

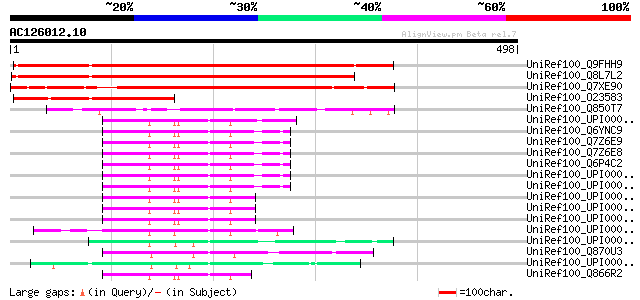
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126012.10 + phase: 0 /pseudo
(498 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FHH9 DNA-binding protein-like [Arabidopsis thaliana] 427 e-118
UniRef100_Q8L7L2 DNA-binding protein-like [Arabidopsis thaliana] 404 e-111
UniRef100_Q7XE90 Putative DNA-binding protein [Oryza sativa] 372 e-101
UniRef100_O23583 Hypothetical protein dl4735w [Arabidopsis thali... 204 5e-51
UniRef100_Q850T7 Putative DNA-binding protein [Oryza sativa] 173 1e-41
UniRef100_UPI00003AADE7 UPI00003AADE7 UniRef100 entry 105 3e-21
UniRef100_Q6YNC9 P53-associated cellular protein PACT [Homo sapi... 104 7e-21
UniRef100_Q7Z6E9 Retinoblastoma binding protein 6 isoform 1 [Hom... 103 1e-20
UniRef100_Q7Z6E8 Retinoblastoma binding protein 6 isoform 2 [Hom... 103 1e-20
UniRef100_Q6P4C2 RBBP6 protein [Homo sapiens] 103 1e-20
UniRef100_UPI000036A6E8 UPI000036A6E8 UniRef100 entry 103 1e-20
UniRef100_UPI000036A6E7 UPI000036A6E7 UniRef100 entry 103 1e-20
UniRef100_UPI00003AADE6 UPI00003AADE6 UniRef100 entry 103 1e-20
UniRef100_UPI00003AADE5 UPI00003AADE5 UniRef100 entry 103 1e-20
UniRef100_UPI00003AADE4 UPI00003AADE4 UniRef100 entry 103 1e-20
UniRef100_UPI00004299A1 UPI00004299A1 UniRef100 entry 102 3e-20
UniRef100_UPI000023F469 UPI000023F469 UniRef100 entry 100 1e-19
UniRef100_Q870U3 Hypothetical protein B11H7.030 [Neurospora crassa] 97 8e-19
UniRef100_UPI000021B32F UPI000021B32F UniRef100 entry 96 2e-18
UniRef100_Q866R2 Retinoblastoma binding protein 6 [Equus caballus] 96 2e-18
>UniRef100_Q9FHH9 DNA-binding protein-like [Arabidopsis thaliana]
Length = 889
Score = 427 bits (1098), Expect = e-118
Identities = 220/377 (58%), Positives = 271/377 (71%), Gaps = 7/377 (1%)
Query: 4 PENLDWDEFGNDLYSIPDQLPVQSSTMIPD-APLTSKADEDSKIKALIDTPALDWQQQGS 62
PE+ ++DEFG DLYSIPD Q P A K DE+SKI+ALIDTPALDWQQQG
Sbjct: 113 PED-EYDEFGTDLYSIPDTQDAQHIIPRPHLATADDKVDEESKIQALIDTPALDWQQQGQ 171
Query: 63 D-FGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDI 121
D FGAGRG+ RG G GRGFG+ERKTPP GYVCHRC + GHFIQHCPTNGD +D+
Sbjct: 172 DTFGAGRGYGRGMPGRM--NGRGFGMERKTPPPGYVCHRCNIPGHFIQHCPTNGDPNYDV 229
Query: 122 KKVRQPTGIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDKEMEGL-SSTRSVGDLPPEL 180
K+V+ PTGIP+SMLM P GSY+L +G+VAVLKPNE AF+KEMEGL S+TRSVG+LPPEL
Sbjct: 230 KRVKPPTGIPKSMLMATPDGSYSLPSGAVAVLKPNEDAFEKEMEGLPSTTRSVGELPPEL 289
Query: 181 HCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNKTLRDAI 240
CPLC VMKDA LTSKCC+KSFCDKCIR++I+SKS CVC +++LADDLLPNKTLRD I
Sbjct: 290 KCPLCKEVMKDAALTSKCCYKSFCDKCIRDHIISKSMCVCGRSDVLADDLLPNKTLRDTI 349
Query: 241 HRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNEGMANIQE 300
+RILE+GN STEN GS + D+ S+RCP PK SPT+S SKGE K N A+ +
Sbjct: 350 NRILEAGNDSTENVGSVGHIPDLESARCPPPKALSPTTSVASKGEKKPVLSNNNDASTLK 409
Query: 301 IVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQGSAKLVEEEVQQKLVP 360
E E+++ + S +V + + E T S+ VKE S+ + + +EE+QQ++
Sbjct: 410 APMEVAEITSAPRASAEVNVEKPVDACESTQGSVIVKE-ATVSKLNTQAPKEEMQQQVAA 468
Query: 361 TDGGKKKKKKKVRMPAN 377
+ KKKKKK R+P N
Sbjct: 469 GEPAGKKKKKKPRVPGN 485
>UniRef100_Q8L7L2 DNA-binding protein-like [Arabidopsis thaliana]
Length = 462
Score = 404 bits (1039), Expect = e-111
Identities = 206/341 (60%), Positives = 250/341 (72%), Gaps = 7/341 (2%)
Query: 2 QYPENLDWDEFGNDLYSIPDQLPVQSSTMIPD-APLTSKADEDSKIKALIDTPALDWQQ- 59
++PE+ ++DEFG DLYSIPD Q P A K DE+SKI+ALIDTPALDWQQ
Sbjct: 114 EFPED-EYDEFGTDLYSIPDTQDAQHIIPRPHLATADDKVDEESKIQALIDTPALDWQQR 172
Query: 60 QGSD-FGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDST 118
QG D FGAGRG+ RG G GRGFG+ERKTPP GYVCHRC + GHFIQHCPTNGD
Sbjct: 173 QGQDTFGAGRGYGRGMPGRM--NGRGFGMERKTPPPGYVCHRCNIPGHFIQHCPTNGDPN 230
Query: 119 FDIKKVRQPTGIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDKEMEGL-SSTRSVGDLP 177
+D+K+V+ PTGIP+SMLM P GSY+L +G+VAVLKPNE AF+KEMEGL S+TRSVG+LP
Sbjct: 231 YDVKRVKPPTGIPKSMLMATPDGSYSLPSGAVAVLKPNEDAFEKEMEGLPSTTRSVGELP 290
Query: 178 PELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNKTLR 237
PEL CPLC VMKDA LTSKCC+KSFCDKCIR++I+SKS CVC +++LADDLLPNKTLR
Sbjct: 291 PELKCPLCKEVMKDAALTSKCCYKSFCDKCIRDHIISKSMCVCGRSDVLADDLLPNKTLR 350
Query: 238 DAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNEGMAN 297
D I+RILE+GN STEN GS + D+ S+RCP PK SPT+S SKGE K N A+
Sbjct: 351 DTINRILEAGNDSTENVGSVGHIPDLESARCPPPKALSPTTSVASKGEKKPVLSNNNDAS 410
Query: 298 IQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKE 338
+ E E+++ + S +V + + E T S+ VKE
Sbjct: 411 TLKAPMEVAEITSAPRASAEVNVEKPVDACESTQGSVIVKE 451
>UniRef100_Q7XE90 Putative DNA-binding protein [Oryza sativa]
Length = 1066
Score = 372 bits (954), Expect = e-101
Identities = 211/380 (55%), Positives = 255/380 (66%), Gaps = 27/380 (7%)
Query: 1 MQYPENLDWD-EFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQ 59
M+YPE +WD EFGNDLY + D +P Q + DA +K DEDSKIKALIDT ALD+ Q
Sbjct: 285 MKYPEESEWDDEFGNDLY-VSDSVPSQLGSQAVDAS-ENKVDEDSKIKALIDTSALDYSQ 342
Query: 60 QGSDFGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTF 119
+G GRG+ RG +GGR+ GGRGFG HFIQHCPTNGD+ F
Sbjct: 343 IPDGYGGGRGYGRG-MGGRMMGGRGFG-------------------HFIQHCPTNGDARF 382
Query: 120 DIKKVRQPTGIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDKEMEGLSSTRSVGDLPPE 179
D+K+++ PTGIP+SMLM P GSYAL +G+ AVLKPNEAAF+KE+EGL +TRS+GDLPPE
Sbjct: 383 DMKRMKPPTGIPKSMLMATPDGSYALPSGAGAVLKPNEAAFEKEIEGLPTTRSLGDLPPE 442
Query: 180 LHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNKTLRDA 239
L CPLC VMKDAVLTSKCCF+SFCDKCIR+YI++KS CVC AT+ILADDLLPNKTLR+
Sbjct: 443 LRCPLCKEVMKDAVLTSKCCFRSFCDKCIRDYIINKSMCVCGATSILADDLLPNKTLRET 502
Query: 240 IHRILES-GNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNEGMANI 298
I RILE+ SSTEN GS QVQDM S+ QPK+ SP SA SK EPK + +
Sbjct: 503 ISRILEAPPTSSTENVGSMVQVQDMESAIPVQPKVRSPAVSAASKEEPKRTPAPVEESPD 562
Query: 299 QEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQGSAKLVEEEVQQKL 358
E +E K + S++ K+P V E T ES +KE P + K E + + +
Sbjct: 563 VESHSEVKTTNVDMSSSDK-KVPALPDVVEGTMESKILKEKTPEATPVVK--ESQEKMPV 619
Query: 359 VPTDGGKKKKKKKVRMPANA 378
V KKKKKKKVR P NA
Sbjct: 620 VGEQVVKKKKKKKVRAPGNA 639
>UniRef100_O23583 Hypothetical protein dl4735w [Arabidopsis thaliana]
Length = 274
Score = 204 bits (519), Expect = 5e-51
Identities = 100/160 (62%), Positives = 119/160 (73%), Gaps = 5/160 (3%)
Query: 4 PENLDWDEFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSD 63
P ++DEFGNDLYSIPD V S+ + D+ DE++K+KALIDTPALDW QQG+D
Sbjct: 112 PVEDEFDEFGNDLYSIPDAPAVHSNNLCHDSAPAD--DEETKLKALIDTPALDWHQQGAD 169
Query: 64 -FGAGRGFRRGAVGGRIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDIK 122
FG GRG+ RG G GGRGFG+ER TPP GYVCHRC VSGHFIQHC TNG+ FD+K
Sbjct: 170 SFGPGRGYGRGMAGRM--GGRGFGMERTTPPPGYVCHRCNVSGHFIQHCSTNGNPNFDVK 227
Query: 123 KVRQPTGIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDK 162
+V+ PTGIP+SMLM P GSY+L +G+VAVLKPNE K
Sbjct: 228 RVKPPTGIPKSMLMATPNGSYSLPSGAVAVLKPNEYVVTK 267
>UniRef100_Q850T7 Putative DNA-binding protein [Oryza sativa]
Length = 697
Score = 173 bits (438), Expect = 1e-41
Identities = 134/388 (34%), Positives = 177/388 (45%), Gaps = 85/388 (21%)
Query: 37 TSKADEDSKIKALIDTPALDWQQQGSDFGAGRGFRRGAVGGRIGGGR-GFG--------- 86
T DED+ I ++ID L W+ + S +RG GGR GR G G
Sbjct: 122 TEVQDEDAAIASIIDAAELKWEDKPS--------KRGQTGGRFTSGRYGHGPVEGETPPP 173
Query: 87 ------------------LERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDIKKVRQPT 128
ERKTPP GYVC+RC++ GHFI HCPT GD FD K
Sbjct: 174 GYVCRSCGVPGHFIQHCQQERKTPPSGYVCYRCRIPGHFIHHCPTIGDPKFDDYKK---- 229
Query: 129 GIPRSMLMVNPQGSYALQNGSVAVLKPNEAAFDKEMEGLSSTRSVGDLPPELHCPLCNNV 188
P +++ P+ S +G + L P +S V DLP ELHC LCN V
Sbjct: 230 --PHTLV---PEVSACPIDGIPSALAP-----------AASVSVVDDLPAELHCRLCNKV 273
Query: 189 MKDAVLTSKCCFKSFCDKCIRNYIMSKSACVCLATNILADDLLPNKTLRDAIHRILESGN 248
M DAVLTSKCCF SFCDKCIR+YI+++S C+C +LADDL+PN+TLR I +L +
Sbjct: 274 MADAVLTSKCCFDSFCDKCIRDYIITQSKCIC-GVKVLADDLIPNQTLRSTISNMLATRA 332
Query: 249 SSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQVNEGMANIQEIVAERKEV 308
SS + ++ S P S T SA + + K ++ + V
Sbjct: 333 SSITSGTGKHRSS---SGSNVDPNSASQTPSAALEKQSKDHHISSAAPDAGLQV------ 383
Query: 309 SATQQVSEQVKIPRAAVVSEVTHESMS----VKEPEPASQGSAKLVEEE----------V 354
AT+ VS V EV E S V++ P + K V + +
Sbjct: 384 -ATEHVSHLEHKLTTGVDLEVKDEGNSAGILVEKIVPTADARLKDVTDSTSKPATISGTM 442
Query: 355 QQKLVPTDGGKKKKKK----KVRMPANA 378
+ K TD KKK+KK K+ P NA
Sbjct: 443 EPKASKTDQLKKKRKKADSTKIVHPNNA 470
>UniRef100_UPI00003AADE7 UPI00003AADE7 UniRef100 entry
Length = 1661
Score = 105 bits (262), Expect = 3e-21
Identities = 66/207 (31%), Positives = 100/207 (47%), Gaps = 22/207 (10%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFD-IKKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ + ++++ TGIPRS +M N +G+
Sbjct: 40 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESVPRIKKSTGIPRSFMMEVKDPNTKGAMLTN 99
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 100 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 158
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRILESGNSSTENA 254
CC S+CD+CIR ++ + C T++ D L+ NK LR A++ + T
Sbjct: 159 CCGNSYCDECIRTALLESEEHTCPTCHQTDVSPDALIANKFLRQAVNNF----KNETGYT 214
Query: 255 GSTYQVQDMLSSRCPQPKIPSPTSSAT 281
++Q P P P P T
Sbjct: 215 KRLRKIQQQQQQPPPPPPPPPPLMRQT 241
>UniRef100_Q6YNC9 P53-associated cellular protein PACT [Homo sapiens]
Length = 1625
Score = 104 bits (259), Expect = 7e-21
Identities = 67/202 (33%), Positives = 100/202 (49%), Gaps = 27/202 (13%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDI-KKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ ++++ TGIPRS +M N +G+
Sbjct: 22 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 81
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 82 TGKYAIPTTDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 140
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRILESGNSSTENA 254
CC S+CD+CIR ++ + C ++ D L+ NK LR A+ N+
Sbjct: 141 CCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAV-------NNFKNET 193
Query: 255 GSTYQVQDMLSSRCPQPKIPSP 276
G T +++ L P P IP P
Sbjct: 194 GYTKRLRKQLPP--PPPPIPPP 213
>UniRef100_Q7Z6E9 Retinoblastoma binding protein 6 isoform 1 [Homo sapiens]
Length = 1792
Score = 103 bits (257), Expect = 1e-20
Identities = 67/202 (33%), Positives = 100/202 (49%), Gaps = 27/202 (13%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDI-KKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ ++++ TGIPRS +M N +G+
Sbjct: 155 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 214
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 215 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 273
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRILESGNSSTENA 254
CC S+CD+CIR ++ + C ++ D L+ NK LR A+ N+
Sbjct: 274 CCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAV-------NNFKNET 326
Query: 255 GSTYQVQDMLSSRCPQPKIPSP 276
G T +++ L P P IP P
Sbjct: 327 GYTKRLRKQLPP--PPPPIPPP 346
>UniRef100_Q7Z6E8 Retinoblastoma binding protein 6 isoform 2 [Homo sapiens]
Length = 1758
Score = 103 bits (257), Expect = 1e-20
Identities = 67/202 (33%), Positives = 100/202 (49%), Gaps = 27/202 (13%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDI-KKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ ++++ TGIPRS +M N +G+
Sbjct: 155 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 214
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 215 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 273
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRILESGNSSTENA 254
CC S+CD+CIR ++ + C ++ D L+ NK LR A+ N+
Sbjct: 274 CCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAV-------NNFKNET 326
Query: 255 GSTYQVQDMLSSRCPQPKIPSP 276
G T +++ L P P IP P
Sbjct: 327 GYTKRLRKQLPP--PPPPIPPP 346
>UniRef100_Q6P4C2 RBBP6 protein [Homo sapiens]
Length = 919
Score = 103 bits (257), Expect = 1e-20
Identities = 67/202 (33%), Positives = 100/202 (49%), Gaps = 27/202 (13%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDI-KKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ ++++ TGIPRS +M N +G+
Sbjct: 155 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 214
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 215 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 273
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRILESGNSSTENA 254
CC S+CD+CIR ++ + C ++ D L+ NK LR A+ N+
Sbjct: 274 CCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAV-------NNFKNET 326
Query: 255 GSTYQVQDMLSSRCPQPKIPSP 276
G T +++ L P P IP P
Sbjct: 327 GYTKRLRKQLPP--PPPPIPPP 346
>UniRef100_UPI000036A6E8 UPI000036A6E8 UniRef100 entry
Length = 1655
Score = 103 bits (256), Expect = 1e-20
Identities = 67/202 (33%), Positives = 99/202 (48%), Gaps = 27/202 (13%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDI-KKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ ++++ TGIPRS +M N +G+
Sbjct: 52 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 111
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C +M DAV+
Sbjct: 112 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEGDDPIPDELVCLICEGIMTDAVVI-P 170
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRILESGNSSTENA 254
CC S+CD+CIR ++ + C ++ D L+ NK LR A+ N+
Sbjct: 171 CCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAV-------NNFKNET 223
Query: 255 GSTYQVQDMLSSRCPQPKIPSP 276
G T +++ L P P IP P
Sbjct: 224 GYTKRLRKQLPP--PPPPIPPP 243
>UniRef100_UPI000036A6E7 UPI000036A6E7 UniRef100 entry
Length = 1689
Score = 103 bits (256), Expect = 1e-20
Identities = 67/202 (33%), Positives = 99/202 (48%), Gaps = 27/202 (13%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDI-KKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ ++++ TGIPRS +M N +G+
Sbjct: 52 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 111
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C +M DAV+
Sbjct: 112 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEGDDPIPDELVCLICEGIMTDAVVI-P 170
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRILESGNSSTENA 254
CC S+CD+CIR ++ + C ++ D L+ NK LR A+ N+
Sbjct: 171 CCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAV-------NNFKNET 223
Query: 255 GSTYQVQDMLSSRCPQPKIPSP 276
G T +++ L P P IP P
Sbjct: 224 GYTKRLRKQLPP--PPPPIPPP 243
>UniRef100_UPI00003AADE6 UPI00003AADE6 UniRef100 entry
Length = 820
Score = 103 bits (256), Expect = 1e-20
Identities = 59/167 (35%), Positives = 90/167 (53%), Gaps = 18/167 (10%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFD-IKKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ + ++++ TGIPRS +M N +G+
Sbjct: 111 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESVPRIKKSTGIPRSFMMEVKDPNTKGAMLTN 170
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 171 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 229
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIH 241
CC S+CD+CIR ++ + C T++ D L+ NK LR A++
Sbjct: 230 CCGNSYCDECIRTALLESEEHTCPTCHQTDVSPDALIANKFLRQAVN 276
>UniRef100_UPI00003AADE5 UPI00003AADE5 UniRef100 entry
Length = 1688
Score = 103 bits (256), Expect = 1e-20
Identities = 59/167 (35%), Positives = 90/167 (53%), Gaps = 18/167 (10%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFD-IKKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ + ++++ TGIPRS +M N +G+
Sbjct: 111 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESVPRIKKSTGIPRSFMMEVKDPNTKGAMLTN 170
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 171 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 229
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIH 241
CC S+CD+CIR ++ + C T++ D L+ NK LR A++
Sbjct: 230 CCGNSYCDECIRTALLESEEHTCPTCHQTDVSPDALIANKFLRQAVN 276
>UniRef100_UPI00003AADE4 UPI00003AADE4 UniRef100 entry
Length = 1654
Score = 103 bits (256), Expect = 1e-20
Identities = 59/167 (35%), Positives = 90/167 (53%), Gaps = 18/167 (10%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFD-IKKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ + ++++ TGIPRS +M N +G+
Sbjct: 111 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESVPRIKKSTGIPRSFMMEVKDPNTKGAMLTN 170
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 171 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 229
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIH 241
CC S+CD+CIR ++ + C T++ D L+ NK LR A++
Sbjct: 230 CCGNSYCDECIRTALLESEEHTCPTCHQTDVSPDALIANKFLRQAVN 276
>UniRef100_UPI00004299A1 UPI00004299A1 UniRef100 entry
Length = 786
Score = 102 bits (253), Expect = 3e-20
Identities = 79/282 (28%), Positives = 126/282 (44%), Gaps = 50/282 (17%)
Query: 24 PVQSSTMIPDAPLT-SKADEDSKIKALIDTPALDWQQQGSDFGAGRGFRRGAVGGRIGGG 82
P+ + +I A L + A E+ KIKA++ Q G + ++ VG
Sbjct: 108 PMSLAQLIETANLAEANASEEDKIKAMMI-------QSGHLYNPINSMKKTPVG------ 154
Query: 83 RGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDIK-KVRQPTGIPRSMLM----V 137
PP Y C C GH+I++CPTN + F+ + ++R+ TGIPRS +M
Sbjct: 155 --------LPPPSYTCFCCGKPGHYIKNCPTNANKNFESRPRIRKSTGIPRSFMMEVKDP 206
Query: 138 NPQGSYALQNGSVAVLKPNEAAF-----------DKEMEGLSSTRSVGDLPPELHCPLCN 186
N +G+ + G A+ N A+ +E SS+ V +P EL C +C
Sbjct: 207 NMKGAMLTKTGQYAIPTLNAEAYAIGKKEKPPFLPEEPSSSSSSEEVDPVPDELLCLICK 266
Query: 187 NVMKDAVLTSKCCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLRDAIHRI 243
+ M DA + CC S+CD+CIR ++ + C ++ D L+ NK LR A++
Sbjct: 267 DTMTDAAII-PCCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALVANKVLRQAVNN- 324
Query: 244 LESGNSSTENAGSTYQVQ-------DMLSSRCPQPKIPSPTS 278
++ ST++ G L + QP + SPTS
Sbjct: 325 FKNQTDSTKSLGKQVTPSPPPLPPPSALIQQNLQPPMKSPTS 366
>UniRef100_UPI000023F469 UPI000023F469 UniRef100 entry
Length = 617
Score = 100 bits (248), Expect = 1e-19
Identities = 81/326 (24%), Positives = 131/326 (39%), Gaps = 55/326 (16%)
Query: 78 RIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDIK-KVRQPTGIPRSMLM 136
+ G R + PP GY+C+RC GH+IQ CPTN D FD + +V++ TGIPRS L
Sbjct: 160 KAGAKRPANVPDHDPPNGYICYRCGNKGHWIQLCPTNDDPEFDNRPRVKRTTGIPRSFLQ 219
Query: 137 ----------------VNPQGSYALQNGSVAVLKPNEAAFD----KEMEGLSSTRSVGDL 176
P G G + +P++A+++ K ++ GD
Sbjct: 220 KVDKSVVLAQTDGDETKRPSGIMVNAEGDFVIAEPDKASWEQFQAKAKSSATTNAPAGDK 279
Query: 177 PPE---LHCPLCNNVMKDAVLTSKCCFKSFCDKCIRN-YIMSKSAC-VCLATNILADDLL 231
+ L C + + + + T CC K+FC+ CI N I S C C + +L DDL
Sbjct: 280 EIQEQGLECSIDKKMFIEPMKT-PCCQKTFCNDCITNALIESDFVCPACQSEGVLIDDLQ 338
Query: 232 PNKTLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKVSQV 291
P++ I L ++ ++ P P P + A + GE +
Sbjct: 339 PDEEASKKIQEYL----------------KEKEIAKSPPPPSPKASEGAKADGESQDKPQ 382
Query: 292 NEGMANIQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQGSAKLVE 351
NE +A+ ++ V E+ P T +S ++K P +G +
Sbjct: 383 NEDIASTEQKVENESHDKTMASAKEKSNSP--------TSQSATIKTPPTGPKGLVSVPH 434
Query: 352 EEVQQKLVPTDGGKKKKKKKVRMPAN 377
+ +Q DG K + PA+
Sbjct: 435 DANKQ----VDGAKTSDDNSKKRPAD 456
>UniRef100_Q870U3 Hypothetical protein B11H7.030 [Neurospora crassa]
Length = 673
Score = 97.4 bits (241), Expect = 8e-19
Identities = 76/296 (25%), Positives = 125/296 (41%), Gaps = 44/296 (14%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDIK-KVRQPTGIPRSMLMV------------- 137
PP+GY+C+RC GH+IQ CPTN + +D + +V++ TGIP+S L
Sbjct: 175 PPQGYICYRCGEKGHWIQLCPTNDNPDYDNRPRVKRTTGIPKSFLKTVDKATALGQTGDG 234
Query: 138 ----NPQGSYALQNGSVAVLKPNEAAFDKEMEGLSSTRSVGDLPPE---------LHCPL 184
P G +G + +P++A++++ S + PE L CP+
Sbjct: 235 DETKTPSGIMVNADGEFVIAEPDKASWEQFQAKAKSNSAAQKATPEGDKEIQERGLECPI 294
Query: 185 CNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACV--CLATNILADDLLPNKTLRDAIHR 242
+ + D + T CC K++C+ CI N ++ C + IL DDL ++ D I
Sbjct: 295 DHKLFIDPMKT-PCCEKTYCNDCITNALIESDFICPGCKSDGILIDDLKADEEAVDKIKA 353
Query: 243 ILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPKV-SQVNEGMANIQEI 301
L NS + S P P+ S TS P V + E +
Sbjct: 354 FLAEKNSKAKEG----------SQSPGSPNSPTAAKSPTSTDAPAVQTTTEETKPKCKSP 403
Query: 302 VAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQGSAKLVEEEVQQK 357
SA+QQ S+Q + S + +S S + P +Q + + ++V +K
Sbjct: 404 TPTDTAPSASQQASQQ---QTSQASSAASMQSGSTGQTTPPTQANREERSQQVSKK 456
>UniRef100_UPI000021B32F UPI000021B32F UniRef100 entry
Length = 671
Score = 95.9 bits (237), Expect = 2e-18
Identities = 88/357 (24%), Positives = 145/357 (39%), Gaps = 48/357 (13%)
Query: 21 DQLPVQSSTMIPDAPLTSKAD---EDSKIKALIDTPALDWQQQGSDFGAGRGFRRGAVGG 77
+Q+ S+T P A L ++ E+ ++ A+ + W Q + G G
Sbjct: 118 EQVAKASTTKAPSAALAQMSNAVTEEERLAAMFQAQSEQWTAQQEELSHQTPIYNGKPGV 177
Query: 78 RIGGGRGFGLERKTPPEGYVCHRCKVSGHFIQHCPTNGDSTFDIK-KVRQPTGIPRSMLM 136
G R + PP GY+C+RC GH+IQ CPTN + FD + +V++ TGIP+S L
Sbjct: 178 ---GKRPANVPDHEPPNGYICYRCGEKGHWIQLCPTNDNPEFDNRPRVKRTTGIPKSFLK 234
Query: 137 V------------------NPQGSYALQNGSVAVLKPNEAAFDK-----EMEGLSSTRSV 173
P G +G + +P++AA++K ++ + +V
Sbjct: 235 TVDKATVLGQAGVDGEEGKVPSGVMVNADGDFVIAQPDKAAWEKFQARTKVSAAAQKVAV 294
Query: 174 GD----LPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRN-YIMSKSACV-CLATNILA 227
D L CP+ + + + T CC K++C+ CI N I S C C A +L
Sbjct: 295 QDEKELQDRGLECPMDKRLFNEPMKT-PCCEKTYCNDCITNALIESDFVCPGCQAEGVLI 353
Query: 228 DDLLPNKTLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSSATSKGEPK 287
D+L P+ I + E + Q PK P+ S ++ +
Sbjct: 354 DNLKPDDETTAKIKQYHEEKEK---------EQQKEKEKEADAPKSPAAAKSPSANNQSA 404
Query: 288 VSQVNEGMANIQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEPEPASQ 344
+ N G A+ +E V + S T V K P + + ++S + PA Q
Sbjct: 405 KTTENNG-ADSKE-VGKASSKSPTTSVKSLAKDPTSPTEISTPNGTVSNPKKRPADQ 459
>UniRef100_Q866R2 Retinoblastoma binding protein 6 [Equus caballus]
Length = 238
Score = 95.9 bits (237), Expect = 2e-18
Identities = 57/163 (34%), Positives = 85/163 (51%), Gaps = 18/163 (11%)
Query: 92 PPEGYVCHRCKVSGHFIQHCPTNGDSTFDI-KKVRQPTGIPRSMLM----VNPQGSYALQ 146
PP Y C RC GH+I++CPTNGD F+ ++++ TGIPRS +M N +G+
Sbjct: 60 PPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTN 119
Query: 147 NGSVAVLKPNEAAF---DKEM------EGLSSTRSVGDLPPELHCPLCNNVMKDAVLTSK 197
G A+ + A+ KE E SS+ +P EL C +C ++M DAV+
Sbjct: 120 TGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVI-P 178
Query: 198 CCFKSFCDKCIRNYIMSK---SACVCLATNILADDLLPNKTLR 237
CC S+CD+CIR ++ + C ++ D L+ NK LR
Sbjct: 179 CCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLR 221
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 848,669,814
Number of Sequences: 2790947
Number of extensions: 36597044
Number of successful extensions: 128801
Number of sequences better than 10.0: 608
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 523
Number of HSP's that attempted gapping in prelim test: 127175
Number of HSP's gapped (non-prelim): 1315
length of query: 498
length of database: 848,049,833
effective HSP length: 132
effective length of query: 366
effective length of database: 479,644,829
effective search space: 175550007414
effective search space used: 175550007414
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC126012.10


