

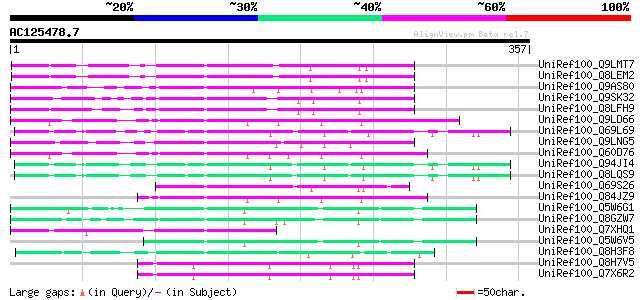
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.7 - phase: 0
(357 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana] 97 7e-19
UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana] 97 7e-19
UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa] 84 5e-15
UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana] 82 3e-14
UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana] 82 3e-14
UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein ... 79 3e-13
UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [... 78 4e-13
UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana] 76 2e-12
UniRef100_Q60D76 Putative polyprotein [Oryza sativa] 75 2e-12
UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa] 74 6e-12
UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa] 74 6e-12
UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [... 70 9e-11
UniRef100_Q84JZ9 Mutator-like transposase [Oryza sativa] 67 8e-10
UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa] 66 2e-09
UniRef100_Q8GZW7 Putative transposable element [Oryza sativa] 60 7e-08
UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sa... 60 9e-08
UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa] 59 2e-07
UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like p... 58 5e-07
UniRef100_Q8H7V5 Putative mutator-like transposase [Oryza sativa] 52 3e-05
UniRef100_Q7X6R2 OSJNBa0001M07.4 protein [Oryza sativa] 52 3e-05
>UniRef100_Q9LMT7 F2H15.15 protein [Arabidopsis thaliana]
Length = 478
Score = 97.1 bits (240), Expect = 7e-19
Identities = 76/285 (26%), Positives = 131/285 (45%), Gaps = 35/285 (12%)
Query: 2 PMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY- 60
P GEMT+TLD+V+ + L+++G+ + G K + + + + LG K+ E
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVV-GVKEKDEDPSQVCLRLLG-------KLPKGELS 135
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRS 120
G ++ L++ + A + +E+E Y R YL+Y+VG +F
Sbjct: 136 GNRVTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTD 181
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRLGHRALGGSVTLLTAWFLA 180
+I + YL D + Y+WGA LA+LY ++ +A + +GG +TLL W
Sbjct: 182 PSKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQCWSYF 240
Query: 181 HFPGFFSVDLNTDYLENYSVAARWK---LQKGHGEGITYRSLLDRIQLDDVCWRSYEEHR 237
H ++D + +A WK + + YR LD + +V W +E
Sbjct: 241 H----LNIDRPKRTTRQFPLALLWKGRQQSRSKNDLFKYRKALDDLDPSNVSWCPFEGDL 296
Query: 238 EI--QDFEE--VFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIP 278
+I Q F++ + SR + G + V H P+R ++Q+G Q IP
Sbjct: 297 DIVPQSFKDNLLLGRSRTKLIGPKVVEWHFPDRCMKQFGLCQVIP 341
>UniRef100_Q8LEM2 Hypothetical protein [Arabidopsis thaliana]
Length = 478
Score = 97.1 bits (240), Expect = 7e-19
Identities = 76/285 (26%), Positives = 131/285 (45%), Gaps = 35/285 (12%)
Query: 2 PMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY- 60
P GEMT+TLD+V+ + L+++G+ + G K + + + + LG K+ E
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVV-GVKEKDEDPSQVCLKLLG-------KLPKGELS 135
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRS 120
G ++ L++ + A + +E+E Y R YL+Y+VG +F
Sbjct: 136 GNRVTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTD 181
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRLGHRALGGSVTLLTAWFLA 180
+I + YL D + Y+WGA LA+LY ++ +A + +GG +TLL W
Sbjct: 182 PSKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQCWSYF 240
Query: 181 HFPGFFSVDLNTDYLENYSVAARWK---LQKGHGEGITYRSLLDRIQLDDVCWRSYEEHR 237
H ++D + +A WK + + YR LD + +V W +E
Sbjct: 241 H----LNIDRPKRTTRQFPLALLWKGRQQSRSKNDLFKYRKALDDLDPSNVSWCPFEGDL 296
Query: 238 EI--QDFEE--VFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIP 278
+I Q F++ + SR + G + V H P+R ++Q+G Q IP
Sbjct: 297 DIVPQSFKDNLLLGRSRTKLIGPKVVEWHFPDRCMKQFGLCQVIP 341
>UniRef100_Q9AS80 P0028E10.12 protein [Oryza sativa]
Length = 792
Score = 84.3 bits (207), Expect = 5e-15
Identities = 89/302 (29%), Positives = 125/302 (40%), Gaps = 48/302 (15%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
MP GE+T+TL DVA + L I G + P++E L+ YLG A
Sbjct: 278 MPCGEITITLQDVAMILGLPIAGHAVTVNPTEPQNE---LVERYLGKAPPPDR------- 327
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRS 120
P LR + RA ED +E E ++ + R Y+L L+ LLF D S
Sbjct: 328 ----PRPGLRVSWV----RAEFNNCPEDADE----ETIKQH-ARAYILSLISGLLFPDAS 374
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRLGHRA--LGGSVTLLTAWF 178
+AD + +YSWG+ TLAYLY + DACR L G + LL W
Sbjct: 375 GDLYTFYPFPLIAD-LENIGSYSWGSATLAYLYRAMCDACRRQSDGSNLTGCLLLLQFWS 433
Query: 179 LAHFP---------GFFSVDLNTDYLENYSVAARWKL---QKGHGEGITYRSLLDRIQL- 225
HFP + +V+ D + +V RW + + Y + L
Sbjct: 434 WEHFPIGRPDLVKLKYPNVEELEDERDRPTVGLRWVVGVCARRAAPARCYEHFTNEFDLL 493
Query: 226 --DDVCWRSYEEHR--EIQ-----DFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQT 276
D V W Y E R E+Q + W +R + V ++PERV+RQ+G Q
Sbjct: 494 TDDQVVWCPYREERMKELQLAPICTQDSHLWLTRAPLLYFFMVEIYMPERVMRQFGLHQV 553
Query: 277 IP 278
P
Sbjct: 554 CP 555
>UniRef100_Q9SK32 Expressed protein [Arabidopsis thaliana]
Length = 509
Score = 81.6 bits (200), Expect = 3e-14
Identities = 75/286 (26%), Positives = 125/286 (43%), Gaps = 33/286 (11%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P+GEMT+TLD+VA + L I+G + K G + M G + N+E
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPIVGSK-----VGDEVAMDMCGRLLGKLPSAANKE- 145
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRS 120
++ R++ ++L R +E PE+ + V+ + R YLLYL+G +F
Sbjct: 146 ---VNCSRVK---LNWLKR----TFSECPEDA-SFDVVKCH-TRAYLLYLIGSTIFATTD 193
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRLGHRALGGSVTLLTAWFLA 180
++ + YL D + Y+WGA LA LY L +A + G +TLL W
Sbjct: 194 GDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCW--- 249
Query: 181 HFPGFFSVDLNTDYLEN--YSVAARWKLQ--KGHGEGITYRSLLDRIQLDDVCWRSYEEH 236
+F +D+ + +A WK + + + YR LD + + W YE
Sbjct: 250 ---SYFHLDIGRPEKSEACFPLALLWKGKGSRSKTDLSEYRRELDDLDPSKITWCPYERF 306
Query: 237 REI----QDFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIP 278
+ + + S+ + ++ H P+R LRQ+G Q IP
Sbjct: 307 ENLIPPHIKAKLILGRSKTTLVCFEKIELHFPDRCLRQFGKRQPIP 352
>UniRef100_Q8LFH9 Hypothetical protein [Arabidopsis thaliana]
Length = 509
Score = 81.6 bits (200), Expect = 3e-14
Identities = 75/286 (26%), Positives = 121/286 (42%), Gaps = 33/286 (11%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P+GEMT+TLD+VA + L I+G + G K+ + LG A K N
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPIV-GSKVDDEVAMDMCGRLLGKLPSAANKEVN--- 147
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRS 120
S +L ++ +E PE+ + V+ + R YLLYL+G +F
Sbjct: 148 ---CSRVKLNWLKRTF---------SECPEDA-SFDVVKCH-TRAYLLYLIGSTIFATTD 193
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRLGHRALGGSVTLLTAWFLA 180
++ + YL D + Y+WGA LA LY L +A + G +TLL W
Sbjct: 194 GDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCW--- 249
Query: 181 HFPGFFSVDLNTDYLEN--YSVAARWKLQ--KGHGEGITYRSLLDRIQLDDVCWRSYEEH 236
+F +D+ + +A WK + + + YR LD + + W YE
Sbjct: 250 ---SYFHLDIGRPEKSEACFPLALLWKGKGSRSKTDLSEYRRELDDLDPSKITWCPYERF 306
Query: 237 REI----QDFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIP 278
+ + + S+ + ++ H P+R LRQ+G Q IP
Sbjct: 307 ENLIPPHIKAKLILGRSKTTLVCFEKIELHFPDRCLRQFGKRQPIP 352
>UniRef100_Q9LD66 Similar to maize transposon MuDR mudrA protein [Oryza sativa]
Length = 1230
Score = 78.6 bits (192), Expect = 3e-13
Identities = 91/335 (27%), Positives = 135/335 (40%), Gaps = 47/335 (14%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRML----AHGKKMPKHEGAALLMTYLGVAQHEAEKIC 56
+P GE+TVTL+DVA + L I G+ + A G + E YLG+ A
Sbjct: 592 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRERVE------EYLGLEPPVAPDGQ 645
Query: 57 NQEYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLF 116
Q + LR + + P E +E V+ YC R Y+LY+ G +LF
Sbjct: 646 RQTKTSGVPLSWLRANFG------------QCPAEADEAT-VQRYC-RAYVLYIFGSILF 691
Query: 117 GDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRL--GHRALGGSVTLL 174
D ++L +AD + +SWG+ LA+LY +L DACR G L G V LL
Sbjct: 692 PDSGGDMASWMWLPLLAD-WDEAGTFSWGSAALAWLYRQLCDACRRQGGDANLAGCVWLL 750
Query: 175 TAWFLAHFP------GFFSVDLNTDYLENYSVAARWKLQKGHGEG------ITYRSLLDR 222
W P + D +VA W+ G Y + +D
Sbjct: 751 QVWMWMRLPVGRPMWRTHQAWPHQDADRRPTVAHLWESVPSPVVGRRNLAYYHYTNEMDY 810
Query: 223 IQLDDVCWRSYEEHREIQ-------DFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQ 275
+Q + V W Y+ ++ E+ R + V + RV+RQ+G +Q
Sbjct: 811 LQPEHVVWMPYQAQEVLELELNPMCHIEDALKTLRCPLICFYAVEFQMCHRVMRQFGRLQ 870
Query: 276 TIPRHPTDVRDLPPP-SIVQMFVDFRTHTLKADAR 309
TIP + DL +I +F HT + D R
Sbjct: 871 TIPHRFSTSIDLHNHFNIYYIFELDILHTCRVDRR 905
>UniRef100_Q69L69 Calcineurin-like phosphoesterase-like protein [Oryza sativa]
Length = 766
Score = 77.8 bits (190), Expect = 4e-13
Identities = 104/387 (26%), Positives = 160/387 (40%), Gaps = 88/387 (22%)
Query: 4 GEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGY 63
GEMTVTL DV+ L L I+GR + H+ A +++ LG H+ + + +
Sbjct: 153 GEMTVTLQDVSMLLALPIDGRPVC---STTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 205
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRSNKR 123
+ Y L+ + + GT+D + +ER VR Y+L L+ +LF D + R
Sbjct: 206 LHYKWLKKHF------YELPEGTDD----QTVER----HVRAYILSLLCGVLFPDGTG-R 250
Query: 124 IELIYLTTMADGYAGMRNYSWGAMTLAYLYGELAD-ACRLGHRALGGSVTLLTAWF---- 178
+ LIYL +AD + + YSWG+ LA+LY L A + +GGS+ LL W
Sbjct: 251 MSLIYLPLIAD-LSLVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQLWSWERS 309
Query: 179 -----LAHFPGFFSVDLNTDYLENYSVAARWKLQKGHGEGIT-----YRSLLDRIQLDDV 228
L P D+ D L V RW + E T YR L+ + D V
Sbjct: 310 HVGRPLVRSPVCPETDIPQDVL---PVGFRWVGARTQSENATRCLKQYRDELNLQRADQV 366
Query: 229 CWRSYEEHREIQDFEEV------FWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIPRHPT 282
W Y H E + W ++ + V +LPERV+RQ+G Q+IP
Sbjct: 367 KWEPY-LHIESASLPLLCTKNADLWLTQAPLINFPIVEMYLPERVMRQFGLRQSIP---- 421
Query: 283 DVRDLPP--PSIVQMFVDFRTHTLKADARGEQAGEDT-------WQVA------------ 321
PP P++ + H + R + E+T W+
Sbjct: 422 -----PPFRPTLQAL------HRISRRGRERENWEETHHEYIQEWEARRQRIFPESEQYD 470
Query: 322 ----DGYVLWYTRVSHPQILPPIPGDL 344
+ Y+ WYT + ++P I D+
Sbjct: 471 PSSYEEYLHWYTGATRRYLVPSISDDV 497
>UniRef100_Q9LNG5 F21D18.16 [Arabidopsis thaliana]
Length = 1340
Score = 75.9 bits (185), Expect = 2e-12
Identities = 81/299 (27%), Positives = 123/299 (41%), Gaps = 46/299 (15%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL DV L L ++G + K + A L LG + +
Sbjct: 101 LPAGEITVTLQDVNILLGLRVDGPAVTGSTK---YNWADLCEDLLGHRPGPKDL-----H 152
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYC-VRCYLLYLVGCLLFGDR 119
G ++S LR+ + + DP+EV C R ++L L+ L+GD+
Sbjct: 153 GSHVSLAWLRENFRNL---------PADPDEVT------LKCHTRAFVLALMSGFLYGDK 197
Query: 120 SNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRLGHRALGGSVTLLTAWFL 179
S + L +L + D + + SWG+ TLA LY EL A + + G + LL W
Sbjct: 198 SKHDVALTFLPLLRD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQLWAW 256
Query: 180 AHF----PGFFSVDLNTDYLENYS------VAARWKLQKGHGEGIT-----YRSLLDRIQ 224
PG D+ Y++ + RW+ H E YR D+ +
Sbjct: 257 ERLHVGRPGRLK-DVGASYMDGIDGPLPDPLGCRWRASLSHKENPRGGLDFYRDQFDQQK 315
Query: 225 LDDVCWRSY-----EEHREIQDFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIP 278
+ V W+ Y + I E W + + V H P+RVLRQ+G QTIP
Sbjct: 316 DEQVIWQPYTPDLLAKIPLICVSGENIWRTVAPLICFDVVEWHRPDRVLRQFGLHQTIP 374
>UniRef100_Q60D76 Putative polyprotein [Oryza sativa]
Length = 1754
Score = 75.5 bits (184), Expect = 2e-12
Identities = 86/312 (27%), Positives = 129/312 (40%), Gaps = 46/312 (14%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRML----AHGKKMPKHEGAALLMTYLGVAQHEAEKIC 56
+P GE+TVTL+DVA + L I G+ + A G + E YLG+ A
Sbjct: 974 LPCGELTVTLEDVAMILGLPIRGQAVTGDTASGNWRERVE------EYLGLEPPVAPDGQ 1027
Query: 57 NQEYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLF 116
Q + LR + + P E +E V+ YC R Y+LY+ G +LF
Sbjct: 1028 RQTKTSGVPLSWLRANFG------------QCPAEADEAT-VQRYC-RAYVLYIFGSILF 1073
Query: 117 GDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRL--GHRALGGSVTLL 174
D ++L +AD + +SWG+ LA+LY +L DACR G L G V LL
Sbjct: 1074 PDSGGDMASWMWLPLLAD-WDEAGTFSWGSAALAWLYRQLCDACRRQGGDANLAGCVWLL 1132
Query: 175 TAW----FLAHFPGFFSVDL--NTDYLENYSVAARWKLQKGHGEG------ITYRSLLDR 222
W L P + + + D +VA W+ G Y + +D
Sbjct: 1133 QVWMWMRLLVGRPMWRTHQAWPHQDADRRPTVAHLWESVPSPVVGRRNLAYCHYTNEMDY 1192
Query: 223 IQLDDVCWRSYEEHREIQ-------DFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQ 275
+Q + V W Y+ ++ E+ R + V + RV+RQ+G +Q
Sbjct: 1193 LQPEHVVWMPYQAQEVLELELNPMCHIEDALKTLRCPLICFYAVEFQMCHRVMRQFGRLQ 1252
Query: 276 TIPRHPTDVRDL 287
TIP + DL
Sbjct: 1253 TIPHRFSTSIDL 1264
>UniRef100_Q94JI4 P0638D12.5 protein [Oryza sativa]
Length = 761
Score = 73.9 bits (180), Expect = 6e-12
Identities = 102/387 (26%), Positives = 155/387 (39%), Gaps = 88/387 (22%)
Query: 4 GEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGY 63
GEM VTL DVA L L I+GR + H+ A +++ LG H+ + + +
Sbjct: 170 GEMAVTLQDVAMLLALPIDGRPVC---STTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 222
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRSNKR 123
+ Y L+ + L D + VE VR Y+L L+ +LF D + R
Sbjct: 223 LHYKWLKKHFYE-------LPEGADDQTVER-------HVRAYILSLLCGVLFPDGTG-R 267
Query: 124 IELIYLTTMADGYAGMRNYSWGAMTLAYLYGELAD-ACRLGHRALGGSVTLLTAWF---- 178
+ LIYL +AD + + YSWG+ LA+LY L A + +GGS+ LL W
Sbjct: 268 MSLIYLPLIAD-LSRVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQLWSWERS 326
Query: 179 -----LAHFPGFFSVDLNTDYLENYSVAARWKLQKGHGEGIT-----YRSLLDRIQLDDV 228
L P D+ D V RW + E T YR L+ + D V
Sbjct: 327 HVGRPLVRSPLCPETDIPQDL---PPVGFRWVGARTQSENATRCLKQYRDELNLQRADQV 383
Query: 229 CWRSYEEHREIQDF------EEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIPRHPT 282
W Y H E + W ++ + V +LPERV+RQ+G Q+IP
Sbjct: 384 KWEPY-LHIESSSLPLLCTKDADLWLTQAPLINFPIVEMYLPERVMRQFGLRQSIP---- 438
Query: 283 DVRDLPP--PSIVQMFVDFRTHTLKADARGEQAGEDT-------WQVA------------ 321
PP P++ + H + R + E+T W+
Sbjct: 439 -----PPFRPTLQAL------HRISRRGRERENWEETHHEYIQEWEARRQRIFPESEQYD 487
Query: 322 ----DGYVLWYTRVSHPQILPPIPGDL 344
+ Y+ WY+ V+ ++P I D+
Sbjct: 488 PSSYEEYLHWYSGVTRRYLVPSISDDV 514
>UniRef100_Q8LQS9 P0702H08.15 protein [Oryza sativa]
Length = 718
Score = 73.9 bits (180), Expect = 6e-12
Identities = 102/387 (26%), Positives = 155/387 (39%), Gaps = 88/387 (22%)
Query: 4 GEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGY 63
GEM VTL DVA L L I+GR + H+ A +++ LG H+ + + +
Sbjct: 184 GEMAVTLQDVAMLLALPIDGRPVC---STTDHDYAQMVIDCLG---HD-PRGPSMPGKSF 236
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRSNKR 123
+ Y L+ + L D + VE VR Y+L L+ +LF D + R
Sbjct: 237 LHYKWLKKHFYE-------LPEGADDQTVER-------HVRAYILSLLCGVLFPDGTG-R 281
Query: 124 IELIYLTTMADGYAGMRNYSWGAMTLAYLYGELAD-ACRLGHRALGGSVTLLTAWF---- 178
+ LIYL +AD + + YSWG+ LA+LY L A + +GGS+ LL W
Sbjct: 282 MSLIYLPLIAD-LSRVGTYSWGSAVLAFLYRSLCSVASSHNIKNIGGSLLLLQLWSWERS 340
Query: 179 -----LAHFPGFFSVDLNTDYLENYSVAARWKLQKGHGEGIT-----YRSLLDRIQLDDV 228
L P D+ D V RW + E T YR L+ + D V
Sbjct: 341 HVGRPLVRSPLCPETDIPQDV---PPVGFRWVGARTQSENATRCLKQYRDELNLQRADQV 397
Query: 229 CWRSYEEHREIQDF------EEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIPRHPT 282
W Y H E + W ++ + V +LPERV+RQ+G Q+IP
Sbjct: 398 KWEPY-LHIESSSLPLLCTKDADLWLTQAPLINFPIVEMYLPERVMRQFGLRQSIP---- 452
Query: 283 DVRDLPP--PSIVQMFVDFRTHTLKADARGEQAGEDT-------WQVA------------ 321
PP P++ + H + R + E+T W+
Sbjct: 453 -----PPFRPTLQAL------HRISRRGREHENWEETHHEYIQEWEARRQRIFPESEQYD 501
Query: 322 ----DGYVLWYTRVSHPQILPPIPGDL 344
+ Y+ WY+ V+ ++P I D+
Sbjct: 502 PSSYEEYLHWYSGVTRRYLVPSISDDV 528
>UniRef100_Q69S26 Serine/threonine phosphatase PP7-related-like [Oryza sativa]
Length = 619
Score = 70.1 bits (170), Expect = 9e-11
Identities = 57/187 (30%), Positives = 87/187 (46%), Gaps = 17/187 (9%)
Query: 101 YCVRCYLLYLVGCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADAC 160
Y R YLLYLVG LF D + YL +AD + +R Y+WG+ LA+LY L+ A
Sbjct: 172 YSTRAYLLYLVGSTLFPDTMRGFVSPRYLPLLAD-FRKIREYAWGSAALAHLYRGLSVAV 230
Query: 161 RLGHRA-LGGSVTLLTAWFLAHFPGFFSVDLNTDYLENYSVAARWKL---QKGHGEGI-T 215
GS TLL AW + P N + L A RW +G + +
Sbjct: 231 TPNATTQFLGSATLLMAWIYEYLPLTQPQQKNQNTL--LPRACRWNFGGATRGQRKKVME 288
Query: 216 YRSLLDRIQLDDVCWRSYEEHRE--IQDF----EEVFWYSRWIMC-GVRRVYRHLPERVL 268
+R + D +Q DV W Y++ I ++ + + + W++ ++ VY +P+R
Sbjct: 289 WRKVFDELQFSDVNWNPYKDMNPAIIPEYCIAADNICYSRTWLISFNIKEVY--VPDRFS 346
Query: 269 RQYGYVQ 275
RQ+G Q
Sbjct: 347 RQFGREQ 353
>UniRef100_Q84JZ9 Mutator-like transposase [Oryza sativa]
Length = 1381
Score = 67.0 bits (162), Expect = 8e-10
Identities = 62/220 (28%), Positives = 92/220 (41%), Gaps = 24/220 (10%)
Query: 89 PEEVEELERVRTYCVRCYLLYLVGCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGAMT 148
P E +E V+ YC R Y+LY+ G +LF D ++L +AD + +SWG+
Sbjct: 804 PAEADEAT-VQRYC-RAYVLYIFGSILFPDSGGDMASWMWLPLLAD-WDEAGTFSWGSAA 860
Query: 149 LAYLYGELADACRL--GHRALGGSVTLLTAWFLAHFP------GFFSVDLNTDYLENYSV 200
LA+LY +L DACR G L G V LL W P + D +V
Sbjct: 861 LAWLYRQLCDACRRQGGDANLAGCVWLLQVWMWMRLPVGRPMWRTHQAWPHQDADRCPTV 920
Query: 201 AARWKLQKGHGEG------ITYRSLLDRIQLDDVCWRSYEEHREIQ-------DFEEVFW 247
A W+ G Y + +D +Q + V W Y+ ++ E+
Sbjct: 921 AHLWESVPSPVVGRRNLAYYHYTNEMDYLQPEHVVWMPYQAQEVLELELNPMCHIEDALK 980
Query: 248 YSRWIMCGVRRVYRHLPERVLRQYGYVQTIPRHPTDVRDL 287
R + V + RV+RQ+G +QTIP + DL
Sbjct: 981 TLRCPLICFYAVEFQMCHRVMRQFGRLQTIPHRFSTSIDL 1020
>UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa]
Length = 1567
Score = 65.9 bits (159), Expect = 2e-09
Identities = 79/334 (23%), Positives = 132/334 (38%), Gaps = 42/334 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAA--LLMTYLGVAQHEAEKICNQ 58
+P GEMT+TL DVA + L + G + + P L L + Q + K +
Sbjct: 1042 LPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWHAQVQQLFGIPLNIEQGQGGK--KK 1099
Query: 59 EYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYC-VRCYLLYLVGCLLFG 117
+ G +S+ ++F S+L ++ E R C R Y+L+L+G +LF
Sbjct: 1100 QNGIPLSWLS-QNF--SHLD--------------DDAEPWRAECYARAYILHLLGGVLFP 1142
Query: 118 DRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADAC--RLGHRALGGSVTLLT 175
D I++ +A+ + +SWG+ LA+ Y +L +AC + + G V L+
Sbjct: 1143 DAGGDIASAIWIPLVAN-LGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGCVLLIQ 1201
Query: 176 AWFLAHFPGFFSVDLNTDYLENYSVAARWKLQKGHGEGITYRSLLDRIQLDDVCWRSYEE 235
W P ++ YL + A + Y + +D +Q + W Y
Sbjct: 1202 MWMWLRLPNEPDMEKTVAYLFESTATAHAHRDVAYKH---YVNEMDYLQPQHIEWLPYHT 1258
Query: 236 HRE--------IQDFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIPRHPTDVRDL 287
+ + F Y ++C V HLP RV+RQ+G Q P V D+
Sbjct: 1259 NEASSLTLNSMCNRDSDYFMYQCPLIC-FWAVEYHLPHRVMRQFGKKQDWP-----VEDI 1312
Query: 288 PPPSIVQMFVDFRTHTLKADARGEQAGEDTWQVA 321
+ + RT +K D W+ A
Sbjct: 1313 STGVELHKYDRVRTKKVKDWGLEHNRYIDEWRTA 1346
>UniRef100_Q8GZW7 Putative transposable element [Oryza sativa]
Length = 663
Score = 60.5 bits (145), Expect = 7e-08
Identities = 82/348 (23%), Positives = 136/348 (38%), Gaps = 55/348 (15%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GEMT+TL DVA + L + G + + P A + G+ + Q
Sbjct: 92 LPSGEMTITLQDVAMILALPLRGHAVTGRTETPGWH--AQVQQLFGIPLN-----IEQGQ 144
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRS 120
GG + S+L + ++ E RV Y R Y+L+L+G +LF D
Sbjct: 145 GGK---KKQNGIPLSWLSQ-----NFSHLDDDAEPWRVECYA-RAYILHLLGGVLFPDAG 195
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADAC--RLGHRALGGSVTLLTAWF 178
I++ +A+ + +SWG+ LA+ Y +L +AC + + G V L+ W
Sbjct: 196 GDIASAIWIPLVAN-LGDLGRFSWGSAVLAWTYRQLCEACCRQAPSSNMSGCVLLIQIWM 254
Query: 179 LAHF------------PGFFS---VDLNTDYL-ENYSVA-ARWKLQKGHGEGITYRSLLD 221
P +++ ++ YL E+ + A A W + H Y + +D
Sbjct: 255 WLRLPVGRPKWRQSFTPWYYNEPDMEKTVAYLFESTATAHAHWDVAYKH-----YVNEMD 309
Query: 222 RIQLDDVCWRSYEEHRE--------IQDFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGY 273
+Q + W Y + + F Y ++C V HLP RV+RQ+G
Sbjct: 310 CLQPQHIEWLPYHTNEASSLTLNLMCNRDSDYFMYQCPLIC-FWAVEYHLPHRVMRQFGK 368
Query: 274 VQTIPRHPTDVRDLPPPSIVQMFVDFRTHTLKADARGEQAGEDTWQVA 321
Q P V D+ + + RT +K D W+ A
Sbjct: 369 KQDWP-----VEDISTGVELHKYDRVRTKKVKDWGLEHNRYIDEWRTA 411
>UniRef100_Q7XHQ1 Calcineurin-like phosphoesterase-like [Oryza sativa]
Length = 327
Score = 60.1 bits (144), Expect = 9e-08
Identities = 47/187 (25%), Positives = 79/187 (42%), Gaps = 15/187 (8%)
Query: 1 MPMGEMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHE----AEKIC 56
+P+GEMT+TL DV+CL L I GR + ++ LG+ E +K
Sbjct: 87 LPVGEMTITLQDVSCLWGLPIHGRPIT---GQADGSWVDMIERLLGIPMEEQHMKQKKRK 143
Query: 57 NQEYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLF 116
++ +SY R + R V+ E+ + R +L ++G ++F
Sbjct: 144 KEDDMTMVSYSRYSISLSKLRDRFRVMPKNATEREI-------NWYTRALVLDIIGSMVF 196
Query: 117 GDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRLGHRALGGSVTLLTA 176
D S + +YL M + + Y+WGA L+ LY +L+ A + + LL
Sbjct: 197 TDTSGDGVPAMYLQFMMN-LSEQTEYNWGAAALSMLYRQLSIASEKERAEISRPLLLLQL 255
Query: 177 WFLAHFP 183
W + P
Sbjct: 256 WSWSRLP 262
>UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa]
Length = 1525
Score = 59.3 bits (142), Expect = 2e-07
Identities = 57/240 (23%), Positives = 95/240 (38%), Gaps = 21/240 (8%)
Query: 93 EELERVRTYC-VRCYLLYLVGCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAY 151
++ E R C R Y+L+L+G +LF D I++ +A+ + +SWG+ LA+
Sbjct: 1075 DDAEPWRAECYARAYILHLLGGVLFPDAGGDIASAIWIPLVAN-LGDLGRFSWGSAVLAW 1133
Query: 152 LYGELADAC--RLGHRALGGSVTLLTAWFLAHFPGFFSVDLNTDYLENYSVAARWKLQKG 209
Y +L +AC + + G V L+ W P ++ YL + A
Sbjct: 1134 TYRQLCEACCRQAPSSNMSGCVLLIQMWMWLRLPNEPDMEKTVAYLFESTATAHAHRDVA 1193
Query: 210 HGEGITYRSLLDRIQLDDVCWRSYEEHRE--------IQDFEEVFWYSRWIMCGVRRVYR 261
+ Y + +D +Q + W Y + + F Y ++C V
Sbjct: 1194 YKH---YVNEMDYLQPQHIEWLPYHTNEASSLTLNSMCNRDSDYFMYQCPLIC-FWAVEY 1249
Query: 262 HLPERVLRQYGYVQTIPRHPTDVRDLPPPSIVQMFVDFRTHTLKADARGEQAGEDTWQVA 321
HLP RV+RQ+G Q P V D+ + + RT +K D W+ A
Sbjct: 1250 HLPHRVMRQFGKKQDWP-----VEDISTGVELHKYDRVRTKKVKDWGLEHNRYIDEWRTA 1304
>UniRef100_Q8H3F8 Serine/threonine phosphatase PP7-related-like protein [Oryza
sativa]
Length = 589
Score = 57.8 bits (138), Expect = 5e-07
Identities = 70/303 (23%), Positives = 119/303 (39%), Gaps = 41/303 (13%)
Query: 5 EMTVTLDDVACLTHLSIEGRMLAHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYGGYI 64
EM +L D A + + + GR++ G + K L YLG C G ++
Sbjct: 83 EMAPSLRDAAYILGIPVTGRVVTTGAVLNKSV-EDLCFQYLGQVPD-----CRDCRGSHV 136
Query: 65 SYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVGCLLFGDRSNKRI 124
L+ ++ E P + + Y R YLL+L+G L +R +
Sbjct: 137 KLSWLQSKFSRI---------PERPTNDQTM-----YGTRAYLLFLIGSALLPERDRGYV 182
Query: 125 ELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADA-CRLGHRALGGSVTLLTAWFLAHFP 183
YL ++D + ++ Y+WGA LA+LY L+ A + L GS LL W + P
Sbjct: 183 SPKYLPLLSD-FDKVQEYAWGAAALAHLYKALSIAVAHSARKRLFGSAALLMGWIYEYIP 241
Query: 184 GFFSVDLNTDYLENYSVAARW---KLQKGHGEGITYRSLLDRIQLDDVCWRSYE------ 234
D+ + +W + + R +Q+ DV W Y+
Sbjct: 242 A-LRPDMYDPPEHIFPRVLKWTGSTISQPAKNVSDIRKAFGLLQVSDVNWEPYKGVDPAS 300
Query: 235 --EHREIQDFEEVFWYSRWIMC-GVRRVYRHLPERVLRQYGYVQTIPRH--PTDVRDLPP 289
+H D + + W++ ++ +Y P+R RQ+G Q P + P R L
Sbjct: 301 IPKHCAAPD--NLCFSRTWLVSFNLKEIY--APDRFARQFGQEQHRPLNDVPAFQRQLWN 356
Query: 290 PSI 292
P++
Sbjct: 357 PAV 359
>UniRef100_Q8H7V5 Putative mutator-like transposase [Oryza sativa]
Length = 1596
Score = 51.6 bits (122), Expect = 3e-05
Identities = 55/223 (24%), Positives = 91/223 (40%), Gaps = 35/223 (15%)
Query: 89 PEEVEELERVRTYCVRCYLLYLVGCLLFGDRSNKRIE---LIYLTTMADGYAG-MRNYSW 144
PEE +E R C+ YLL+L G ++F ++ + Y ++AD G + +SW
Sbjct: 1054 PEEADEYSFSR--CLEAYLLWLFGWVMFCGSHGHSVDRGLVHYARSIADAAVGEVPQWSW 1111
Query: 145 GAMTLAYLYGELADAC-RLGHRAL-GGSVTLLTAW----------FLAHFPGFFSVDLNT 192
G+ LA Y L +AC R A+ GG ++ W + H P S+ N
Sbjct: 1112 GSALLAAQYRGLCEACTRTDPGAIFGGCPLFISLWAAERIAIGRPLVEHHPYDASLYENR 1171
Query: 193 DYLENYSVAARWKLQKGHGEGITYRSL-------LDRIQLDDVCWRSYEE---HRE---- 238
++ ++ W ++ + R DR+ DV W Y H
Sbjct: 1172 PAVDYPTMGTLWCRRQRRWAHVQVRRSYAEFVMEFDRLLPTDVVWEPYSALLTHSRAPLG 1231
Query: 239 ---IQDFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIP 278
+ ++ +W + M V H P RV+RQ+G+ Q+ P
Sbjct: 1232 LSTLCTRDQAYWMTTVPMVFDICVEPHAPYRVMRQFGFRQSFP 1274
>UniRef100_Q7X6R2 OSJNBa0001M07.4 protein [Oryza sativa]
Length = 1031
Score = 51.6 bits (122), Expect = 3e-05
Identities = 55/223 (24%), Positives = 91/223 (40%), Gaps = 35/223 (15%)
Query: 89 PEEVEELERVRTYCVRCYLLYLVGCLLFGDRSNKRIE---LIYLTTMADGYAG-MRNYSW 144
PEE +E R C+ YLL+L G ++F ++ + Y ++AD G + +SW
Sbjct: 524 PEEADEYSFSR--CLEAYLLWLFGWVMFCGSHGHSVDRGLVHYARSIADAAVGEVPQWSW 581
Query: 145 GAMTLAYLYGELADAC-RLGHRAL-GGSVTLLTAW----------FLAHFPGFFSVDLNT 192
G+ LA Y L +AC R A+ GG ++ W + H P S+ N
Sbjct: 582 GSALLAAQYRGLCEACTRTDPGAIFGGCPLFISLWAAERIAIGRPLVEHHPYDASLYENR 641
Query: 193 DYLENYSVAARWKLQKGHGEGITYRSL-------LDRIQLDDVCWRSYEE---HRE---- 238
++ ++ W ++ + R DR+ DV W Y H
Sbjct: 642 PAVDYPTMGTLWCRRQRRWAHVQVRRSYAEFVMEFDRLLPTDVVWEPYSALLTHSRAPLG 701
Query: 239 ---IQDFEEVFWYSRWIMCGVRRVYRHLPERVLRQYGYVQTIP 278
+ ++ +W + M V H P RV+RQ+G+ Q+ P
Sbjct: 702 LSTLCTRDQAYWMTTVPMVFDICVEPHAPYRVMRQFGFRQSFP 744
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.140 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 646,503,059
Number of Sequences: 2790947
Number of extensions: 27925235
Number of successful extensions: 55039
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 75
Number of HSP's that attempted gapping in prelim test: 54904
Number of HSP's gapped (non-prelim): 155
length of query: 357
length of database: 848,049,833
effective HSP length: 128
effective length of query: 229
effective length of database: 490,808,617
effective search space: 112395173293
effective search space used: 112395173293
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC125478.7


