

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125480.15 - phase: 0
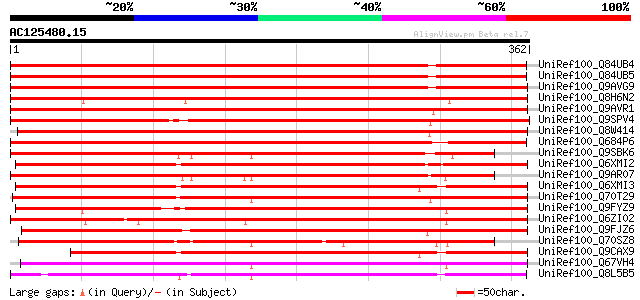
(362 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84UB4 S-adenosyl-L-methionine:benzoic acid/salicylic ... 444 e-123
UniRef100_Q84UB5 S-adenosyl-L-methionine:benzoic acid/salicylic ... 444 e-123
UniRef100_Q9AVG9 S-adenosyl-L-methionine:salicylic acid carboxyl... 429 e-119
UniRef100_Q8H6N2 S-adenosyl-L-methionine:salicylic acid methyltr... 426 e-118
UniRef100_Q9AVR1 S-adenosyl-L-methionine:salicylic acid carboxyl... 419 e-116
UniRef100_Q9SPV4 S-adenosyl-L-methionine:salicylic acid carboxyl... 416 e-115
UniRef100_Q8W414 S-adenosyl-L-methionine:salicylic acid calboxyl... 392 e-107
UniRef100_Q684P6 S-adenosyl-L-methionine:benzoic acid/salicylic ... 360 4e-98
UniRef100_Q9SBK6 Jasmonate O-methyltransferase [Brassica rapa su... 323 6e-87
UniRef100_Q6XMI2 Methyl transferase [Arabidopsis lyrata subsp. l... 310 5e-83
UniRef100_Q9AR07 Jasmonate O-methyltransferase [Arabidopsis thal... 305 1e-81
UniRef100_Q6XMI3 Methyl transferase [Arabidopsis thaliana] 304 3e-81
UniRef100_Q70T29 Carboxyl methyltransferase [Bixa orellana] 301 2e-80
UniRef100_Q9FYZ9 SAM:benzoic acid carboxyl methyltransferase [An... 295 2e-78
UniRef100_Q6ZI02 Putative S-adenosyl-L-methionine:salicylic acid... 292 9e-78
UniRef100_Q9FJZ6 S-adenosyl-L-methionine:salicylic acid carboxyl... 291 2e-77
UniRef100_Q70SZ8 Carboxyl methyltransferase [Crocus sativus] 291 3e-77
UniRef100_Q9CAX9 Hypothetical protein F24K9.15 [Arabidopsis thal... 282 1e-74
UniRef100_Q67VH4 Putative benzothiadiazole-induced S-adenosyl-L-... 282 1e-74
UniRef100_Q8L5B5 Putative S-adenosyl-L-methionine:salicylic acid... 281 3e-74
>UniRef100_Q84UB4 S-adenosyl-L-methionine:benzoic acid/salicylic acid carboxyl
methyltransferase [Petunia hybrida]
Length = 357
Score = 444 bits (1143), Expect = e-123
Identities = 212/360 (58%), Positives = 286/360 (78%), Gaps = 5/360 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
MEV +VLHMNGG G++SYANNSLVQ+KVI +TKP+ ++A+ +Y++ ++L IADLGCS
Sbjct: 1 MEVVEVLHMNGGNGDSSYANNSLVQQKVILMTKPITEQAMIDLYSSLFPETLCIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
G+NT LV+ ++K+VEK +K KSPE+ + NDLPGNDFNT+F SL F+E L +
Sbjct: 61 LGANTFLVVSQLVKIVEKERKKHGFKSPEFYFHFNDLPGNDFNTLFQSLGAFQEDLRKHI 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G GPCFFSGVPGSF+ R+FP +SLHFV+SSYSL WLS+VP G +NNKGNIY++ TSPL
Sbjct: 121 GESFGPCFFSGVPGSFYTRLFPSKSLHFVYSSYSLMWLSQVPNGIENNKGNIYMARTSPL 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V+KAYYKQ++IDFS FLK R+EE+++GG M++T +GR+S++PTSKECCYIWELLAMALN
Sbjct: 181 SVIKAYYKQYEIDFSNFLKYRSEELMKGGKMVLTLLGRESEDPTSKECCYIWELLAMALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESES 300
+V EG+IKEEK++ FNIP Y PSP+EVK V EGSF +N+LE S V+WNA S +
Sbjct: 241 KLVEEGLIKEEKVDAFNIPQYTPSPAEVKYIVEKEGSFTINRLETSRVHWNA-----SNN 295
Query: 301 FGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ GYNV++CMRA+AEPLLVSHF + ++ +F++Y++ ++D SK +T+FIN+ + L K
Sbjct: 296 EKNGGYNVSRCMRAVAEPLLVSHFDKELMDLVFHKYEEIISDCMSKEKTEFINVIVSLTK 355
>UniRef100_Q84UB5 S-adenosyl-L-methionine:benzoic acid/salicylic acid carboxyl
methyltransferase [Petunia hybrida]
Length = 357
Score = 444 bits (1141), Expect = e-123
Identities = 213/360 (59%), Positives = 285/360 (79%), Gaps = 5/360 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
MEV +VLHMNGG G++SYANNSLVQ+KVI +TKP+ ++A+ +Y++ ++L IADLGCS
Sbjct: 1 MEVVEVLHMNGGNGDSSYANNSLVQQKVILMTKPITEQAMIDLYSSLFPETLCIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
G+NT LV+ ++K+VEK +K KSPE+ + NDLPGNDFNT+F SL F+E L +
Sbjct: 61 LGANTFLVVSQLVKIVEKERKKHGFKSPEFYFHFNDLPGNDFNTLFQSLGAFQEDLRKHI 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G GPCFFSGVPGSF+ R+FP +SLHFV+SSYSL WLS+VP G +NNKGNIY++ TSPL
Sbjct: 121 GESFGPCFFSGVPGSFYTRLFPSKSLHFVYSSYSLMWLSQVPNGIENNKGNIYMARTSPL 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V+KAYYKQ++IDFS FLK R+EE+++GG M++T +GR+S++PTSKECCYIWELLAMALN
Sbjct: 181 SVIKAYYKQYEIDFSNFLKYRSEELMKGGKMVLTLLGRESEDPTSKECCYIWELLAMALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESES 300
+V EG+IKEEK++ FNIP Y PSP+EVK V EGSF +N+LE S V+WNA S +
Sbjct: 241 KLVEEGLIKEEKVDAFNIPQYTPSPAEVKYIVEKEGSFTINRLETSRVHWNA-----SNN 295
Query: 301 FGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ GYNV++CMRA+AEPLLVSHF + ++ +F++Y++ ++D SK T+FIN+ I L K
Sbjct: 296 EKNGGYNVSRCMRAVAEPLLVSHFDKELMDLVFHKYEEIVSDCMSKENTEFINVIISLTK 355
>UniRef100_Q9AVG9 S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase
[Atropa belladonna]
Length = 357
Score = 429 bits (1102), Expect = e-119
Identities = 207/361 (57%), Positives = 285/361 (78%), Gaps = 5/361 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
M+V +VLHMNGG G+ SYANNSLVQRKVI +TKP+ ++AI+ +Y + ++L IADLGCS
Sbjct: 1 MKVVEVLHMNGGNGDISYANNSLVQRKVILMTKPITEQAISDLYCSFFPETLCIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
SG+NT LV+ +++K+VEK + N +S + + NDLPGNDFNTIF SL F++ L ++
Sbjct: 61 SGANTFLVVSELVKIVEKERKIHNLQSAGNLFHFNDLPGNDFNTIFQSLGKFQQDLRKQI 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G E GPCFFSGVPGSF+ R+FP +SLHFVHSSYSL WLS+VP+ + NK NIYI+STSP
Sbjct: 121 GEEFGPCFFSGVPGSFYTRLFPSESLHFVHSSYSLMWLSQVPDLIEKNKENIYIASTSPP 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V+KAYYKQ++ DFS FLK R+EE+++GG M++TF+GR+S++P+SKECCYIWELL+MALN
Sbjct: 181 SVIKAYYKQYEKDFSNFLKYRSEELMKGGKMVLTFLGRESEDPSSKECCYIWELLSMALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESES 300
++VLEG+I+EEK+++FNIP Y PSP EVK V EGSF++N+LE + V+WN S
Sbjct: 241 ELVLEGLIEEEKVDSFNIPQYTPSPEEVKYIVEKEGSFIINRLEATRVHWNV-----SNE 295
Query: 301 FGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ GYNVA+CMRA+AEPLLVS F + ++ +F +Y++ ++D SK +T+FIN+ + L K
Sbjct: 296 GINGGYNVAKCMRAVAEPLLVSQFDQKLMNLVFQKYEEIISDCISKEKTEFINVIVSLTK 355
Query: 361 R 361
+
Sbjct: 356 K 356
>UniRef100_Q8H6N2 S-adenosyl-L-methionine:salicylic acid methyltransferase
[Antirrhinum majus]
Length = 383
Score = 426 bits (1096), Expect = e-118
Identities = 214/371 (57%), Positives = 287/371 (76%), Gaps = 10/371 (2%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSK--SLTIADLG 58
M++A+VLHMNGG+G++SYA+NSLVQRKVI +TKP+ +EA+T Y L +++IADLG
Sbjct: 12 MKLAQVLHMNGGLGKSSYASNSLVQRKVISITKPIIEEAMTEFYTRMLPSPHTISIADLG 71
Query: 59 CSSGSNTLLVILDIIKVVEKLCRKLNHKSP-EYMIYLNDLPGNDFNTIFTSL-DIFKEKL 116
CS G NTLLV +++K++ KL +KL+ + P E+ I+LNDLPGNDFN+IF L +F+E+L
Sbjct: 72 CSCGPNTLLVAAELVKIIVKLRQKLDREPPPEFQIHLNDLPGNDFNSIFRYLLPMFREEL 131
Query: 117 LDEMG--TEMGP-CFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIY 173
+E+G E G CF SGVPGSF+GR+FP +SLHFVHSSYSL WLSKVPEG NK NIY
Sbjct: 132 REEIGGGEEAGRRCFVSGVPGSFYGRLFPTKSLHFVHSSYSLMWLSKVPEGVKMNKENIY 191
Query: 174 ISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWE 233
I+STSP NV+ AYY+QFQ DFS FL CR+EE++ GG M++TF+GRKS + SKECCYIWE
Sbjct: 192 IASTSPQNVINAYYEQFQRDFSSFLICRSEEVIGGGRMVLTFLGRKSASARSKECCYIWE 251
Query: 234 LLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW-NA 292
LL++AL +VLEG+I++EKL++F+IP Y PSP+EVK EV EGSF +N+LE+SE+ W +
Sbjct: 252 LLSLALKQLVLEGVIEKEKLHSFHIPQYTPSPTEVKAEVEKEGSFTVNRLEVSEITWASC 311
Query: 293 RDDFESESFGDDG--YNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTK 350
+DF G YNVA+CMR++AEPLL+ HFGE V+ +F +Y++ + DR S+ TK
Sbjct: 312 GNDFHLPELVASGNEYNVAKCMRSVAEPLLIEHFGESVIDRLFEKYREIIFDRMSREETK 371
Query: 351 FINITILLAKR 361
F N+TI + +R
Sbjct: 372 FFNVTISMTRR 382
>UniRef100_Q9AVR1 S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase
[Stephanotis floribunda]
Length = 366
Score = 419 bits (1078), Expect = e-116
Identities = 203/365 (55%), Positives = 274/365 (74%), Gaps = 4/365 (1%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
MEV +VLHMNGG G+ SYA+NSL+Q+KVI LTKP+ +EAIT +Y KS+ IAD+GCS
Sbjct: 1 MEVVEVLHMNGGTGDASYASNSLLQKKVILLTKPITEEAITELYTRLFPKSICIADMGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
SG NT L + ++IK VEK L H+SPEY I+LNDLP NDFNTIF SL F++ +M
Sbjct: 61 SGPNTFLAVSELIKNVEKKRTSLGHESPEYQIHLNDLPSNDFNTIFRSLPSFQKSFSKQM 120
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPL 180
G+ G CFF+GVPGSF+GR+FP +SLHFVHSSYSL WLS+VP+ + NKGNIY+SSTSPL
Sbjct: 121 GSGFGHCFFTGVPGSFYGRLFPNKSLHFVHSSYSLMWLSRVPDLEEVNKGNIYLSSTSPL 180
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALN 240
+V++AY KQFQ DF+ FL+CRAEE+V GG M++T +GRK ++ + KE Y ELLA ALN
Sbjct: 181 SVIRAYLKQFQRDFTTFLQCRAEELVPGGVMVLTLMGRKGEDHSGKESGYALELLARALN 240
Query: 241 DMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARD----DF 296
++V EG I+EE+L+ FN+P Y PSP+EVK V EGSF + +LE + ++W A D
Sbjct: 241 ELVSEGQIEEEQLDCFNVPQYTPSPAEVKYFVEEEGSFSITRLEATTIHWTAYDHDHVTG 300
Query: 297 ESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITI 356
+F D GY+++ C+RA+ EPLLV HFGE ++ E+F+RY++ LT+ +K + +FIN+T+
Sbjct: 301 HHHAFKDGGYSLSNCVRAVVEPLLVRHFGEAIMDEVFHRYREILTNCMTKEKIEFINVTV 360
Query: 357 LLAKR 361
+ +R
Sbjct: 361 SMKRR 365
>UniRef100_Q9SPV4 S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase
[Clarkia breweri]
Length = 359
Score = 416 bits (1068), Expect = e-115
Identities = 201/366 (54%), Positives = 281/366 (75%), Gaps = 12/366 (3%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYN-NTLSKSLTIADLGC 59
M+V +VLHM GG GE SYA NS +QR+VI +TKP+ + AIT++Y+ +T++ L IADLGC
Sbjct: 1 MDVRQVLHMKGGAGENSYAMNSFIQRQVISITKPITEAAITALYSGDTVTTRLAIADLGC 60
Query: 60 SSGSNTLLVILDIIKVVEKLCRKLNHK-SPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLD 118
SSG N L + ++IK VE+L +K+ + SPEY I+LNDLPGNDFN IF SL I E +D
Sbjct: 61 SSGPNALFAVTELIKTVEELRKKMGRENSPEYQIFLNDLPGNDFNAIFRSLPI--ENDVD 118
Query: 119 EMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTS 178
G CF +GVPGSF+GR+FP +LHF+HSSYSL WLS+VP G ++NKGNIY+++T
Sbjct: 119 ------GVCFINGVPGSFYGRLFPRNTLHFIHSSYSLMWLSQVPIGIESNKGNIYMANTC 172
Query: 179 PLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMA 238
P +V+ AYYKQFQ D +LFL+CRA+E+V GG M++T +GR+S++ S ECC IW+LLAMA
Sbjct: 173 PQSVLNAYYKQFQEDHALFLRCRAQEVVPGGRMVLTILGRRSEDRASTECCLIWQLLAMA 232
Query: 239 LNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNA--RDDF 296
LN MV EG+I+EEK++ FNIP Y PSP+EV+ E++ EGSF+++ +E SE+ W++ +D
Sbjct: 233 LNQMVSEGLIEEEKMDKFNIPQYTPSPTEVEAEILKEGSFLIDHIEASEIYWSSCTKDGD 292
Query: 297 ESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITI 356
S ++GYNVA+CMRA+AEPLL+ HFGE +++++F+RYK + +R SK +TKFIN+ +
Sbjct: 293 GGGSVEEEGYNVARCMRAVAEPLLLDHFGEAIIEDVFHRYKLLIIERMSKEKTKFINVIV 352
Query: 357 LLAKRS 362
L ++S
Sbjct: 353 SLIRKS 358
>UniRef100_Q8W414 S-adenosyl-L-methionine:salicylic acid calboxyl
methyltransferase-like protein [Cucumis sativus]
Length = 370
Score = 392 bits (1006), Expect = e-107
Identities = 195/368 (52%), Positives = 269/368 (72%), Gaps = 12/368 (3%)
Query: 6 VLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNT 65
+LH+NGG+G TSYANNS +QR++I +T + EA+T+ YN + S+TIADLGCSSG NT
Sbjct: 1 MLHVNGGMGNTSYANNSRLQREIISMTCSIAKEALTNFYNQHIPTSITIADLGCSSGQNT 60
Query: 66 LLVILDIIKVVEKLCRKLNHKSP-EYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEM 124
L+++ +IK VE++ +KL+ + P EY I+LNDL GNDFN +FTSL F E L ++G +
Sbjct: 61 LMLVSYLIKQVEEIRQKLHQRLPLEYQIFLNDLHGNDFNAVFTSLPRFLEDLGTQIGGDF 120
Query: 125 GPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVK 184
GPCFF+GVPGSF+ R+FP +S+HF HSS SLHWLS+VP G +NNKGNIYI STSP +V +
Sbjct: 121 GPCFFNGVPGSFYARLFPTKSVHFFHSSSSLHWLSRVPVGIENNKGNIYIGSTSPKSVGE 180
Query: 185 AYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALNDMVL 244
AYYKQFQ DFS+FLKCRAEE+V GG M++T VGR S++P+ YIWELL +ALN MV
Sbjct: 181 AYYKQFQKDFSMFLKCRAEELVMGGGMVLTLVGRTSEDPSKSGGYYIWELLGLALNTMVA 240
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWN---------ARDD 295
EGI++E+K ++FNIP Y PSP EV+ EV+ EGSF++NQL+ S +N N +
Sbjct: 241 EGIVEEKKADSFNIPYYIPSPKEVEAEVVKEGSFILNQLKASSINLNHTVHKTEEESSTP 300
Query: 296 FESESFGD-DGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGR-TKFIN 353
+ S D Y+ A+C+++++EPLL+ HFGE ++ E+F R++ + +K R + IN
Sbjct: 301 LINNSLADATDYDFAKCIQSVSEPLLIRHFGEAIMDELFIRHRNIVAGCMAKHRIMECIN 360
Query: 354 ITILLAKR 361
+TI L K+
Sbjct: 361 LTISLTKK 368
>UniRef100_Q684P6 S-adenosyl-L-methionine:benzoic acid/salicylic acid carboxyl
methyltransferase [Nicotiana suaveolens]
Length = 355
Score = 360 bits (923), Expect = 4e-98
Identities = 183/363 (50%), Positives = 257/363 (70%), Gaps = 13/363 (3%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
MEVAKVLHMN G+G+ SYA NSL Q+KVI +TK +RDEAI ++Y + +++ IADLGCS
Sbjct: 1 MEVAKVLHMNEGIGKASYAKNSLFQQKVILMTKSIRDEAIYALYRSLSPEAICIADLGCS 60
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHK-SPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDE 119
SG NT L I ++IK + + + K SPE+ ++LNDLPGNDFNTIF SL E L
Sbjct: 61 SGPNTFLTISELIKTIYEESKINGQKQSPEFQVFLNDLPGNDFNTIFRSLPALYEDLRKH 120
Query: 120 MGTEMGP-CFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTS 178
MG G CF +GV GSF+ R+FP S+HFVHSS+SLHWLS+VP G +NNKGNI ++STS
Sbjct: 121 MGDGFGTNCFVAGVAGSFYNRLFPSNSVHFVHSSFSLHWLSRVPHGIENNKGNIQVASTS 180
Query: 179 PLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMA 238
P +VV+AYY+Q++ DF FLK R+ E+V+GG M++T +GR +++ SK CYI E + MA
Sbjct: 181 PQDVVEAYYEQYERDFVNFLKLRSIELVKGGRMVLTVMGRNNEDRFSKASCYILEPMVMA 240
Query: 239 LNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFES 298
LN+++ EG I+EEK+ FNIP+YYPSP+EVK V EGSF ++ L+ SE++ ++ ++
Sbjct: 241 LNELIAEGSIEEEKVAAFNIPVYYPSPAEVKYIVEKEGSFAIDVLKTSEIHMDSSNE--- 297
Query: 299 ESFGDDGYNVAQCMRALAEPLLVSHFGEGV-VKEIFNRYKKYLTDRNSKGRTKFINITIL 357
YNV QCMRA EPL+V+HFG+ + + ++F++ + + +K +T IN+ +
Sbjct: 298 -------YNVTQCMRAFIEPLVVNHFGDELNMDQVFHKCGEIFDNIIAKEKTTSINVVVS 350
Query: 358 LAK 360
L K
Sbjct: 351 LTK 353
>UniRef100_Q9SBK6 Jasmonate O-methyltransferase [Brassica rapa subsp. pekinensis]
Length = 392
Score = 323 bits (827), Expect = 6e-87
Identities = 178/374 (47%), Positives = 236/374 (62%), Gaps = 42/374 (11%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITS-MYNNTLSKSLTIADLGC 59
MEV ++LHMN G GETSYA NS+VQ +I L + + DEA+ M N+ S IADLGC
Sbjct: 1 MEVMRILHMNKGNGETSYAKNSIVQSNIISLGRRVMDEALKKLMIRNSEILSFGIADLGC 60
Query: 60 SSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKL--- 116
SSG N+LL I +I++ ++ LC L+ PE + LNDLP NDFN IF SL F +++
Sbjct: 61 SSGPNSLLSISNIVETIQNLCHDLDRPVPELSLSLNDLPSNDFNYIFASLPEFYDRVKKR 120
Query: 117 ---LDEMGTEMG---PCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN--- 167
+ +G E G PCF S VPGSF+GR+FP +SLHFVHSS SLHWLS+VP G N
Sbjct: 121 DNNYESLGFEHGSGGPCFVSAVPGSFYGRLFPRRSLHFVHSSSSLHWLSQVPCGEVNKKD 180
Query: 168 ---------NKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGR 218
N+G IY+S TSP + K Y QFQ DFS+FL+ R+EE+V GG M+++F+GR
Sbjct: 181 GVVITADLDNRGKIYLSKTSPKSAHKVYALQFQTDFSVFLRSRSEELVPGGRMVLSFLGR 240
Query: 219 KSDNPTSKECCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSF 278
S +PT++E CY WELLA AL + EGII+EE ++ FN P Y SP E+K+ + EGSF
Sbjct: 241 SSPDPTTEESCYQWELLAQALMSLAKEGIIEEENIDAFNAPYYAASPEELKMAIEKEGSF 300
Query: 279 VMNQLEISEVNWNARDDFESESFGDDGYN--------------VAQCMRALAEPLLVSHF 324
+++LEIS V+W E S DD Y+ VA+ +RA+ EP+L F
Sbjct: 301 SIDRLEISPVDW------EGGSISDDSYDIVRFKPEALASGRRVAKTIRAVVEPMLEPTF 354
Query: 325 GEGVVKEIFNRYKK 338
G+ V+ E+F RY K
Sbjct: 355 GQKVMDELFERYAK 368
>UniRef100_Q6XMI2 Methyl transferase [Arabidopsis lyrata subsp. lyrata]
Length = 380
Score = 310 bits (793), Expect = 5e-83
Identities = 154/359 (42%), Positives = 231/359 (63%), Gaps = 7/359 (1%)
Query: 5 KVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNT-LSKSLTIADLGCSSGS 63
K L M+GG G SY+ NS +QRKV+ + KP+ + M + + +A+LGCSSG
Sbjct: 27 KALCMSGGDGTNSYSANSRLQRKVLTMAKPVLVKTTEEMMMSLDFPTYIKVAELGCSSGQ 86
Query: 64 NTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTE 123
NT L I +II + LC+ +N PE LNDLP NDFNT F + F ++L M T
Sbjct: 87 NTFLAISEIINTISVLCQHVNKNPPEIDCCLNDLPENDFNTTFKFVPFFNKEL---MITS 143
Query: 124 MGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVV 183
CF G PGSF+ R+F SLH +HSSY+LHWLSKVPE +NNKGN+YI+S+SP +
Sbjct: 144 KASCFVYGAPGSFYSRLFSRNSLHIIHSSYALHWLSKVPEKLENNKGNVYITSSSPQSAY 203
Query: 184 KAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-DNPTSKECCYIWELLAMALNDM 242
KAY QFQ DFS+FL+ R+EEIV G M++TF+GR + ++P ++CC+ W LL+ +L D+
Sbjct: 204 KAYLNQFQKDFSMFLRLRSEEIVSNGRMVLTFIGRNTLNDPLYRDCCHFWTLLSKSLRDL 263
Query: 243 VLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFG 302
V EG++ E KL+ FN+P Y P+ E+K + EGSF +N+LE + ++E + +
Sbjct: 264 VFEGLVSESKLDAFNMPFYDPNVQELKQVIRNEGSFEINELETHGFDL-GHSNYEEDDY- 321
Query: 303 DDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAKR 361
+ G++ A C+RA++EP+LV+HFGE ++ +F++Y ++T + +++ + L K+
Sbjct: 322 EAGHDEANCIRAVSEPMLVAHFGEDIIDTLFDKYAHHVTQHANCRNKTTVSLVVSLTKK 380
>UniRef100_Q9AR07 Jasmonate O-methyltransferase [Arabidopsis thaliana]
Length = 389
Score = 305 bits (782), Expect = 1e-81
Identities = 174/366 (47%), Positives = 228/366 (61%), Gaps = 29/366 (7%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITS-MYNNTLSKSLTIADLGC 59
MEV +VLHMN G GETSYA NS Q +I L + + DEA+ M +N+ S+ IADLGC
Sbjct: 1 MEVMRVLHMNKGNGETSYAKNSTAQSNIISLGRRVMDEALKKLMMSNSEISSIGIADLGC 60
Query: 60 SSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDE 119
SSG N+LL I +I+ + LC L+ PE + LNDLP NDFN I SL F +++ +
Sbjct: 61 SSGPNSLLSISNIVDTIHNLCPDLDRPVPELRVSLNDLPSNDFNYICASLPEFYDRVNNN 120
Query: 120 ---MGTEMG---PCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP----EGADN-- 167
+G G CF S VPGSF+GR+FP +SLHFVHSS SLHWLS+VP E D
Sbjct: 121 KEGLGFGRGGGESCFVSAVPGSFYGRLFPRRSLHFVHSSSSLHWLSQVPCREAEKEDRTI 180
Query: 168 -----NKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDN 222
N G IYIS TSP + KAY QFQ DF +FL+ R+EE+V GG M+++F+GR+S +
Sbjct: 181 TADLENMGKIYISKTSPKSAHKAYALQFQTDFLVFLRSRSEELVPGGRMVLSFLGRRSLD 240
Query: 223 PTSKECCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQ 282
PT++E CY WELLA AL M EGII+EEK++ FN P Y S E+K+ + EGSF +++
Sbjct: 241 PTTEESCYQWELLAQALMSMAKEGIIEEEKIDAFNAPYYAASSEELKMVIEKEGSFSIDR 300
Query: 283 LEISEVNWNARDDFESESFG----------DDGYNVAQCMRALAEPLLVSHFGEGVVKEI 332
LEIS ++W ES+ G V+ +RA+ EP+L FGE V+ E+
Sbjct: 301 LEISPIDWEG-GSISEESYDLVIRSKPEALASGRRVSNTIRAVVEPMLEPTFGENVMDEL 359
Query: 333 FNRYKK 338
F RY K
Sbjct: 360 FERYAK 365
>UniRef100_Q6XMI3 Methyl transferase [Arabidopsis thaliana]
Length = 379
Score = 304 bits (778), Expect = 3e-81
Identities = 156/362 (43%), Positives = 229/362 (63%), Gaps = 14/362 (3%)
Query: 5 KVLHMNGGVGETSYANNSLVQRKVIYLTKP-LRDEAITSMYNNTLSKSLTIADLGCSSGS 63
K L M+GG G SY+ NS +Q+KV+ + KP L M N + +A+LGCSSG
Sbjct: 27 KALCMSGGDGANSYSANSRLQKKVLSMAKPVLVRNTEEMMMNLDFPTYIKVAELGCSSGQ 86
Query: 64 NTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTE 123
N+ L I +II + LC+ +N SPE LNDLP NDFNT F + F ++L M T
Sbjct: 87 NSFLAIFEIINTINVLCQHVNKNSPEIDCCLNDLPENDFNTTFKFVPFFNKEL---MITN 143
Query: 124 MGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVV 183
CF G PGSF+ R+F SLH +HSSY+LHWLSKVPE +NNKGN+YI+S+SP +
Sbjct: 144 KSSCFVYGAPGSFYSRLFSRNSLHLIHSSYALHWLSKVPEKLENNKGNLYITSSSPQSAY 203
Query: 184 KAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-DNPTSKECCYIWELLAMALNDM 242
KAY QFQ DF++FL+ R+EEIV G M++TF+GR + ++P ++CC+ W LL+ +L D+
Sbjct: 204 KAYLNQFQKDFTMFLRLRSEEIVSNGRMVLTFIGRNTLNDPLYRDCCHFWTLLSNSLRDL 263
Query: 243 VLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLE---ISEVNWNARDDFESE 299
V EG++ E KL+ FN+P Y P+ E+K + EGSF +N+LE ++ DDFE+
Sbjct: 264 VFEGLVSESKLDAFNMPFYDPNVQELKEVIQKEGSFEINELESHGFDLGHYYEEDDFEA- 322
Query: 300 SFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLA 359
G N A +RA++EP+L++HFGE ++ +F++Y ++T + +++ + L
Sbjct: 323 -----GRNEANGIRAVSEPMLIAHFGEEIIDTLFDKYAYHVTQHANCRNKTTVSLVVSLT 377
Query: 360 KR 361
K+
Sbjct: 378 KK 379
>UniRef100_Q70T29 Carboxyl methyltransferase [Bixa orellana]
Length = 375
Score = 301 bits (770), Expect = 2e-80
Identities = 167/372 (44%), Positives = 234/372 (62%), Gaps = 15/372 (4%)
Query: 3 VAKVLHMNGGVGETSYANNSLVQRKVIY-LTKPLRDEAITSMYNNTLSKS-LTIADLGCS 60
V L M GG G+ SYA NSL+QRK + +TKPL + AI +Y ++ L IADLGCS
Sbjct: 4 VRSTLCMTGGTGDASYAQNSLLQRKALSKITKPLTEAAIKELYATIKPQTRLVIADLGCS 63
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
SG NT L + +++ V + +K SPE LNDLP NDFNT+F S+D F +K +
Sbjct: 64 SGPNTFLAVSELVDAVGEFRKKATRNSPEIQTNLNDLPRNDFNTLFRSVDKFNQKA--KA 121
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NKGNIYI 174
E F SGVPGSF+ R+FP +S+HF+HSSY+ HWLS+VP+G N NKGNIYI
Sbjct: 122 VDEDNIYFVSGVPGSFYNRLFPSESIHFIHSSYARHWLSQVPKGRTNDAGLERNKGNIYI 181
Query: 175 SSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGR-KSDNPTSKECCYIWE 233
+++SP +V KAY +QFQ DF+ FLK R+ E GG M++ FVG+ +S + +ECC ++
Sbjct: 182 ANSSPQSVWKAYLRQFQTDFANFLKIRSRENKPGGRMVLAFVGKDESPLASRQECCAVYN 241
Query: 234 LLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNA- 292
LLAMAL+ +V EG++ + K++ FN+P Y PSP E+ V GSF + +LE E W +
Sbjct: 242 LLAMALSGLVAEGLLADSKVDQFNLPKYNPSPQEIMPLVRKVGSFEIAKLENHERQWESC 301
Query: 293 --RDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTK 350
D + + G NVAQ +RA+AEP L HFG+ +++E+F RY K + + + K
Sbjct: 302 PQDADGRTSNALQSGQNVAQTIRAVAEPALEKHFGDAIMEELFTRYAKLVAKHLTAEKRK 361
Query: 351 FI-NITILLAKR 361
F+ N+ L KR
Sbjct: 362 FVLNVMQLTKKR 373
>UniRef100_Q9FYZ9 SAM:benzoic acid carboxyl methyltransferase [Antirrhinum majus]
Length = 364
Score = 295 bits (754), Expect = 2e-78
Identities = 163/370 (44%), Positives = 235/370 (63%), Gaps = 24/370 (6%)
Query: 5 KVLHMN-GGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLS--KSLTIADLGCSS 61
K+L MN G GETSYANNS +Q+ ++ + + DE + + + + K + D+GCSS
Sbjct: 6 KLLCMNIAGDGETSYANNSGLQKVMMSKSLHVLDETLKDIIGDHVGFPKCFKMMDMGCSS 65
Query: 62 GSNTLLVILDIIKVVEKLCRKLN-HKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
G N LLV+ II +E L + N ++ PE+ ++LNDLP NDFN +F KLL
Sbjct: 66 GPNALLVMSGIINTIEDLYTEKNINELPEFEVFLNDLPDNDFNNLF--------KLLSH- 116
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGA-DNNKGNIYISSTSP 179
E G CF G+PGSF+GR+ P +SLHF +SSYS+HWLS+VPEG DNN+ NIY+++ SP
Sbjct: 117 --ENGNCFVYGLPGSFYGRLLPKKSLHFAYSSYSIHWLSQVPEGLEDNNRQNIYMATESP 174
Query: 180 LNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMAL 239
V KAY KQ++ DFS FLK R EEIV GG M++TF GR ++P+SK+ I+ LLA L
Sbjct: 175 PEVYKAYAKQYERDFSTFLKLRGEEIVPGGRMVLTFNGRSVEDPSSKDDLAIFTLLAKTL 234
Query: 240 NDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESE 299
DMV EG++K + L +FNIPIY P EV+ +++EGSF +++LE+ V W+A D + +
Sbjct: 235 VDMVAEGLVKMDDLYSFNIPIYSPCTREVEAAILSEGSFTLDRLEVFRVCWDASDYTDDD 294
Query: 300 SFGD--------DGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKF 351
D G VA C+RA+ EP+L SHFG ++ +F +Y K + + S + +
Sbjct: 295 DQQDPSIFGKQRSGKFVADCVRAITEPMLASHFGSTIMDLLFGKYAKKIVEHLSVENSSY 354
Query: 352 INITILLAKR 361
+I + L++R
Sbjct: 355 FSIVVSLSRR 364
>UniRef100_Q6ZI02 Putative S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase [Oryza sativa]
Length = 380
Score = 292 bits (748), Expect = 9e-78
Identities = 159/380 (41%), Positives = 233/380 (60%), Gaps = 20/380 (5%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKS-------LT 53
M+V + LHM+ G GETSYA NS +Q K I T+PL +A+ + + S +
Sbjct: 1 MKVEQDLHMSRGDGETSYAANSRLQEKAILKTRPLLHKAVEEAHASLSGLSRAPAGGKMV 60
Query: 54 IADLGCSSGSNTLLVILDIIKVVEKLCRKLNHKSP---EYMIYLNDLPGNDFNTIFTSLD 110
+ADLGCSSG NTLLV+ +++ V +HKS + +LNDLPGNDFN +F SL+
Sbjct: 61 VADLGCSSGPNTLLVVSEVLSAVANRS-SCDHKSSLVADVQFFLNDLPGNDFNLVFQSLE 119
Query: 111 IFKEKLLDEMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPE----GAD 166
+FK+ E G + P + +G+PGSF+ R+FP +S+H HSSY L W SKVP+ G
Sbjct: 120 LFKKLAEMEFGKALPPYYIAGLPGSFYTRLFPDRSVHLFHSSYCLMWRSKVPDKLASGEV 179
Query: 167 NNKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSK 226
N GN+YI T+P +VVK Y +QFQ DFS FL R +E+V GG M++TF+GRK+ +
Sbjct: 180 LNAGNMYIWETTPPSVVKLYQRQFQEDFSQFLALRHDELVSGGQMVLTFLGRKNRDVLRG 239
Query: 227 ECCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEIS 286
E Y++ LLA AL +V EG ++EEKL++FN+P Y PS EVK + G F ++ +++
Sbjct: 240 EVSYMYGLLAQALQSLVQEGRVEEEKLDSFNLPFYSPSVDEVKAVIRQSGLFDISHIQLF 299
Query: 287 EVNWNARDDFESESFGD-----DGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLT 341
E NW+ +DD + + G NVA+C+RA+ EPL+ HFG +V ++F+ Y + +
Sbjct: 300 ESNWDPQDDSDDDDVATLDSVRSGVNVARCIRAVLEPLIARHFGRCIVDDLFDMYARNVA 359
Query: 342 DRNSKGRTKFINITILLAKR 361
+ +TK+ I + L R
Sbjct: 360 QHLEQVKTKYPVIVLSLKAR 379
>UniRef100_Q9FJZ6 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein [Arabidopsis thaliana]
Length = 354
Score = 291 bits (745), Expect = 2e-77
Identities = 149/359 (41%), Positives = 221/359 (61%), Gaps = 11/359 (3%)
Query: 9 MNGGVGETSYANNSLVQRKVIYLTKP-LRDEAITSMYNNTLSKSLTIADLGCSSGSNTLL 67
M+GG G+ SY+ NSL+Q+KV+ KP L M N + +ADLGC++G NT L
Sbjct: 1 MSGGDGDNSYSTNSLLQKKVLSKAKPVLVKNTKGMMINLNFPNYIKVADLGCATGENTFL 60
Query: 68 VILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGPC 127
+ +I+ + LC++ N K PE LNDLP NDFNT F + F +++ + C
Sbjct: 61 TMAEIVNTINVLCQQCNQKPPEIDCCLNDLPDNDFNTTFKFVPFFNKRVKSKR-----LC 115
Query: 128 FFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNVVKAYY 187
F SGVPGSF+ R+FP +SLHFVHSSYSLHWLSKVP+G + N ++YI+++SP N KAY
Sbjct: 116 FVSGVPGSFYSRLFPRKSLHFVHSSYSLHWLSKVPKGLEKNSSSVYITTSSPPNAYKAYL 175
Query: 188 KQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-DNPTSKECCYIWELLAMALNDMVLEG 246
QFQ DF FL+ R+EE+V G M++TF+GRK+ D+P ++CC+ W LL+ +L D+V EG
Sbjct: 176 NQFQSDFKSFLEMRSEEMVSNGRMVLTFIGRKTLDDPLHRDCCHFWTLLSTSLRDLVYEG 235
Query: 247 IIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW---NARDDFESES-FG 302
++ K+++FNIP Y PS EV + EGSF +N LEI N +D+ S
Sbjct: 236 LVSASKVDSFNIPFYDPSKEEVMEMIRNEGSFEINDLEIHGFELGLSNHDEDYMLHSQIS 295
Query: 303 DDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAKR 361
G A C+RA++E +LV+ FG ++ +F ++ +++ S + + + L ++
Sbjct: 296 KAGQREANCIRAVSESMLVADFGVDIMDTLFKKFAYHVSQHASCTNKTTVTLVVSLIRK 354
>UniRef100_Q70SZ8 Carboxyl methyltransferase [Crocus sativus]
Length = 372
Score = 291 bits (744), Expect = 3e-77
Identities = 169/347 (48%), Positives = 220/347 (62%), Gaps = 19/347 (5%)
Query: 7 LHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTL 66
L M GG GET+YA NSL+Q K I TKP+ +EAI +YN+ KSL +ADLGCSSG NT
Sbjct: 8 LCMGGGDGETNYAKNSLIQDKAISRTKPIVEEAIKEVYNSLNPKSLVVADLGCSSGPNTF 67
Query: 67 LVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGP 126
LVI +I++ + CRKL H PE LNDLPGNDFNT+F + FKEK L E+ E+ P
Sbjct: 68 LVISEIVEAIGDHCRKLGHNPPEIQYILNDLPGNDFNTLFDYSEKFKEK-LKEVEEEVVP 126
Query: 127 CFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NKGNIYISSTSPL 180
+ GVPGSF+GR+FP S+HF+HSSYSLHWLS+VP+G + NK NIYI+ +SP
Sbjct: 127 -YVVGVPGSFYGRLFPQSSVHFIHSSYSLHWLSQVPQGLKSDTGLPLNKRNIYIAKSSPQ 185
Query: 181 NVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYI---WELLAM 237
V ++Y KQFQ+ + +GG MI+ F G+ D+ T C + + LLA
Sbjct: 186 IVAESYLKQFQMGLFSISHVKILITRDGGPMILIFFGK--DDRTKAPCGELSSCFGLLAD 243
Query: 238 ALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDF- 296
ALN MVLEGI+ E K+ FN+PIY S EV V T G F + Q+EI E NW+ DD
Sbjct: 244 ALNAMVLEGIMNEAKVEDFNLPIYAASMEEVMTIVETIGLFHVEQVEIFETNWDPFDDSS 303
Query: 297 -ESESFGDD---GYNVAQC-MRALAEPLLVSHFGEGVVKEIFNRYKK 338
+ ES D+ G NV C +RA+ EP+ +FGE ++ E+F+RY K
Sbjct: 304 DDDESAFDNFASGKNVVNCSIRAVVEPMFEKYFGEAIMDELFSRYAK 350
>UniRef100_Q9CAX9 Hypothetical protein F24K9.15 [Arabidopsis thaliana]
Length = 327
Score = 282 bits (721), Expect = 1e-74
Identities = 140/323 (43%), Positives = 207/323 (63%), Gaps = 13/323 (4%)
Query: 43 MYNNTLSKSLTIADLGCSSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDF 102
M N + +A+LGCSSG N+ L I +II + LC+ +N SPE LNDLP NDF
Sbjct: 14 MMNLDFPTYIKVAELGCSSGQNSFLAIFEIINTINVLCQHVNKNSPEIDCCLNDLPENDF 73
Query: 103 NTIFTSLDIFKEKLLDEMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP 162
NT F + F ++L M T CF G PGSF+ R+F SLH +HSSY+LHWLSKVP
Sbjct: 74 NTTFKFVPFFNKEL---MITNKSSCFVYGAPGSFYSRLFSRNSLHLIHSSYALHWLSKVP 130
Query: 163 EGADNNKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKS-D 221
E +NNKGN+YI+S+SP + KAY QFQ DF++FL+ R+EEIV G M++TF+GR + +
Sbjct: 131 EKLENNKGNLYITSSSPQSAYKAYLNQFQKDFTMFLRLRSEEIVSNGRMVLTFIGRNTLN 190
Query: 222 NPTSKECCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMN 281
+P ++CC+ W LL+ +L D+V EG++ E KL+ FN+P Y P+ E+K + EGSF +N
Sbjct: 191 DPLYRDCCHFWTLLSNSLRDLVFEGLVSESKLDAFNMPFYDPNVQELKEVIQKEGSFEIN 250
Query: 282 QLE---ISEVNWNARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKK 338
+LE ++ DDFE+ G N A +RA++EP+L++HFGE ++ +F++Y
Sbjct: 251 ELESHGFDLGHYYEEDDFEA------GRNEANGIRAVSEPMLIAHFGEEIIDTLFDKYAY 304
Query: 339 YLTDRNSKGRTKFINITILLAKR 361
++T + +++ + L K+
Sbjct: 305 HVTQHANCRNKTTVSLVVSLTKK 327
>UniRef100_Q67VH4 Putative benzothiadiazole-induced S-adenosyl-L-methionine:salicylic
acid carboxyl methyltransferase 1 [Oryza sativa]
Length = 375
Score = 282 bits (721), Expect = 1e-74
Identities = 155/365 (42%), Positives = 220/365 (59%), Gaps = 11/365 (3%)
Query: 8 HMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNTLL 67
HM GG GE SYA NS VQ K + K + D+AI +Y L+ + +ADLGCSSG NTL
Sbjct: 10 HMVGGEGEISYAKNSRVQAKAMIEAKFVLDKAIRELYATLLANIMVVADLGCSSGQNTLH 69
Query: 68 VILDIIKVVEKLCRKLNHKSP-EYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMGP 126
+ ++I + K L + +LNDLPGNDFN +F L+ F K ++ P
Sbjct: 70 FVSEVINIFTKHQNNLGQSDTVDLQFFLNDLPGNDFNHLFRILNTFTFKGASNHKGDILP 129
Query: 127 CF-FSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NKGNIYISSTSP 179
+ G PGS++ R+FP Q++H HSS SLHW S+VPE + N+ NIYI+ T+P
Sbjct: 130 AYHIYGAPGSYYTRLFPPQAVHLFHSSLSLHWRSQVPEQLNGKQKSYLNEENIYITKTTP 189
Query: 180 LNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMAL 239
L+VVK + +QF D SLFLK R EE+V+GG M++T GRKS++P S + I+ LL +L
Sbjct: 190 LHVVKLFQEQFIKDVSLFLKLRHEELVDGGRMVLTIYGRKSEDPYSGDVNDIFGLLGKSL 249
Query: 240 NDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDDFESE 299
+V EG++++EKL++FN+P+Y PS E++ V F M+ + + E NW+ DD + +
Sbjct: 250 QSLVAEGLVEKEKLDSFNLPVYGPSVGELEEIVNRVNLFDMDHMHLFECNWDPYDDSQGD 309
Query: 300 SFGD---DGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITI 356
D G NVA C+RA+ EPL+ SHFGEG++ +F Y + K +TKF I I
Sbjct: 310 IVHDSALSGINVANCVRAVTEPLIASHFGEGILSALFTDYAHRVASHLEKEKTKFAWIVI 369
Query: 357 LLAKR 361
L KR
Sbjct: 370 SLKKR 374
>UniRef100_Q8L5B5 Putative S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase [Oryza sativa]
Length = 375
Score = 281 bits (718), Expect = 3e-74
Identities = 154/371 (41%), Positives = 224/371 (59%), Gaps = 22/371 (5%)
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
M V VLHM G+GETSYA NS + + K L + +Y + + + +ADLGCS
Sbjct: 15 MNVETVLHMKEGLGETSYAQNS----RGMDTLKSLITNSAADVYLSQMPERFAVADLGCS 70
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLL--- 117
SG N L + DII + ++C + + PE+ + LNDLP NDFNTIF SL F ++L
Sbjct: 71 SGPNALCLAEDIIGSIGRICCRSSRPPPEFSVLLNDLPTNDFNTIFFSLPEFTDRLKAAA 130
Query: 118 --DEMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NK 169
DE G M F SGVPGSF+GR+FP +S+HFV S SLHWLS+VP G + NK
Sbjct: 131 KSDEWGRPM--VFLSGVPGSFYGRLFPAKSVHFVCSCSSLHWLSQVPSGLLDEMNRPINK 188
Query: 170 GNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECC 229
G +YISSTSPL V AY +QFQ DFSLFLK RA E+ GG M++ +GR++D +
Sbjct: 189 GKMYISSTSPLAVPVAYLRQFQRDFSLFLKSRAAEVFSGGRMVLAMLGRQADGYIDRRTT 248
Query: 230 YIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVN 289
++WELL+ + +V +G+++E+K++ +N+P Y PS E++ EV EGSF M+ ++ E+N
Sbjct: 249 FLWELLSESFASLVAQGLVEEDKVDAYNVPFYAPSIGEIEEEVRREGSFRMDYVQTYEIN 308
Query: 290 WNARDDFESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRT 349
++ D DG V+ +RA+ E +L HFG +V +F +Y + +T +
Sbjct: 309 LSSSGDARR-----DGRTVSMAIRAIQESMLSHHFGPEIVDALFAKYTELVTASMEREEV 363
Query: 350 KFINITILLAK 360
K + I ++L +
Sbjct: 364 KSVQIGVVLTR 374
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 620,652,862
Number of Sequences: 2790947
Number of extensions: 26852227
Number of successful extensions: 61803
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 61247
Number of HSP's gapped (non-prelim): 145
length of query: 362
length of database: 848,049,833
effective HSP length: 128
effective length of query: 234
effective length of database: 490,808,617
effective search space: 114849216378
effective search space used: 114849216378
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC125480.15


