

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125479.4 - phase: 0
(110 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
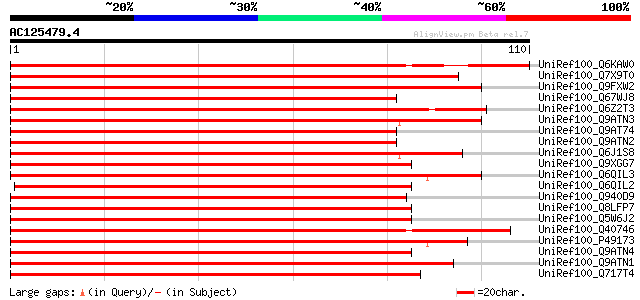
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6KAW0 Nod26-like major intrinsic protein [Cicer ariet... 167 7e-41
UniRef100_Q7X9T0 Nod26-like protein [Cucurbita pepo] 130 7e-30
UniRef100_Q9FXW2 MIP [Adiantum capillus-veneris] 119 2e-26
UniRef100_Q67WJ8 Putative NOD26-like membrane integral protein [... 117 8e-26
UniRef100_Q6Z2T3 Putative major intrinsic protein [Oryza sativa] 117 8e-26
UniRef100_Q9ATN3 NOD26-like membrane integral protein ZmNIP2-1 [... 116 1e-25
UniRef100_Q9AT74 NOD26-like membrane integral protein ZmNIP2-3 [... 115 2e-25
UniRef100_Q9ATN2 NOD26-like membrane integral protein ZmNIP2-2 [... 115 3e-25
UniRef100_Q6J1S8 Nodulin 26-like protein [Medicago truncatula] 110 8e-24
UniRef100_Q9XGG7 Nodulin26-like major intrinsic protein [Pisum s... 109 1e-23
UniRef100_Q6QIL3 NIP3 [Medicago truncatula] 109 1e-23
UniRef100_Q6QIL2 NIP2 [Medicago truncatula] 109 1e-23
UniRef100_Q940D9 Early embryogenesis aquaglyceroporin [Pinus taeda] 108 2e-23
UniRef100_Q8LFP7 Aquaporin NIP1.2 [Arabidopsis thaliana] 108 2e-23
UniRef100_Q5W6J2 Hypothetical protein OSJNBb0115F21.2 [Oryza sat... 108 3e-23
UniRef100_Q40746 Major intrinsic protein [Oryza sativa] 108 3e-23
UniRef100_P49173 Probable aquaporin NIP-type [Nicotiana alata] 106 1e-22
UniRef100_Q9ATN4 NOD26-like membrane integral protein ZmNIP1-1 [... 105 2e-22
UniRef100_Q9ATN1 NOD26-like membrane integral protein ZmNIP3-1 [... 105 2e-22
UniRef100_Q717T4 Nodulin-like intrinsic protein NIP1-1 [Atriplex... 105 2e-22
>UniRef100_Q6KAW0 Nod26-like major intrinsic protein [Cicer arietinum]
Length = 273
Score = 167 bits (422), Expect = 7e-41
Identities = 89/110 (80%), Positives = 96/110 (86%), Gaps = 6/110 (5%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
MV ISTAV+TD KA GEL+GVAVGSSV IASIVAGPISGGSMNPARTLGPAIATSSYKGI
Sbjct: 160 MVLISTAVSTDPKAIGELSGVAVGSSVCIASIVAGPISGGSMNPARTLGPAIATSSYKGI 219
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHKQDLATSQSPLSVKIHNEMNGIEL 110
W+YMVGPITGALLG WSYVVIQET +KQ L T S+K+H+EM GIEL
Sbjct: 220 WVYMVGPITGALLGTWSYVVIQET-NKQALTT-----SLKLHHEMKGIEL 263
Score = 39.3 bits (90), Expect = 0.022
Identities = 35/108 (32%), Positives = 56/108 (51%), Gaps = 11/108 (10%)
Query: 1 MVFISTAVAT-----DSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATS 55
+VF+ + A ++K + A +A G VT+ G ISG MNPA +L A AT
Sbjct: 44 LVFVGSGSAAMNAIDENKVSKLGASMAGGFIVTVMIYAIGHISGAHMNPAVSL--AFATV 101
Query: 56 S---YKGIWIYMVGPITGALLGAWSYVVIQETDHKQDLATSQSPLSVK 100
S +K + Y+ +TGA+ +++ V+ E KQ ATS S +++
Sbjct: 102 SHFPWKQVPFYIAAQLTGAISASYTLKVLLEPS-KQLGATSPSGSNIQ 148
>UniRef100_Q7X9T0 Nod26-like protein [Cucurbita pepo]
Length = 288
Score = 130 bits (327), Expect = 7e-30
Identities = 60/95 (63%), Positives = 77/95 (80%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
M+F++ AVATD+KA GELAG+AVGS+V I SI+AGP+SGGSMNP RTLGPA+A+ +YKG+
Sbjct: 176 MMFVTCAVATDTKAVGELAGLAVGSAVCITSILAGPVSGGSMNPVRTLGPAMASDNYKGL 235
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHKQDLATSQS 95
W+Y VGP+TG LLGAWSY I+ +D L + S
Sbjct: 236 WVYFVGPVTGTLLGAWSYKFIRASDKPVHLISPHS 270
Score = 34.3 bits (77), Expect = 0.71
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 6/83 (7%)
Query: 1 MVFISTAVA----TDSKATGEL-AGVAVGSSVTIASIVAGPISGGSMNPARTLG-PAIAT 54
+VF++ A +D++ +L A VA G VT+ G ISG MNPA T A
Sbjct: 60 LVFVTCGAAALNGSDAQRVSQLGASVAGGLIVTVMIYAVGHISGAHMNPAVTTAFAATRH 119
Query: 55 SSYKGIWIYMVGPITGALLGAWS 77
+K + +Y ++GA A++
Sbjct: 120 FPWKQVPLYAAAQLSGATCAAFT 142
>UniRef100_Q9FXW2 MIP [Adiantum capillus-veneris]
Length = 282
Score = 119 bits (298), Expect = 2e-26
Identities = 55/100 (55%), Positives = 76/100 (76%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ +AVATD++A GELAG+AVGS+V + +I AGPI G SMNPAR++GPA+A+ +K +
Sbjct: 178 LMFVVSAVATDTRAIGELAGLAVGSAVALDAIFAGPICGASMNPARSIGPAVASYDFKSL 237
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHKQDLATSQSPLSVK 100
W+Y+VGPI G LLGAWSY +I+ + QDLA S K
Sbjct: 238 WVYIVGPILGCLLGAWSYTMIKLPEQPQDLAMISQSKSFK 277
>UniRef100_Q67WJ8 Putative NOD26-like membrane integral protein [Oryza sativa]
Length = 298
Score = 117 bits (292), Expect = 8e-26
Identities = 53/82 (64%), Positives = 67/82 (81%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
M+F++ AVATDS+A GELAG+AVGS+V I SI AGP+SGGSMNPARTL PA+A++ Y G+
Sbjct: 180 MMFVTCAVATDSRAVGELAGLAVGSAVCITSIFAGPVSGGSMNPARTLAPAVASNVYTGL 239
Query: 61 WIYMVGPITGALLGAWSYVVIQ 82
WIY +GP+ G L GAW Y I+
Sbjct: 240 WIYFLGPVVGTLSGAWVYTYIR 261
Score = 31.2 bits (69), Expect = 6.0
Identities = 26/82 (31%), Positives = 36/82 (43%), Gaps = 6/82 (7%)
Query: 1 MVFISTAVAT----DSKATGELAGVAVGSSVTIASIVA-GPISGGSMNPARTLGPAIATS 55
+VF++ A+ D K +L VG + I A G ISG MNPA TL A
Sbjct: 64 LVFVTCGAASIYGEDMKRISQLGQSVVGGLIVTVMIYATGHISGAHMNPAVTLSFAFFRH 123
Query: 56 -SYKGIWIYMVGPITGALLGAW 76
+ + Y TGA+ A+
Sbjct: 124 FPWIQVPFYWAAQFTGAMCAAF 145
>UniRef100_Q6Z2T3 Putative major intrinsic protein [Oryza sativa]
Length = 298
Score = 117 bits (292), Expect = 8e-26
Identities = 57/101 (56%), Positives = 77/101 (75%), Gaps = 1/101 (0%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
M+F++ AVATD++A GELAG+AVGS+V I SI AG ISGGSMNPARTLGPA+A++ + G+
Sbjct: 177 MMFVTLAVATDTRAVGELAGLAVGSAVCITSIFAGAISGGSMNPARTLGPALASNKFDGL 236
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHKQDLATSQSPLSVKI 101
WIY +GP+ G L GAW+Y I+ D ++ +SQ S K+
Sbjct: 237 WIYFLGPVMGTLSGAWTYTFIRFEDTPKE-GSSQKLSSFKL 276
Score = 33.1 bits (74), Expect = 1.6
Identities = 24/84 (28%), Positives = 39/84 (45%), Gaps = 4/84 (4%)
Query: 21 VAVGSSVTIASIVAGPISGGSMNPARTLGPAIATS-SYKGIWIYMVGPITGALLGAWSYV 79
+A G VT+ G ISG MNPA TL A+ + + Y TGA+ ++
Sbjct: 86 IAGGLIVTVMIYAVGHISGAHMNPAVTLAFAVFRHFPWIQVPFYWAAQFTGAICASF--- 142
Query: 80 VIQETDHKQDLATSQSPLSVKIHN 103
V++ H D+ + +P+ H+
Sbjct: 143 VLKAVIHPVDVIGTTTPVGPHWHS 166
>UniRef100_Q9ATN3 NOD26-like membrane integral protein ZmNIP2-1 [Zea mays]
Length = 295
Score = 116 bits (291), Expect = 1e-25
Identities = 55/102 (53%), Positives = 76/102 (73%), Gaps = 2/102 (1%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
M+F++ AVATD++A GELAG+AVGS+V I SI AG +SGGSMNPARTLGPA+A++ Y G+
Sbjct: 175 MMFVTLAVATDTRAVGELAGLAVGSAVCITSIFAGAVSGGSMNPARTLGPALASNLYTGL 234
Query: 61 WIYMVGPITGALLGAWSYVVI--QETDHKQDLATSQSPLSVK 100
WIY +GP+ G L GAW+Y I +E +D++ S ++
Sbjct: 235 WIYFLGPVLGTLSGAWTYTYIRFEEAPSHKDMSQKLSSFKLR 276
>UniRef100_Q9AT74 NOD26-like membrane integral protein ZmNIP2-3 [Zea mays]
Length = 301
Score = 115 bits (288), Expect = 2e-25
Identities = 52/82 (63%), Positives = 67/82 (81%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
M+F++ AVATDS+A GELAG+AVGS+V I SI AGP+SGGSMNPARTL PA+A++ + G+
Sbjct: 183 MMFVTCAVATDSRAVGELAGLAVGSAVCITSIFAGPVSGGSMNPARTLAPAVASNVFTGL 242
Query: 61 WIYMVGPITGALLGAWSYVVIQ 82
WIY +GP+ G L GAW Y I+
Sbjct: 243 WIYFLGPVVGTLSGAWVYTYIR 264
>UniRef100_Q9ATN2 NOD26-like membrane integral protein ZmNIP2-2 [Zea mays]
Length = 294
Score = 115 bits (287), Expect = 3e-25
Identities = 52/82 (63%), Positives = 67/82 (81%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
M+F++ AVATDS+A GELAG+AVGS+V I SI AGP+SGGSMNPARTL PA+A++ + G+
Sbjct: 180 MMFVTCAVATDSRAVGELAGLAVGSAVCITSIFAGPVSGGSMNPARTLAPAVASNVFTGL 239
Query: 61 WIYMVGPITGALLGAWSYVVIQ 82
WIY +GP+ G L GAW Y I+
Sbjct: 240 WIYFLGPVIGTLSGAWVYTYIR 261
Score = 31.6 bits (70), Expect = 4.6
Identities = 26/82 (31%), Positives = 38/82 (45%), Gaps = 6/82 (7%)
Query: 1 MVFISTAVAT----DSKATGELA-GVAVGSSVTIASIVAGPISGGSMNPARTLGPA-IAT 54
+VF++ A+ D++ +L VA G VT+ G ISG MNPA TL A
Sbjct: 64 LVFVTCGAASIYGEDNRRISQLGQSVAGGLIVTVMIYATGHISGAHMNPAVTLSFACFRH 123
Query: 55 SSYKGIWIYMVGPITGALLGAW 76
+ + Y TGA+ A+
Sbjct: 124 FPWIQVPFYWAAQFTGAMCAAF 145
>UniRef100_Q6J1S8 Nodulin 26-like protein [Medicago truncatula]
Length = 310
Score = 110 bits (275), Expect = 8e-24
Identities = 52/99 (52%), Positives = 75/99 (75%), Gaps = 3/99 (3%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ TAVATD++A GELAG+AVG++V + ++AGPI+G SMNP RTLGPAIA ++YK I
Sbjct: 208 LMFVVTAVATDTRAVGELAGIAVGATVMLNILIAGPITGASMNPVRTLGPAIAANNYKAI 267
Query: 61 WIYMVGPITGALLGAWSYVVI---QETDHKQDLATSQSP 96
W+Y++ PI GAL GA +Y + +E D+ + A+S P
Sbjct: 268 WVYLLAPILGALGGAGTYTAVKLPEEDDNAKTNASSNHP 306
>UniRef100_Q9XGG7 Nodulin26-like major intrinsic protein [Pisum sativum]
Length = 270
Score = 109 bits (273), Expect = 1e-23
Identities = 50/85 (58%), Positives = 67/85 (78%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++FI + VATD++A GELAG+AVGS+V + + AGPI+G SMNPAR++GPA + Y+GI
Sbjct: 168 LMFIISGVATDNRAIGELAGIAVGSTVLLNVMFAGPITGASMNPARSIGPAFVHNEYRGI 227
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
WIYM+ PI GA+ GAW Y VI+ TD
Sbjct: 228 WIYMISPIVGAVSGAWVYNVIRYTD 252
>UniRef100_Q6QIL3 NIP3 [Medicago truncatula]
Length = 305
Score = 109 bits (273), Expect = 1e-23
Identities = 51/102 (50%), Positives = 76/102 (74%), Gaps = 2/102 (1%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ TAVATD++A GELAG+AVG++V + ++AGP +G SMNP RTLGPAIA ++YKGI
Sbjct: 203 LMFVVTAVATDTRAVGELAGIAVGATVMLNILIAGPATGASMNPVRTLGPAIAANNYKGI 262
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHK--QDLATSQSPLSVK 100
W+Y++ PI GAL GA +Y ++ D + ++ S +P S +
Sbjct: 263 WLYLIAPILGALGGAGAYTAVKLPDEEFNSEVKASSAPGSFR 304
>UniRef100_Q6QIL2 NIP2 [Medicago truncatula]
Length = 269
Score = 109 bits (273), Expect = 1e-23
Identities = 51/84 (60%), Positives = 67/84 (79%)
Query: 2 VFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGIW 61
+FI + VATD++A GELAG+AVGS+V + + AGPI+G SMNPAR++GPA+ S Y+GIW
Sbjct: 168 MFIISGVATDNRAIGELAGIAVGSTVLLNVMFAGPITGASMNPARSIGPALLHSEYRGIW 227
Query: 62 IYMVGPITGALLGAWSYVVIQETD 85
IY+V PI GA+ GAW Y VI+ TD
Sbjct: 228 IYLVSPILGAVAGAWVYNVIRYTD 251
>UniRef100_Q940D9 Early embryogenesis aquaglyceroporin [Pinus taeda]
Length = 264
Score = 108 bits (271), Expect = 2e-23
Identities = 48/84 (57%), Positives = 68/84 (80%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
+VF+ +AVATD+KA GEL G+A+G+++ + ++GPISG SMNPART+G A+A + Y I
Sbjct: 157 LVFVVSAVATDTKAVGELGGLAIGATIAMNVAISGPISGASMNPARTIGSAVAGNKYTSI 216
Query: 61 WIYMVGPITGALLGAWSYVVIQET 84
W+YMVGP+ GAL+GA SY +I+ET
Sbjct: 217 WVYMVGPVIGALMGAMSYNMIRET 240
>UniRef100_Q8LFP7 Aquaporin NIP1.2 [Arabidopsis thaliana]
Length = 294
Score = 108 bits (271), Expect = 2e-23
Identities = 48/85 (56%), Positives = 68/85 (79%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATD++A GELAG+AVGS+V + I+AGP+SG SMNP R+LGPA+ S Y+G+
Sbjct: 188 LMFVISGVATDNRAIGELAGLAVGSTVLLNVIIAGPVSGASMNPGRSLGPAMVYSCYRGL 247
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
WIY+V PI GA+ GAW Y +++ TD
Sbjct: 248 WIYIVSPIVGAVSGAWVYNMVRYTD 272
>UniRef100_Q5W6J2 Hypothetical protein OSJNBb0115F21.2 [Oryza sativa]
Length = 286
Score = 108 bits (270), Expect = 3e-23
Identities = 48/85 (56%), Positives = 67/85 (78%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATD++A GELAG+AVG++V + + AGPISG SMNPART+GPAI Y GI
Sbjct: 183 LMFVVSGVATDNRAIGELAGLAVGATVLVNVLFAGPISGASMNPARTIGPAIILGRYTGI 242
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
W+Y+ GP+ GA+ GAW+Y +I+ TD
Sbjct: 243 WVYIAGPVFGAVAGAWAYNLIRFTD 267
>UniRef100_Q40746 Major intrinsic protein [Oryza sativa]
Length = 284
Score = 108 bits (270), Expect = 3e-23
Identities = 51/106 (48%), Positives = 78/106 (73%), Gaps = 1/106 (0%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATD++A GELAG+AVG+++ + ++AGPISG SMNPAR+LGPA+ Y+ I
Sbjct: 177 LMFVISGVATDNRAIGELAGLAVGATILLNVLIAGPISGASMNPARSLGPAMIGGEYRSI 236
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHKQDLATSQSPLSVKIHNEMN 106
W+Y+VGP+ GA+ GAW+Y +I+ T +K ++S +K N MN
Sbjct: 237 WVYIVGPVAGAVAGAWAYNIIRFT-NKPLREITKSGSFLKSMNRMN 281
>UniRef100_P49173 Probable aquaporin NIP-type [Nicotiana alata]
Length = 270
Score = 106 bits (264), Expect = 1e-22
Identities = 51/98 (52%), Positives = 72/98 (73%), Gaps = 1/98 (1%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATD +A G++AG+AVG ++T+ VAGPISG SMNPAR++GPAI Y G+
Sbjct: 171 LMFVISGVATDDRAIGQVAGIAVGMTITLNVFVAGPISGASMNPARSIGPAIVKHVYTGL 230
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHK-QDLATSQSPL 97
W+Y+VGPI G L GA+ Y +I+ TD ++LA S S L
Sbjct: 231 WVYVVGPIIGTLAGAFVYNLIRSTDKPLRELAKSASSL 268
>UniRef100_Q9ATN4 NOD26-like membrane integral protein ZmNIP1-1 [Zea mays]
Length = 282
Score = 105 bits (263), Expect = 2e-22
Identities = 43/85 (50%), Positives = 69/85 (80%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ + VATD++A GELAG+AVG+++ + ++AGP+SG SMNPAR++GPA+ + Y I
Sbjct: 173 LMFVISGVATDNRAIGELAGLAVGATILLNVLIAGPVSGASMNPARSVGPALVSGEYTSI 232
Query: 61 WIYMVGPITGALLGAWSYVVIQETD 85
W+Y+VGP+ GA+ GAW+Y +I+ T+
Sbjct: 233 WVYVVGPVVGAVAGAWAYNLIRFTN 257
>UniRef100_Q9ATN1 NOD26-like membrane integral protein ZmNIP3-1 [Zea mays]
Length = 302
Score = 105 bits (263), Expect = 2e-22
Identities = 48/94 (51%), Positives = 69/94 (73%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ TAVATD++A GELAG+AVG++VT+ +VAGP +GGSMNP RTLGPA+A +Y+ +
Sbjct: 204 LLFVVTAVATDTRAVGELAGIAVGAAVTLNILVAGPTTGGSMNPVRTLGPAVAAGNYRQL 263
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHKQDLATSQ 94
WIY++ P GAL GA Y ++ D + +Q
Sbjct: 264 WIYLLAPTLGALAGASVYKAVKLRDENGETPRTQ 297
>UniRef100_Q717T4 Nodulin-like intrinsic protein NIP1-1 [Atriplex nummularia]
Length = 300
Score = 105 bits (263), Expect = 2e-22
Identities = 48/87 (55%), Positives = 67/87 (76%)
Query: 1 MVFISTAVATDSKATGELAGVAVGSSVTIASIVAGPISGGSMNPARTLGPAIATSSYKGI 60
++F+ TAVATD++A GELAG+AVG++V + +VAGP SG SMNP RTLGPA+A +Y+ +
Sbjct: 202 LMFVVTAVATDTRAVGELAGIAVGATVMLNILVAGPSSGASMNPVRTLGPAVAAGNYRAV 261
Query: 61 WIYMVGPITGALLGAWSYVVIQETDHK 87
WIY+V P GAL GA Y ++Q +H+
Sbjct: 262 WIYLVAPTLGALGGAAIYKLVQLQEHE 288
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.129 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 163,537,421
Number of Sequences: 2790947
Number of extensions: 5582547
Number of successful extensions: 17254
Number of sequences better than 10.0: 880
Number of HSP's better than 10.0 without gapping: 620
Number of HSP's successfully gapped in prelim test: 260
Number of HSP's that attempted gapping in prelim test: 15549
Number of HSP's gapped (non-prelim): 1555
length of query: 110
length of database: 848,049,833
effective HSP length: 86
effective length of query: 24
effective length of database: 608,028,391
effective search space: 14592681384
effective search space used: 14592681384
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC125479.4


