

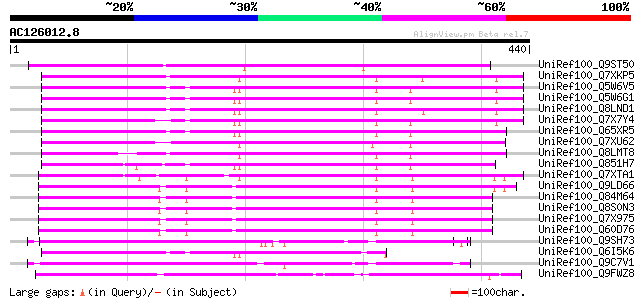
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126012.8 + phase: 0
(440 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ST50 Transposase related protein [Zea mays] 234 4e-60
UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa] 226 1e-57
UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa] 222 1e-56
UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa] 222 1e-56
UniRef100_Q8LND1 Putative transposable element [Oryza sativa] 221 3e-56
UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa] 218 3e-55
UniRef100_Q65XR5 Putative polyprotein [Oryza sativa] 216 8e-55
UniRef100_Q7XU62 OSJNBb0091E11.13 protein [Oryza sativa] 203 9e-51
UniRef100_Q8LMT8 Putative retroelement [Oryza sativa] 201 3e-50
UniRef100_Q851H7 Putative mutator-like transposase [Oryza sativa] 193 7e-48
UniRef100_Q7XTA1 OSJNBa0008A08.9 protein [Oryza sativa] 184 3e-45
UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein ... 175 3e-42
UniRef100_Q84M64 Putative mutator-like transposase [Oryza sativa] 170 6e-41
UniRef100_Q8S0N3 Putative mutator-like transposase [Oryza sativa] 169 1e-40
UniRef100_Q7X975 Putative Mutator protein [Oryza sativa] 169 2e-40
UniRef100_Q60D76 Putative polyprotein [Oryza sativa] 168 2e-40
UniRef100_Q9SH73 F22C12.1 [Arabidopsis thaliana] 167 4e-40
UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sa... 165 3e-39
UniRef100_Q9C7V1 Hypothetical protein F15H21.15 [Arabidopsis tha... 163 8e-39
UniRef100_Q9FWZ8 F11O6.3 protein [Arabidopsis thaliana] 163 1e-38
>UniRef100_Q9ST50 Transposase related protein [Zea mays]
Length = 863
Score = 234 bits (597), Expect = 4e-60
Identities = 142/399 (35%), Positives = 210/399 (52%), Gaps = 10/399 (2%)
Query: 17 KNADWREWRAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKT 76
K +WR R F R FW F C+ F++C+PV+ +DGT+L KYKGTLL+A++ D +N
Sbjct: 464 KPNEWRNGREIFFRAFWCFSQCVEAFRHCRPVLSIDGTFLLGKYKGTLLVAISCDADNTL 523
Query: 77 IPIAFALVEGETKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGS 135
+P+AFALVE E +E W++FL+ +R HV G V ++SDRH+ I +A + G+
Sbjct: 524 VPLAFALVERENRESWAWFLRLVRIHVVGPGREVGVISDRHQGILNAVQEQIPGY--APM 581
Query: 136 AHVYCIRHIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEA-NQRALDWVDNI 194
H +C RH+ +N +R + + + L V F +++A N +W+ +
Sbjct: 582 HHRWCTRHLAENLLRKDRSKANFPLFEEICQQLEVSFFEDKLKELKDATNAEGKNWIAGL 641
Query: 195 PKE--KWTQAYDEGR-RWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRA 251
+E KWT AYDEG R+ TSN+ E +NSV KG R +PV IV T+YRL F DR
Sbjct: 642 LREPQKWTSAYDEGGWRFEFQTSNMAELFNSVLKGIRGMPVNAIVTFTFYRLVAWFNDRH 701
Query: 252 QKAFARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRE---TVNYREGRPM 308
+A A G ++ + + ++N H V+ FD + + VR T + E RP
Sbjct: 702 AQAKAMQTRGLRWAPKPTAHLNNAKERANRHEVQCFDEDLGKYEVRTMGGTTSDGEVRPS 761
Query: 309 GTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFK 368
T V L A C CGK + H PCSH +AA + ++IP F ES+ ++ F+
Sbjct: 762 RTHVVLLDAFSCGCGKPRQYHFPCSHYVAAARHRNFAFESMIPSEFSVESLVRTWSPRFE 821
Query: 369 VVHDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEM 407
D+S W Y GP+ +P R K G +R M
Sbjct: 822 PFLDESQWPTYTGPIYIADPAHRWDKCGTRKRSRCNMVM 860
>UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa]
Length = 1495
Score = 226 bits (575), Expect = 1e-57
Identities = 138/430 (32%), Positives = 212/430 (49%), Gaps = 24/430 (5%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+P++ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 446 FMRAFWCFGPSIEAFKHCRPILAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 505
Query: 88 TKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH I +A P G H +C+RH
Sbjct: 506 NSRDWCWFIDLVRRVVVGPHWEVCIISDRHAGIMNAMTTPVPGLPPV--HHRWCMRHFSA 563
Query: 147 NFMRTIKD-GDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDN--IPKEKWTQAY 203
NF + D K+++ V Q + W+++ + K+KW++AY
Sbjct: 564 NFHKAGADKHQTKELLRICQIDEKWIFERDVEAVRQRIPEGPRKWLEDELLDKDKWSRAY 623
Query: 204 D-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
D GRRWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV G
Sbjct: 624 DRNGRRWGYMTTNMAEQFNSVLVGVRKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQ 683
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDLRAGW 319
+S + ++ K+N H FD+++ T+ V E R G G FKV+
Sbjct: 684 RWSTKIDSKMKVQKNKANEHTARCFDQQKKTYEVTERGGITRGGVRFGARAFKVEGEGNS 743
Query: 320 CDCGKFQALHLPCSHVIAACSSFRHDYTT--LIPHVFKNESVYSIYNTAFKVVHDKSYWL 377
C C + H+PCSH+I + D + +P+ F + +V + + + F+ D + W
Sbjct: 744 CSCQRPLLYHMPCSHLIHVYLIYAIDEESPNRMPYQFSSRAVVNTWASRFEPYLDPTQWP 803
Query: 378 PYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEV------------VERTPTPRRCGLCR 425
PYDG +PN++ +G+ S R + EMD + + P RC LC
Sbjct: 804 PYDGEEFVADPNLKIKTRGKRRSKRFKNEMDSGLGGSGRKPPSCVQLYAAPVQNRCSLCH 863
Query: 426 HTGHIRRNCP 435
GH + CP
Sbjct: 864 QEGHKKTKCP 873
>UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa]
Length = 1525
Score = 222 bits (566), Expect = 1e-56
Identities = 137/433 (31%), Positives = 214/433 (48%), Gaps = 30/433 (6%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 521 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 580
Query: 88 TKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH I +A P G + H +C+RH
Sbjct: 581 NSRDWCWFIDLVRRVVVGPHREVCIISDRHAGIMNAMMTPVPGLPSV--HHRWCMRHFSA 638
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALD----WVDN--IPKEKWT 200
NF K G K + + + ++ QR + W+++ + K+KW+
Sbjct: 639 NFH---KAGADKHQTKELLRICQIDEKWIFERDVEALRQRIPEGPRKWLEDELLDKDKWS 695
Query: 201 QAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
+AYD GRRWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV
Sbjct: 696 RAYDRNGRRWGYMTTNMAEQFNSVLVGVRKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQ 755
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDLR 316
G +S + ++ K+N H FD+++ T+ V E R G G FKV+
Sbjct: 756 LGQRWSTKVDSKMKVQESKANKHTARCFDKQKKTYEVTERGGITRGGVRFGARAFKVEGE 815
Query: 317 AGWCDCGKFQALHLPCSHVIAA--CSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKS 374
C C + H+PCSH+I + + +P+ F + +V + + + F+ D +
Sbjct: 816 GNSCSCQRPLLYHMPCSHLIHVYLIHAIDKESPNRMPYQFSSRAVVNTWASRFEPYLDPT 875
Query: 375 YWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEV------------VERTPTPRRCG 422
W Y+G +PN++ +G+ S R + EMD + ++ P RC
Sbjct: 876 QWPSYNGEEFVADPNLKIKTRGKRRSKRFKNEMDSGLGGSGRKPPSCVQLDAAPVQNRCS 935
Query: 423 LCRHTGHIRRNCP 435
LC GH + CP
Sbjct: 936 LCHQEGHKKTKCP 948
>UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa]
Length = 1567
Score = 222 bits (566), Expect = 1e-56
Identities = 137/433 (31%), Positives = 214/433 (48%), Gaps = 30/433 (6%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 536 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 595
Query: 88 TKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH I +A P G + H +C+RH
Sbjct: 596 NSRDWCWFIDLVRRVVVGPHREVCIISDRHAGIMNAMMTPVPGLPSV--HHRWCMRHFSA 653
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALD----WVDN--IPKEKWT 200
NF K G K + + + ++ QR + W+++ + K+KW+
Sbjct: 654 NFH---KAGADKHQTKELLRICQIDEKWIFERDVEALRQRIPEGPRKWLEDELLDKDKWS 710
Query: 201 QAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
+AYD GRRWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV
Sbjct: 711 RAYDRNGRRWGYMTTNMAEQFNSVLVGVRKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQ 770
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDLR 316
G +S + ++ K+N H FD+++ T+ V E R G G FKV+
Sbjct: 771 LGQRWSTKVDSKMKVQESKANKHTARCFDKQKKTYEVTERGGITRGGVRFGARAFKVEGE 830
Query: 317 AGWCDCGKFQALHLPCSHVIAA--CSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKS 374
C C + H+PCSH+I + + +P+ F + +V + + + F+ D +
Sbjct: 831 GNSCSCQRPLLYHMPCSHLIHVYLIHAIDKESPNRMPYQFSSRAVVNTWASRFEPYLDPT 890
Query: 375 YWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEV------------VERTPTPRRCG 422
W Y+G +PN++ +G+ S R + EMD + ++ P RC
Sbjct: 891 QWPSYNGEEFVADPNLKIKTRGKRRSKRFKNEMDSGLGGSGRKPPSCVQLDAAPVQNRCS 950
Query: 423 LCRHTGHIRRNCP 435
LC GH + CP
Sbjct: 951 LCHQEGHKKTKCP 963
>UniRef100_Q8LND1 Putative transposable element [Oryza sativa]
Length = 1592
Score = 221 bits (563), Expect = 3e-56
Identities = 137/433 (31%), Positives = 214/433 (48%), Gaps = 30/433 (6%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 521 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 580
Query: 88 TKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH I +A P G + H +C+RH
Sbjct: 581 NSRDWCWFIDLVRRVVVGPHREVCIISDRHAGIMNAMMTPVPGLPSV--HHRWCMRHFSA 638
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALD----WVDN--IPKEKWT 200
NF K G K + + + ++ QR + W+++ + K+KW+
Sbjct: 639 NFH---KAGADKHQTKELLRICQIDEKWIFERDVEALRQRIPEGPRKWLEDELLDKDKWS 695
Query: 201 QAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
+AYD GRRWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV
Sbjct: 696 RAYDRNGRRWGYMTTNMAEQFNSVLVGVRKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQ 755
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDLR 316
G +S + ++ K+N H FD+++ T+ V E R G G FKV+
Sbjct: 756 LGQRWSTKVDSKMKVQESKANKHTARCFDKQKKTYEVTERGGITRGGVRFGARAFKVEGE 815
Query: 317 AGWCDCGKFQALHLPCSHVIAACSSFRHDYTTL--IPHVFKNESVYSIYNTAFKVVHDKS 374
C C + H+PCSH+I D +L +P+ F + ++ + + + F+ D +
Sbjct: 816 GNSCSCQRPLLYHMPCSHLIHVYLIHAIDKESLNRMPYQFSSRAIVNTWASRFEPYLDPT 875
Query: 375 YWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEV------------VERTPTPRRCG 422
W Y+G +PN++ +G+ R + EMD + ++ P RC
Sbjct: 876 QWPSYNGEEFVADPNLKIKTRGKRRLKRFKNEMDSGLGGSGRKPPSCVQLDAAPVQNRCS 935
Query: 423 LCRHTGHIRRNCP 435
LC GH + CP
Sbjct: 936 LCHQEGHKKTKCP 948
>UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa]
Length = 1575
Score = 218 bits (555), Expect = 3e-55
Identities = 135/433 (31%), Positives = 210/433 (48%), Gaps = 41/433 (9%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 489 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 548
Query: 88 TKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH + S H +C+RH
Sbjct: 549 NSRDWCWFIDLVRRVVVGPHREVCIISDRHAGLPSVH-------------HRWCMRHFSA 595
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALD----WVDN--IPKEKWT 200
NF K G K + + + ++ QR + W+++ + K+KW+
Sbjct: 596 NFH---KAGADKHQTKELLRICQIDEKWIFERDVEALRQRIPEGPRKWLEDELLDKDKWS 652
Query: 201 QAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
+AYD GRRWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV
Sbjct: 653 RAYDRNGRRWGYMTTNMAEQFNSVLVGVRKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQ 712
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDLR 316
G +S + ++ K+N H FD+++ T+ V E R G G FKV+
Sbjct: 713 LGQRWSTKVDSKMKVQESKANKHTARCFDKQKKTYEVTERGGITRGGVRFGARAFKVEGE 772
Query: 317 AGWCDCGKFQALHLPCSHVIAA--CSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKS 374
C C + H+PCSH+I + + +P+ F + +V + + + F+ D +
Sbjct: 773 GNSCSCQRPLLYHMPCSHLIHVYLIHAIDKESPNRMPYQFSSRAVVNTWASRFEPYLDPT 832
Query: 375 YWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEV------------VERTPTPRRCG 422
W YDG +PN++ +G+ S R + EMD + ++ P RC
Sbjct: 833 QWPSYDGEEFVADPNLKIKTRGKRRSKRFKNEMDSGLGGSGRKPPSCVQLDAAPVQNRCS 892
Query: 423 LCRHTGHIRRNCP 435
LC GH + CP
Sbjct: 893 LCHQEGHKKTKCP 905
>UniRef100_Q65XR5 Putative polyprotein [Oryza sativa]
Length = 1378
Score = 216 bits (551), Expect = 8e-55
Identities = 132/407 (32%), Positives = 205/407 (49%), Gaps = 18/407 (4%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 331 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 390
Query: 88 TKEGWSFFLKNLRR-HVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH I +A P G H +C+RH
Sbjct: 391 NSRDWCWFIDLVRRVMVGPHREVCIISDRHAGIMNAMTTPVPGLPPV--HHRWCMRHFSA 448
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALD----WVDN--IPKEKWT 200
NF K G K + + + ++ QR + W+++ + K+KW+
Sbjct: 449 NFH---KAGADKHQTKELLRICQIDEKWIFERDVEALRQRIPEGPRKWLEDELVDKDKWS 505
Query: 201 QAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
+AYD GRRWG+MT+N+ E +NSV G R LP T IV T+ + F +R +A RV
Sbjct: 506 RAYDRNGRRWGYMTTNMAEQFNSVLVGVRKLPATAIVSFTFMKCNDYFVNRHDEALKRVQ 565
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDLR 316
G +S + ++ K+N H FD+++ T+ V E R G G FKV++
Sbjct: 566 LGQRWSTKVDSKMKVQKSKANKHTARCFDKQKKTYEVTERGGVTRGGVRFGARAFKVEVE 625
Query: 317 AGWCDCGKFQALHLPCSHVIAA--CSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKS 374
C C + H+PCSH+I + + +P+ F + +V + + + F+ D +
Sbjct: 626 GNSCSCQRPLLYHMPCSHLIHVYLIHAIDEESPNRMPYQFSSRAVVNTWASRFEPYLDPT 685
Query: 375 YWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRC 421
WLPYDG +PN++ +G+ S R + EMD + P C
Sbjct: 686 QWLPYDGEEFVADPNLKIKTRGKRRSKRFKNEMDSGLGGSGRKPPSC 732
>UniRef100_Q7XU62 OSJNBb0091E11.13 protein [Oryza sativa]
Length = 1107
Score = 203 bits (516), Expect = 9e-51
Identities = 122/403 (30%), Positives = 200/403 (49%), Gaps = 23/403 (5%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 536 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 595
Query: 88 TKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH + S H +C+RH
Sbjct: 596 NSRDWCWFIDLVRRVVVGPHREVCIISDRHAGLPSVH-------------HRWCMRHFSA 642
Query: 147 NFMRTIKD-GDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDN--IPKEKWTQAY 203
NF + D K+++ + Q + + W+++ + K+KW++AY
Sbjct: 643 NFHKAGADKHQTKELLRICQIDEKWIFERDVEALRQHIPEGSRKWLEDELLDKDKWSRAY 702
Query: 204 D-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
D GRRWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV G
Sbjct: 703 DRNGRRWGYMTTNMAEQFNSVLVGVRKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQ 762
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGR---PMGTFKVDLRAGW 319
+S + ++ K+N H FD+++ T+ V E G FKV+
Sbjct: 763 RWSTKVDSKMKVQKSKANKHTARCFDKQKKTYEVTERGGITHGGVRFGARAFKVEGEGNS 822
Query: 320 CDCGKFQALHLPCSHVIAA--CSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWL 377
C C + H+PCSH+I + + +P+ F + +V + + + F+ D + W
Sbjct: 823 CSCQRPLLYHMPCSHLIHVYLIHAIDEESPNRMPYQFSSRAVVNTWASRFEPYLDPTQWP 882
Query: 378 PYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRR 420
YDG +PN++ +G+ S R + E + ++++ T R+
Sbjct: 883 SYDGEEFVADPNLKIKTRGKRRSKRFKNEEETQILKCTLRARK 925
>UniRef100_Q8LMT8 Putative retroelement [Oryza sativa]
Length = 1460
Score = 201 bits (512), Expect = 3e-50
Identities = 125/403 (31%), Positives = 194/403 (48%), Gaps = 26/403 (6%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 594 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYGGALMTALSADAEDQLVPLAFALVEKE 653
Query: 88 TKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQN 147
W VC++SDRH I +A P G H +C+RH N
Sbjct: 654 NSRDW---------------EVCIISDRHAGIMNAMTTPIPGLPPV--HHRWCMRHFSAN 696
Query: 148 FMRTIKDG-DLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDN--IPKEKWTQAYD 204
F + D K+++ + Q + W+++ + K+KW++AYD
Sbjct: 697 FHKAGADKYQTKELLRICQIDEKWIFERDVEALRQRIPEGPRKWLEDELLDKDKWSRAYD 756
Query: 205 -EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDL 263
GRRWG+MT+N+ E +NSV G LPVT IV T+ + F +R +A RV G
Sbjct: 757 RNGRRWGYMTTNMAEQFNSVLVGVHKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQR 816
Query: 264 FSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDLRAGWC 320
+S + ++ K+N H FD+++ T+ V E R G G FKV+ C
Sbjct: 817 WSTKVDSKMKVQKSKANKHTARCFDKQKKTYEVTERGGITRGGVRFGARAFKVEGEGNSC 876
Query: 321 DCGKFQALHLPCSHVIAA--CSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLP 378
C + H+PCSH+I + + +P+ F + +V + + + F+ D + W P
Sbjct: 877 SCQRPLLYHMPCSHLIHVYMIHAIDEESPNRMPYQFSSRAVVNTWASRFEPYLDPTQWPP 936
Query: 379 YDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPTPRRC 421
YDG +PN++ +G+ S R + EMD + P C
Sbjct: 937 YDGEEFVADPNLKIKTRGKRRSKRFKNEMDSGLGGSGRKPPSC 979
>UniRef100_Q851H7 Putative mutator-like transposase [Oryza sativa]
Length = 1360
Score = 193 bits (491), Expect = 7e-48
Identities = 123/399 (30%), Positives = 198/399 (48%), Gaps = 20/399 (5%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY + A++ D ++ +P+AFA VE E
Sbjct: 514 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTGKYSDAFMTALSADAEDQLVPLAFASVEKE 573
Query: 88 TKEGWSFFLKNLRRHVTKG--ISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
W +F+ +L R V G VC++SDRH I +A P G + H +C+RH
Sbjct: 574 NSRDWCWFI-DLVRQVVVGPHREVCIISDRHAGIMNAMRAPVPG--LSPIHHRWCMRHFS 630
Query: 146 QNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALD----WVDN--IPKEKW 199
NF K G K + + + ++ QR + W+++ + K+KW
Sbjct: 631 ANFH---KAGADKHQTEELLRICKIDKKWIFERDVEALRQRIPEGPRKWLEDELLDKDKW 687
Query: 200 TQAYDEGR-RWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARV 258
++AYD RWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV
Sbjct: 688 SRAYDRNSCRWGYMTTNMAEQFNSVLVGVRKLPVTAIVAFTFMKCNHYFVNRQNEAVKRV 747
Query: 259 GSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFKVDL 315
SG+ +S + ++ K+N H FD+++ T+ V E R G G FKV+
Sbjct: 748 QSGERWSAKVASKMKVQKSKANKHTSRCFDKQKKTYEVTEKGGITRSGVRFGARAFKVEG 807
Query: 316 RAGWCDCGKFQALHLPCSHVIAA--CSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDK 373
C C + H+PCSH++ + + I + F + +V + + + F+ D
Sbjct: 808 EGNSCSCQRPLLYHMPCSHLVHVYLIHAIDEESPNRISYQFSSRAVVNTWASRFEPYLDP 867
Query: 374 SYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDEEVV 412
+ W PYDG +P+++ +G+ R + E +V
Sbjct: 868 TQWPPYDGEEFVADPDLKIKTRGKRRLKRFKNEWTRVLV 906
>UniRef100_Q7XTA1 OSJNBa0008A08.9 protein [Oryza sativa]
Length = 1560
Score = 184 bits (468), Expect = 3e-45
Identities = 136/439 (30%), Positives = 204/439 (45%), Gaps = 34/439 (7%)
Query: 25 RAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
R F R +W F I FK+ +PV+ +DGT+L KY+GTLL+A+ D +P+AFALV
Sbjct: 365 RRVFGRAYWIFGQSIEAFKHLRPVLAIDGTFLTGKYQGTLLMAIGVDAWLHLVPLAFALV 424
Query: 85 EGETKEGWSFFLKNLRRHVTKGIS--VCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIR 142
E E W +F+ N+ RH G + VC++SDRH I ++ N N H +C+R
Sbjct: 425 EKENTSNWEWFI-NMLRHKLIGPNREVCIISDRHPGILNSIN--HNMPHHLTIHHRWCMR 481
Query: 143 HIKQNF----MRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDN--IPK 196
H NF T + DL+ + AL + GV+ E R W+++ K
Sbjct: 482 HFCANFYTAGATTDQMKDLERICQINEKALFLDEIKRLLGVVGE---RPKKWLEDHMALK 538
Query: 197 EKWTQAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAF 255
KW +A+D GRR MTSN+ ES+N+V +G R LPVT IV T+ + F DR ++A
Sbjct: 539 VKWARAFDTNGRRHSIMTSNMAESFNNVLRGIRKLPVTAIVAYTFSKCNSWFVDRHKEAT 598
Query: 256 ARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFK 312
+ SG + ++ +++ ++ FD + V E G G +
Sbjct: 599 VDILSGKKWPTKVKDMLEEQQRRTLGQRTACFDFPSMKYEVSEQGGVTAAGVQWGGRHYV 658
Query: 313 VDLRAGWCDCGKFQALHLPCSHVIAACS--SFRHDYTTLIPHVFKNESVYSIYNTAFKVV 370
V R C C Q HLPCSH+I C + + + N +V Y+ F+
Sbjct: 659 VVARDNTCSCQFPQLHHLPCSHMITVCKLRGLDVEVAPRMCYEASNRAVQDTYSPRFEPY 718
Query: 371 HDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDE----EVVERTPTP-------- 418
D S W YDG + +++ +GR + R + +MD+ R+ P
Sbjct: 719 LDPSQWPSYDGEFFVSDLSLKNNTRGRRRTRRFKNDMDKVYKGSANRRSQKPNNTNSEPV 778
Query: 419 --RRCGLCRHTGHIRRNCP 435
RC LC GH + CP
Sbjct: 779 MQNRCSLCHELGHKKTRCP 797
>UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein [Oryza sativa]
Length = 1230
Score = 175 bits (443), Expect = 3e-42
Identities = 131/433 (30%), Positives = 201/433 (46%), Gaps = 34/433 (7%)
Query: 25 RAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
R F R +W F I FK+ +PV+ +DGT+L KY+GTLL A+ D +P+AFALV
Sbjct: 74 RRVFGRAYWIFGQSIEAFKHLRPVLAIDGTFLTGKYQGTLLTAIGVDAGLHLVPLAFALV 133
Query: 85 EGETKEGWSFFLKNLR-RHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCI 141
E E W +F+ LR + + VC++SDRH I ++ P + H +C+
Sbjct: 134 EKENTSNWEWFINMLRNKLIGPNREVCIISDRHPGILNSIIHIMPHH----LTIHHRWCM 189
Query: 142 RHIKQNF----MRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP-K 196
RH NF T + DL+ + AL + GV+ E ++ L+ D++P K
Sbjct: 190 RHFCANFYTAGATTDQMKDLERICQINEKALFLDEIKRLMGVVGERPKKWLE--DHMPLK 247
Query: 197 EKWTQAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAF 255
KW +A+D GRR MTSN+ ES+N+V +G R LPVT IV T+ + F DR ++A
Sbjct: 248 VKWARAFDTNGRRHSIMTSNMAESFNNVLRGIRKLPVTAIVAYTFSKCNSWFVDRHKEAT 307
Query: 256 ARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFK 312
+ G + ++ +++ ++ FD + V E G G +
Sbjct: 308 VDILCGKKWPTKVKDMLEEQQRRTLGQRAACFDFPSMKYEVSEQGGVTAAGVQWGGRHYV 367
Query: 313 VDLRAGWCDCGKFQALHLPCSHVIAACS--SFRHDYTTLIPHVFKNESVYSIYNTAFKVV 370
V R C C Q HLPCSH+I C + + + N++V Y+ F+
Sbjct: 368 VVARDNTCSCQFPQLHHLPCSHMITVCKLRGLDVEVAPRMCYEASNKAVQDSYSPRFEPY 427
Query: 371 HDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDE----EVVERTPTP-------- 418
D S W YDG + +++ +GR + R + +MD+ R+ P
Sbjct: 428 LDPSQWPSYDGEFFVPDLSLKNNTRGRRRTRRFKNDMDKAYKGSANRRSQKPNNTNSEPV 487
Query: 419 --RRCGLCRHTGH 429
RC LC GH
Sbjct: 488 MQNRCSLCHELGH 500
>UniRef100_Q84M64 Putative mutator-like transposase [Oryza sativa]
Length = 1204
Score = 170 bits (431), Expect = 6e-41
Identities = 123/399 (30%), Positives = 192/399 (47%), Gaps = 20/399 (5%)
Query: 25 RAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
R F R +W F I FK+ +PV+ +DGT+L KY+GTLL A+ D +P+AFALV
Sbjct: 362 RRVFGRAYWIFGQSIEAFKHLRPVLAIDGTFLTGKYQGTLLTAIGVDAGLHLVPLAFALV 421
Query: 85 EGETKEGWSFFLKNLR-RHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCI 141
E E W +F+ LR + + VC++SDRH I ++ P + H +C+
Sbjct: 422 EKENTSNWEWFINMLRNKLIGPNREVCIISDRHPGILNSIIHIMPHH----LTIHHRWCM 477
Query: 142 RHIKQNF----MRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP-K 196
RH NF T + DL+ + AL + GV+ E ++ L+ D++P K
Sbjct: 478 RHFCANFYTAGATTDQMKDLERICQINEKALFLDEIKRLMGVVGERPKKWLE--DHMPLK 535
Query: 197 EKWTQAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAF 255
KW +A+D GRR MTSN+ ES+N+V +G R LPVT IV T+ + F DR ++A
Sbjct: 536 VKWARAFDTNGRRHSIMTSNMAESFNNVLRGIRKLPVTAIVAYTFSKCNSWFVDRHKEAT 595
Query: 256 ARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFK 312
+ G + ++ +++ ++ FD + V E G G +
Sbjct: 596 VDILCGKKWPTKVKDILEEQQRRTLGQRAACFDFPSMKYEVSEQGGVTAAGVQWGGRHYV 655
Query: 313 VDLRAGWCDCGKFQALHLPCSHVIAACS--SFRHDYTTLIPHVFKNESVYSIYNTAFKVV 370
V R C C Q HLPCSH+I C + + + N++V Y+ F+
Sbjct: 656 VVARDNTCSCQFPQLHHLPCSHMITVCKLRGLDVEVAPRMCYEASNKAVQDSYSPRFEPY 715
Query: 371 HDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDE 409
D S W YDG + +++ +GR + R + +MD+
Sbjct: 716 LDPSQWPSYDGEFFVPDLSLKNNTRGRRRTRRFKNDMDK 754
>UniRef100_Q8S0N3 Putative mutator-like transposase [Oryza sativa]
Length = 1556
Score = 169 bits (428), Expect = 1e-40
Identities = 123/399 (30%), Positives = 192/399 (47%), Gaps = 20/399 (5%)
Query: 25 RAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
R F R +W F I FK+ +PV+ +DGT+L KY+GTLL A+ D +P+AFALV
Sbjct: 493 RRVFGRAYWIFGQSIEAFKHLRPVLAIDGTFLTGKYQGTLLTAIGVDAGLHLVPLAFALV 552
Query: 85 EGETKEGWSFFLKNLR-RHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCI 141
E E W +F+ LR + + VC++SDRH I ++ P + H +C+
Sbjct: 553 EKENTSNWEWFINMLRNKLIGPNREVCIISDRHPGILNSIIHIMPHH----LTIHHRWCM 608
Query: 142 RHIKQNF----MRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP-K 196
RH NF T + DL+ + AL + GV+ E ++ L+ D++P K
Sbjct: 609 RHFCANFYTAGATTDQMKDLERICQINEKALFLDEIKRLMGVVGERPKKWLE--DHMPLK 666
Query: 197 EKWTQAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAF 255
KW +A+D GRR MTSN+ ES+N+V +G R LPVT IV T+ + F DR ++A
Sbjct: 667 VKWARAFDTNGRRHSIMTSNMDESFNNVLRGIRKLPVTAIVAYTFSKCNSWFVDRHKEAT 726
Query: 256 ARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFK 312
+ G + ++ +++ ++ FD + V E G G +
Sbjct: 727 VDILCGKKWPTKVKDILEEQQRRTLGQRAACFDFPSMKYEVSEQGGVTAAGVQWGGRHYV 786
Query: 313 VDLRAGWCDCGKFQALHLPCSHVIAACS--SFRHDYTTLIPHVFKNESVYSIYNTAFKVV 370
V R C C Q HLPCSH+I C + + + N++V Y+ F+
Sbjct: 787 VVARDNTCSCQFPQLHHLPCSHMITVCKLRGLDVEVAPRMCYEASNKAVQDSYSPRFEPY 846
Query: 371 HDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDE 409
D S W YDG + +++ +GR + R + +MD+
Sbjct: 847 LDPSQWPSYDGEFFVPDLSLKNNTRGRRRTRRFKNDMDK 885
>UniRef100_Q7X975 Putative Mutator protein [Oryza sativa]
Length = 1456
Score = 169 bits (427), Expect = 2e-40
Identities = 122/399 (30%), Positives = 191/399 (47%), Gaps = 20/399 (5%)
Query: 25 RAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
R F R +W F I FK+ +PV+ +DGT+L KY+GTLL A+ D +P+AFALV
Sbjct: 424 RRVFGRAYWIFGQSIEAFKHLRPVLAIDGTFLTGKYQGTLLTAIGVDAGLHLVPLAFALV 483
Query: 85 EGETKEGWSFFLKNLR-RHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCI 141
E E W +F+ LR + + VC++SDRH I ++ P + H +C+
Sbjct: 484 EKENTSNWEWFINMLRNKLIGPNREVCIISDRHPGILNSIIHIMPHH----LTIHHRWCM 539
Query: 142 RHIKQNF----MRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP-K 196
RH NF T + DL+ + A + GV+ E ++ L+ D++P K
Sbjct: 540 RHFCANFYTAGATTDQMKDLERICQINEKAFFLDEIKRLMGVVGERPKKWLE--DHMPLK 597
Query: 197 EKWTQAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAF 255
KW +A+D GRR MTSN+ ES+N+V +G R LPVT IV T+ + F DR ++A
Sbjct: 598 VKWARAFDTNGRRHSIMTSNMAESFNNVLRGIRKLPVTAIVAYTFSKCNSWFVDRHKEAT 657
Query: 256 ARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFK 312
+ SG + ++ +++ ++ FD + V E G G +
Sbjct: 658 VDILSGKKWPTKVKDMLEEQQRRTLGQRAACFDFPSMKYEVSEQGGVTAAGVQWGGRHYV 717
Query: 313 VDLRAGWCDCGKFQALHLPCSHVIAACS--SFRHDYTTLIPHVFKNESVYSIYNTAFKVV 370
V R C C Q HLPC H+I C + + + N++V Y+ F+
Sbjct: 718 VVARDNTCSCQFPQLHHLPCPHMITVCKLRGLDVEVAPRMCYEASNKAVQDSYSPRFEPY 777
Query: 371 HDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDE 409
D S W YDG + +++ +GR + R + +MD+
Sbjct: 778 LDPSQWPSYDGEFFVPDLSLKNNTRGRRRTRRFKNDMDK 816
>UniRef100_Q60D76 Putative polyprotein [Oryza sativa]
Length = 1754
Score = 168 bits (426), Expect = 2e-40
Identities = 122/399 (30%), Positives = 191/399 (47%), Gaps = 20/399 (5%)
Query: 25 RAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALV 84
R F R +W F I FK+ +PV+ +DGT+L KY+GTLL A+ D +P+ FALV
Sbjct: 485 RRVFGRAYWIFGQSIEAFKHLRPVLAIDGTFLTGKYQGTLLTAIGVDAGLHLVPLVFALV 544
Query: 85 EGETKEGWSFFLKNLR-RHVTKGISVCMVSDRHESIKSAFND--PRNGWQATGSAHVYCI 141
E E W +F+ LR + + VC++SDRH I ++ P + H +C+
Sbjct: 545 EKENTSNWEWFINMLRNKLIGPNREVCIISDRHPGILNSIIHIMPHH----LTIHHRWCM 600
Query: 142 RHIKQNF----MRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP-K 196
RH NF T + DL+ + AL + GV+ E ++ L+ D++P K
Sbjct: 601 RHFCANFYTAGATTDQMKDLERICQINEKALFLDEIKRLMGVVGERPKKWLE--DHMPLK 658
Query: 197 EKWTQAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAF 255
KW +A+D GRR MTSN+ ES+N+V +G R LPVT IV T+ + F DR ++A
Sbjct: 659 VKWARAFDTNGRRHSIMTSNMAESFNNVLRGIRKLPVTAIVAYTFSKCNSWFVDRHKEAT 718
Query: 256 ARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNY-REGRPMG--TFK 312
+ G + ++ +++ ++ FD + V E G G +
Sbjct: 719 VDILCGKKWPTKVKDMLEEQQRRTLGQRAACFDFPSIKYEVSEQGGVTAAGVQWGGRHYI 778
Query: 313 VDLRAGWCDCGKFQALHLPCSHVIAACS--SFRHDYTTLIPHVFKNESVYSIYNTAFKVV 370
V R C C Q HLPCSH+I C + + + N++V Y+ F+
Sbjct: 779 VVARDNTCSCQFPQLHHLPCSHMITVCKLRGLDVEVAPRMCYEASNKAVQDSYSPRFEPY 838
Query: 371 HDKSYWLPYDGPVICHNPNMRRLKKGRPNSTRIRTEMDE 409
D S W YDG + +++ +GR + R + +MD+
Sbjct: 839 LDPSQWPSYDGEFFVPDLSLKNNTRGRRRTRRFKNDMDK 877
>UniRef100_Q9SH73 F22C12.1 [Arabidopsis thaliana]
Length = 3290
Score = 167 bits (424), Expect = 4e-40
Identities = 106/377 (28%), Positives = 171/377 (45%), Gaps = 25/377 (6%)
Query: 26 AQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVE 85
A F +FWAF I GF++C+P++ VD L KY L++A + ++ P+AFA
Sbjct: 1430 ASFRSVFWAFPQSIEGFQHCRPLIVVDTKDLKGKYPMKLMIASGVEADDCYFPLAFAFTT 1489
Query: 86 GETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
+ + W +FL +R VT+ +C++S H I N+P + WQ + H++C+ I
Sbjct: 1490 EVSSDTWRWFLSGIREKVTQRKDICLISRPHPDILDVINEPGSQWQEPWAYHMFCLDDIC 1549
Query: 146 QNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDE 205
F +D LK++V G F+ Y I++ N A W+D P+ +W QA+D
Sbjct: 1550 TQFHYVFQDDYLKNLVYEAGSTSEKEEFDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDS 1609
Query: 206 GRRWGHMTSN------IVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
GRR+ MT + ES+ S+ LPVT + + F + RV
Sbjct: 1610 GRRYRVMTIDAENLFAFCESFQSL-----GLPVTATALLLFDEMRFFFYSGLCDSSGRVN 1664
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGW 319
GD++++ + ++ + S H V E+ F V E + E + V L
Sbjct: 1665 RGDMYTKPVMDQLEKLMTDSIPHVV--MPLEKGLFQVTEPLQEDE------WIVQLSEWS 1716
Query: 320 CDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLPY 379
C CG+FQ PC HV+A C + + + + + +Y Y F V + + W
Sbjct: 1717 CTCGEFQLKKFPCLHVLAVCEKLKINPLQYVDDCYSLDRLYKTYAATFSPVPEVAAWPEA 1776
Query: 380 DG------PVICHNPNM 390
G PVI PN+
Sbjct: 1777 SGVPTLFPPVILPPPNV 1793
Score = 163 bits (413), Expect = 8e-39
Identities = 102/381 (26%), Positives = 174/381 (44%), Gaps = 28/381 (7%)
Query: 16 FKNADWREWRAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNK 75
F N D+ A F +FW+F I GF++C+P++ VD L KY+ L++A D NK
Sbjct: 2921 FPNPDF----ASFRGVFWSFSQSIEGFQHCRPLIVVDTKSLNGKYQLKLMIASGVDAANK 2976
Query: 76 TIPIAFALVEGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGS 135
P+AFA+ + + + W +F +R VT+ +C++S I + N+P + WQ +
Sbjct: 2977 FFPLAFAVTKEVSTDSWRWFFTKIREKVTQRKDLCLISSPLRDIVAVVNEPGSLWQEPWA 3036
Query: 136 AHVYCIRHIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP 195
H +C+ H++ F+ +D +L+ +V G F+ Y I+E N A W+D IP
Sbjct: 3037 HHKFCLNHLRSQFLGVFRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIP 3096
Query: 196 KEKWTQAYDEGRRWGHMTSNIVE-SWNSVFKGTRNLP-----VTPIVQSTYYRLACLFAD 249
+ KW A+D G R+G I+E ++F R P +T V + L F
Sbjct: 3097 RHKWALAHDSGLRYG-----IIEIDREALFAVCRGFPYCTVAMTGGVMLMFDELRSSFDK 3151
Query: 250 RAQKAFARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMG 309
++ + G +++E + +++ + S + + Q +R+ +F V E+ E
Sbjct: 3152 SLSSIYSSLNRGVVYTEPFMDKLEEFMTDSIPYVITQLERD--SFKVSESSEKEE----- 3204
Query: 310 TFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKV 369
+ V L C C KFQ+ PC H +A + + + + E Y F
Sbjct: 3205 -WIVQLNVSTCTCRKFQSYKFPCLHALAVFEKLKINPLQYVDECYTVEQYCKTYAATFSP 3263
Query: 370 VHDKSYWLPYDGPVICHNPNM 390
V D + W P C P +
Sbjct: 3264 VPDVAAW-----PEDCRVPTL 3279
Score = 148 bits (373), Expect = 3e-34
Identities = 92/354 (25%), Positives = 158/354 (43%), Gaps = 13/354 (3%)
Query: 26 AQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVE 85
A F +FWAF I GF +C+P++ VD L KY+ L++A A D + +AFA
Sbjct: 602 ASFRGVFWAFSQSIEGFHHCRPLIIVDTKNLNCKYQWKLMIASAVDAADNFFLLAFAFTT 661
Query: 86 GETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
+ + W +FL +R VT+ +C++S + + N+ + WQ + + +C+RH+
Sbjct: 662 ELSTDSWRWFLSGIRERVTQRKGLCLISSPDPDLLAVINESGSQWQEPWAYNRFCLRHLL 721
Query: 146 QNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDE 205
F +D L+D+V G F+ Y I++ N A W+D P+ +W A+D
Sbjct: 722 SQFSGIFRDYYLEDLVKRAGSTSQKEEFDSYMKDIEKKNSEARKWLDQFPQNQWALAHDN 781
Query: 206 GRRWGHM---TSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGD 262
GRR+G M T+ + E +N N +T V + L F + + G+
Sbjct: 782 GRRYGIMEIETTTLFEDFN--VSHLDNHVLTGYVLRLFDELRHSFDEFFCFSRGSRKCGN 839
Query: 263 LFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDC 322
+++E + + S T+ V D + + + + + V L C C
Sbjct: 840 VYTEPVTEKLAESRKDSVTYDVMPLDNNAFQVTAPQEND--------EWTVQLSDCSCTC 891
Query: 323 GKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYW 376
G+FQ+ PC H +A C + + + + E +Y Y F V + S W
Sbjct: 892 GEFQSCKFPCLHALAVCKLLKINPLEYVDDCYTLERLYKTYTATFSPVPELSAW 945
Score = 141 bits (356), Expect = 3e-32
Identities = 98/371 (26%), Positives = 160/371 (42%), Gaps = 15/371 (4%)
Query: 26 AQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVE 85
A F +FWAF I GF++C+P++ VD L +Y+ L++A D NK P+AFA+ +
Sbjct: 2204 ASFCGVFWAFPQSIEGFQHCRPLIVVDTKNLNCEYQLKLMIASGVDAANKYFPLAFAVTK 2263
Query: 86 GETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIK 145
+ + W +FL +R VT+ +C++S H I + N+ + WQ + H + + H
Sbjct: 2264 EVSTDIWRWFLTGIREKVTQRKGLCLISSPHPDIIAVVNESGSQWQEPWAYHRFSLNHFY 2323
Query: 146 QNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQAYDE 205
F R L + G F Y I+E N A W+D P+ +W A+D
Sbjct: 2324 SQFSRVFPSFCLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDN 2383
Query: 206 GRRWGHMTSNIVESWN--SVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSGDL 263
GRR+G M N + + F+ ++ VT V + L F + + + GD+
Sbjct: 2384 GRRYGIMEINTKALFAVCNAFEQAGHV-VTGSVLLLFDELRSKFDKSFSCSRSSLNCGDV 2442
Query: 264 FSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCDCG 323
++E + +++ T+ + F V ++ G V L C CG
Sbjct: 2443 YTEPVMDKLEEFRTTFVTYSYIVTPLDNNAFQVATALD------KGECIVQLSDCSCTCG 2496
Query: 324 KFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKVVHDKSYWLPYDG-- 381
FQ PC H +A C + + + + E + Y T F V + S W G
Sbjct: 2497 DFQRYKFPCLHALAVCKKLKFNPLQYVDDCYTLERLKRTYATIFSHVPEMSAWPEASGVP 2556
Query: 382 ----PVICHNP 388
PVI +P
Sbjct: 2557 RLLPPVIPPSP 2567
>UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sativa]
Length = 1564
Score = 165 bits (417), Expect = 3e-39
Identities = 104/304 (34%), Positives = 156/304 (51%), Gaps = 21/304 (6%)
Query: 28 FHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFALVEGE 87
F R FW F P I FK+C+PV+ +D T+L KY G L+ A++ D ++ +P+AFALVE E
Sbjct: 593 FMRAFWCFGPSIEAFKHCRPVLAIDATFLTRKYGGALMTALSADAEDQLVPLAFALVEKE 652
Query: 88 TKEGWSFFLKNLRRHVT-KGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIRHIKQ 146
W +F+ +RR V VC++SDRH I +A P G H +C+RH
Sbjct: 653 NLRDWCWFIDLVRRVVVGPHREVCIISDRHAGIMNAMTTPVPGLPLV--HHRWCMRHFSA 710
Query: 147 NFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALD----WVDN--IPKEKWT 200
NF K G K + + ++ ++ QR + W+++ + K+KW+
Sbjct: 711 NFH---KAGADKHQTKELLRICQIDEKWIFKRDVEALRQRIPEGPRKWLEDELLDKDKWS 767
Query: 201 QAYD-EGRRWGHMTSNIVESWNSVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVG 259
+AYD GRRWG+MT+N+ E +NSV G R LPVT IV T+ + F +R +A RV
Sbjct: 768 RAYDRNGRRWGYMTTNMAEQFNSVLVGVRKLPVTAIVSFTFMKCNDYFVNRHDEALKRVQ 827
Query: 260 SGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRA-- 317
G +S + ++ K+N H FD+++ T+ E R G G + RA
Sbjct: 828 LGQRWSTKVDSKMKVQKSKANKHTARCFDKQKKTYEFTE----RGGITRGGVRFGARAFK 883
Query: 318 --GW 319
GW
Sbjct: 884 VEGW 887
>UniRef100_Q9C7V1 Hypothetical protein F15H21.15 [Arabidopsis thaliana]
Length = 719
Score = 163 bits (413), Expect = 8e-39
Identities = 102/381 (26%), Positives = 174/381 (44%), Gaps = 28/381 (7%)
Query: 16 FKNADWREWRAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNK 75
F N D+ A F +FW+F I GF++C+P++ VD L KY+ L++A D NK
Sbjct: 350 FPNPDF----ASFRGVFWSFSQSIEGFQHCRPLIVVDTKSLNGKYQLKLMIASGVDAANK 405
Query: 76 TIPIAFALVEGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGS 135
P+AFA+ + + + W +F +R VT+ +C++S I + N+P + WQ +
Sbjct: 406 FFPLAFAVTKEVSTDSWRWFFTKIREKVTQRKDLCLISSPLRDIVAVVNEPGSLWQEPWA 465
Query: 136 AHVYCIRHIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIP 195
H +C+ H++ F+ +D +L+ +V G F+ Y I+E N A W+D IP
Sbjct: 466 HHKFCLNHLRSQFLGVFRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIP 525
Query: 196 KEKWTQAYDEGRRWGHMTSNIVE-SWNSVFKGTRNLP-----VTPIVQSTYYRLACLFAD 249
+ KW A+D G R+G I+E ++F R P +T V + L F
Sbjct: 526 RHKWALAHDSGLRYG-----IIEIDREALFAVCRGFPYCTVAMTGGVMLMFDELRSSFDK 580
Query: 250 RAQKAFARVGSGDLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMG 309
++ + G +++E + +++ + S + + Q +R+ +F V E+ E
Sbjct: 581 SLSSIYSSLNRGVVYTEPFMDKLEEFMTDSIPYVITQLERD--SFKVSESSEKEE----- 633
Query: 310 TFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTAFKV 369
+ V L C C KFQ+ PC H +A + + + + E Y F
Sbjct: 634 -WIVQLNVSTCTCRKFQSYKFPCLHALAVFEKLKINPLQYVDECYTVEQYCKTYAATFSP 692
Query: 370 VHDKSYWLPYDGPVICHNPNM 390
V D + W P C P +
Sbjct: 693 VPDVAAW-----PEDCRVPTL 708
>UniRef100_Q9FWZ8 F11O6.3 protein [Arabidopsis thaliana]
Length = 884
Score = 163 bits (412), Expect = 1e-38
Identities = 127/416 (30%), Positives = 189/416 (44%), Gaps = 23/416 (5%)
Query: 23 EWRAQFHRLFWAFYPCINGFKYCKPVVHVDGTWLYDKYKGTLLLAVAQDGNNKTIPIAFA 82
E + +F +F AF I GFK+ + V+ VDGT L KYKG LL A QD N + P+AFA
Sbjct: 484 ERKERFLYMFLAFGASIQGFKHLRRVLVVDGTHLKGKYKGVLLTASGQDANFQVYPLAFA 543
Query: 83 LVEGETKEGWSFFLKNLRRHVTKGISVCMVSDRHESIKSAFNDPRNGWQATGSAHVYCIR 142
+V+ E + W++F L R + ++ ++SDRHESIK + H CI
Sbjct: 544 VVDSENDDAWTWFFTKLERIIADNNTLTILSDRHESIKVGVKK-----VFPQAHHGACII 598
Query: 143 HIKQNFMRTIKDGDLKDVVNNMGYALNVPLFNYYRGVIQEANQRALDWVDNIPKEKWTQA 202
H+ +N K+ L +V N GY F I N + ++ ++ WT+
Sbjct: 599 HLCRNIQARFKNRGLTQLVKNAGYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRL 658
Query: 203 YDEGRRWGHMTSNIVESWN-SVFKGTRNLPVTPIVQSTYYRLACLFADRAQKAFARVGSG 261
Y G+R+ MTSNI E+ N ++FKG R+ + +++ L F +K+ A SG
Sbjct: 659 YFPGQRFNLMTSNIAETLNKALFKG-RSSHIVELLRFIRSMLTRWFNAHRKKSQAH--SG 715
Query: 262 DLFSEYCQNAIQDDIVKSNTHHVEQFDRERYTFSVRETVNYREGRPMGTFKVDLRAGWCD 321
+ E I ++ S+ V + Y E V G+ G+ VDL C
Sbjct: 716 PVPPE-VDKQISKNLTTSSGSKVGRVTSWSY-----EVV----GKLGGSNVVDLEKKQCT 765
Query: 322 CGKFQALHLPCSHVIAACSSFRHDYTTLIPHVFKNESVYSIYNTA-FKVVHDKSYWLPYD 380
C ++ L +PC H + A +S Y L+ FK S + Y A F + K +P +
Sbjct: 766 CKRYDKLKIPCGHALVAANSINLSYKALVDDYFKPHSWVASYKGAVFPEANGKEEDIPEE 825
Query: 381 GPVICHNPNMRRLKKGRPNSTRIRT--EMDEEVVERTPTPRRCGLCRHTGHIRRNC 434
P P R GRP RI + E + RT RC C+ GH R +C
Sbjct: 826 LPHRSMLPPYTRRPSGRPKVARIPSTGEYKKPKTGRT-QQNRCSRCKGLGHNRTSC 880
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 809,736,763
Number of Sequences: 2790947
Number of extensions: 35245001
Number of successful extensions: 76317
Number of sequences better than 10.0: 337
Number of HSP's better than 10.0 without gapping: 287
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 74995
Number of HSP's gapped (non-prelim): 552
length of query: 440
length of database: 848,049,833
effective HSP length: 130
effective length of query: 310
effective length of database: 485,226,723
effective search space: 150420284130
effective search space used: 150420284130
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC126012.8


