+ V D K +T+I+ EA+
Sbjct: 533 DWGDNIDVVGFSTLPTAQDYKPPQDLQSFLDAGPAPVYIGFGSIVVDDSKALTDIVFEAV 592
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
E G R +ISKGWG +G DSI ++D PHDWLF H VVHHGGAGTTAAGL
Sbjct: 593 EKAGVRAVISKGWGNIGANHAASDSIMMIDKCPHDWLFQHVSCVVHHGGAGTTAAGLALG 652
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
PT ++PFFGDQ FWG V G GP PIP + KL AI L + K A E+ +
Sbjct: 653 KPTIVIPFFGDQAFWGSIVSRAGAGPDPIPWKRLTAEKLAEAIEMALKDETKRKAEEIGE 712
Query: 208 AMENEDGVTGAVKVFFKQL 226
M +E G AV F++ L
Sbjct: 713 QMRSEQGARNAVCSFYRHL 731
>UniRef100_UPI000021A76A UPI000021A76A UniRef100 entry
Length = 1271
Score = 204 bits (518), Expect = 2e-51
Identities = 99/212 (46%), Positives = 126/212 (58%), Gaps = 1/212 (0%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DW P+I + GF FL+LAS+Y P L +L DG P+Y+GFGS+ V DP +T++I EA+
Sbjct: 385 DWNPEISISGFYFLNLASSYTPEPDLAAFLADGPPPVYIGFGSIVVDDPNALTKMIFEAV 444
Query: 88 ETTGQRGIISKGWGGLG-DLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKA 146
TG R ++SKGWGG+G D D +++L N PHDWLF H VVHHGGAGTTAAG+K
Sbjct: 445 TKTGVRALVSKGWGGIGADSIGIPDGVFMLGNCPHDWLFQHVSCVVHHGGAGTTAAGIKL 504
Query: 147 ACPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELA 206
PT +VPFFGDQPFWG + G GP P+P L AAI L+P A EL
Sbjct: 505 GKPTVVVPFFGDQPFWGSMIARAGAGPEPVPYKKLDAESLAAAIRKALEPATLAKAKELG 564
Query: 207 KAMENEDGVTGAVKVFFKQLPKNKPEPNTEPS 238
+ + E G K F L + + PS
Sbjct: 565 EKIREEQGCDVGGKSFHDFLDTDSLRCSLAPS 596
>UniRef100_UPI000023F6DB UPI000023F6DB UniRef100 entry
Length = 748
Score = 203 bits (517), Expect = 3e-51
Identities = 102/210 (48%), Positives = 127/210 (59%), Gaps = 1/210 (0%)
Query: 28 DWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEAL 87
DW IDV GF F D+ S YEP L K+L G P+Y+GFGS+ + +P+ +T I EA+
Sbjct: 326 DWADNIDVCGFFFRDMPS-YEPDADLKKFLSSGTTPVYIGFGSIVIDNPEELTATIREAV 384
Query: 88 ETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKAA 147
TG R I+S+GW LG + DSI+ L + PH+WLF +AVVHHGGAGTTA GL A
Sbjct: 385 RATGTRAIVSRGWSKLGGDSPSDDSIFFLGDCPHEWLFQQVRAVVHHGGAGTTACGLLNA 444
Query: 148 CPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELAK 207
PT IVPFFGDQPFWG V+ G GP PIP + L AI F L P+ + A ++A+
Sbjct: 445 KPTAIVPFFGDQPFWGHMVNAGGAGPAPIPFKSLNKDNLADAIRFCLTPEASDSARKIAQ 504
Query: 208 AMENEDGVTGAVKVFFKQLPKNKPEPNTEP 237
M E GV AV F LP NK + P
Sbjct: 505 KMSREAGVRRAVASFHANLPLNKMRHQSHP 534
>UniRef100_UPI000023EA4E UPI000023EA4E UniRef100 entry
Length = 898
Score = 201 bits (510), Expect = 2e-50
Identities = 97/192 (50%), Positives = 126/192 (65%), Gaps = 1/192 (0%)
Query: 27 ADWGPKIDVVGFCFLDLASNYEPPESLVKWLEDGDKPIYVGFGSLPVQDPKTMTEIIVEA 86
+DW +++ GF FL LAS+Y PP LV +L++G PIY+GFGS+ V +P+ +T +I EA
Sbjct: 362 SDWDSHLNITGFSFLPLASSYTPPPDLVSFLDNGSTPIYIGFGSIVVDNPQALTTMIFEA 421
Query: 87 LETTGQRGIISKGWGGLGDLTEPKDSIYLLDNVPHDWLFLHCKAVVHHGGAGTTAAGLKA 146
++ G R I+SKGWGG+G P DS+YL+ N PHDWLF AVVHHGGAGTTAAG+ A
Sbjct: 422 IKIAGVRAILSKGWGGVGTGDVP-DSVYLIGNCPHDWLFQRVAAVVHHGGAGTTAAGIAA 480
Query: 147 ACPTTIVPFFGDQPFWGERVHDRGVGPPPIPIDVFSLPKLIAAINFMLDPKVKEHAIELA 206
PT IVPFFGDQPFWG+ + G GP +P + L +I F L P V A ++A
Sbjct: 481 GRPTVIVPFFGDQPFWGQMMARAGAGPVAVPYKDLTAEILAESITFALQPGVMAVAKDMA 540
Query: 207 KAMENEDGVTGA 218
+ EDG GA
Sbjct: 541 LQIGEEDGSGGA 552
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.142 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,041,483
Number of Sequences: 2790947
Number of extensions: 21454426
Number of successful extensions: 47956
Number of sequences better than 10.0: 410
Number of HSP's better than 10.0 without gapping: 240
Number of HSP's successfully gapped in prelim test: 170
Number of HSP's that attempted gapping in prelim test: 47542
Number of HSP's gapped (non-prelim): 422
length of query: 253
length of database: 848,049,833
effective HSP length: 124
effective length of query: 129
effective length of database: 501,972,405
effective search space: 64754440245
effective search space used: 64754440245
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC126019.1


