

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.4 + phase: 0
(260 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
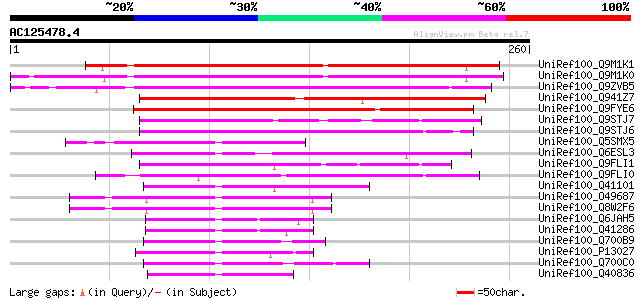
UniRef100_Q9M1K1 Hypothetical protein F24I3.50 [Arabidopsis thal... 175 1e-42
UniRef100_Q9M1K0 Hypothetical protein F24I3.60 [Arabidopsis thal... 173 4e-42
UniRef100_Q9ZVB5 Hypothetical protein At2g41240 [Arabidopsis tha... 162 8e-39
UniRef100_Q941Z7 P0431G06.12 protein [Oryza sativa] 137 4e-31
UniRef100_Q9FYE6 Myc-like protein [Arabidopsis thaliana] 134 3e-30
UniRef100_Q9STJ7 Hypothetical protein T30C3.70 [Arabidopsis thal... 56 8e-07
UniRef100_Q9STJ6 Hypothetical protein T30C3.80 [Arabidopsis thal... 56 8e-07
UniRef100_Q5SMX5 Basic helix-loop-helix protein-like [Oryza sativa] 54 4e-06
UniRef100_Q6ESL3 DNA binding protein-like [Oryza sativa] 52 1e-05
UniRef100_Q9FLI1 Arabidopsis thaliana genomic DNA, chromosome 5,... 52 2e-05
UniRef100_Q9FLI0 Arabidopsis thaliana genomic DNA, chromosome 5,... 52 2e-05
UniRef100_Q41101 Phaseolin G-box binding protein PG1 [Phaseolus ... 51 3e-05
UniRef100_O49687 BHLH protein-like [Arabidopsis thaliana] 51 3e-05
UniRef100_Q8W2F6 Putative transcription factor BHLH4 [Arabidopsi... 51 3e-05
UniRef100_Q6JAH5 B1-1 [Sorghum bicolor] 50 4e-05
UniRef100_Q41286 Myc-like regulatory R gene product [Sorghum bic... 50 7e-05
UniRef100_Q700B9 MYC transcription factor [Solanum tuberosum] 50 7e-05
UniRef100_P13027 Anthocyanin regulatory R-S protein [Zea mays] 50 7e-05
UniRef100_Q700C0 MYC transcription factor [Solanum tuberosum] 49 1e-04
UniRef100_Q40836 Myc-like regulatory R gene product [Pennisetum ... 49 1e-04
>UniRef100_Q9M1K1 Hypothetical protein F24I3.50 [Arabidopsis thaliana]
Length = 253
Score = 175 bits (443), Expect = 1e-42
Identities = 95/213 (44%), Positives = 145/213 (67%), Gaps = 11/213 (5%)
Query: 39 YSLPYHQ----FSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPG 94
Y + +HQ S E EI+ P ++ KKLNHNASERDRRKKIN+L SSLRS LP
Sbjct: 44 YEVTHHQNSLGVSVSSEGNEIDNNP---VVVKKLNHNASERDRRKKINTLFSSLRSCLPA 100
Query: 95 EDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPN 154
DQ+KK+SIP T+S+ LKYIP+LQ+QV+ L +KKEE+L R+S ++++ + + Q K + +
Sbjct: 101 SDQSKKLSIPETVSKSLKYIPELQQQVKRLIQKKEEILVRVSGQRDFELYDKQQPKAVAS 160
Query: 155 YNSAFVVSTSRLNDTELVIHISSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFY 214
Y S VS +RL D E+++ +SS + + +S +L +E +G +L++ SSS++ G RLFY
Sbjct: 161 YLS--TVSATRLGDNEVMVQVSSSKIHNFSISNVLGGIEEDGFVLVDVSSSRSQGERLFY 218
Query: 215 NLHFQVDKTQRYE--CDDLIQKLSSIYEKQQNN 245
LH QV+ Y+ C++L +++ +YEK +N+
Sbjct: 219 TLHLQVENMDDYKINCEELSERMLYLYEKCENS 251
>UniRef100_Q9M1K0 Hypothetical protein F24I3.60 [Arabidopsis thaliana]
Length = 258
Score = 173 bits (438), Expect = 4e-42
Identities = 109/265 (41%), Positives = 155/265 (58%), Gaps = 25/265 (9%)
Query: 1 MVAFCPPQFSYSNMGW-LLEELEPESLISHKEKNYASLEYSLPYHQF------------- 46
M A PP F N GW E + L N L++ +P +
Sbjct: 1 MCALVPPLFP--NFGWPSTGEYDSYYLAGDILNNGGFLDFPVPEETYGAVTAVTQHQNSF 58
Query: 47 --SSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIP 104
S E EI+ P ++ KKLNHNASERDRR+KINSL SSLRS LP Q+KK+SIP
Sbjct: 59 GVSVSSEGNEIDNNP---VVVKKLNHNASERDRRRKINSLFSSLRSCLPASGQSKKLSIP 115
Query: 105 VTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTS 164
T+SR LKYIP+LQ+QV+ L KKKEELL +IS ++ + K + NY S VS +
Sbjct: 116 ATVSRSLKYIPELQEQVKKLIKKKEELLVQISGQRNTECYVKQPPKAVANYIS--TVSAT 173
Query: 165 RLNDTELVIHISSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQ 224
RL D E+++ ISS + + +S +L LE + +L++ SSS++ G RLFY LH QV+K +
Sbjct: 174 RLGDNEVMVQISSSKIHNFSISNVLSGLEEDRFVLVDMSSSRSQGERLFYTLHLQVEKIE 233
Query: 225 RYE--CDDLIQKLSSIYEKQQNNHL 247
Y+ C++L Q++ +YE+ N+++
Sbjct: 234 NYKLNCEELSQRMLYLYEECGNSYI 258
>UniRef100_Q9ZVB5 Hypothetical protein At2g41240 [Arabidopsis thaliana]
Length = 242
Score = 162 bits (410), Expect = 8e-39
Identities = 95/246 (38%), Positives = 149/246 (59%), Gaps = 15/246 (6%)
Query: 1 MVAFCPPQFSYSNMGWLLEELEPESLISHKEKNYASLEYSLP-----YHQFSSPKEHVEI 55
M A PP Y N GW + S + + L++ LP + SS +
Sbjct: 1 MCALVPPL--YPNFGWPCGD---HSFYETDDVSNTFLDFPLPDLTVTHENVSSENNRTLL 55
Query: 56 ERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIP 115
+ P ++ KKLNHNASER+RRKKIN++ SSLRS LP +QTKK+S+ T+S+ LKYIP
Sbjct: 56 DNP----VVMKKLNHNASERERRKKINTMFSSLRSCLPPTNQTKKLSVSATVSQALKYIP 111
Query: 116 DLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHI 175
+LQ+QV+ L KKKEEL +IS +++ ++ + + + A VS++RL++TE+++ I
Sbjct: 112 ELQEQVKKLMKKKEELSFQISGQRDLVYTDQNSKSEEGVTSYASTVSSTRLSETEVMVQI 171
Query: 176 SSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKL 235
SS + K +L +E +GL+L+ +SSS++ G RLFY++H Q+ K + ++L +L
Sbjct: 172 SSLQTEKCSFGNVLSGVEEDGLVLVGASSSRSHGERLFYSMHLQI-KNGQVNSEELGDRL 230
Query: 236 SSIYEK 241
+YEK
Sbjct: 231 LYLYEK 236
>UniRef100_Q941Z7 P0431G06.12 protein [Oryza sativa]
Length = 248
Score = 137 bits (344), Expect = 4e-31
Identities = 76/177 (42%), Positives = 114/177 (63%), Gaps = 8/177 (4%)
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
+KL+HNA ERDRRK++N L SSLR+LLP D TKK+SIP T+SRVLKYIP+LQKQV+ L
Sbjct: 69 RKLSHNAYERDRRKQLNELYSSLRALLPDADHTKKLSIPTTVSRVLKYIPELQKQVENLE 128
Query: 126 KKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHI---SSYEANK 182
+KK+EL + + + V ++ + A +VS + +ND E+++ + S+ +
Sbjct: 129 RKKKELTTTSTTNCKPGV----LGSQLMSEGMAPIVSATCINDMEIMVQVSLLSNVAGSV 184
Query: 183 IPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQ-VDKTQRYECDDLIQKLSSI 238
+PLS+ + LEN GL ++SS+S FG R FY++H Q + T EC ++L +
Sbjct: 185 LPLSKCIKVLENEGLHFISSSTSSGFGNRTFYSIHLQRSEGTINEECPAFCERLEKV 241
>UniRef100_Q9FYE6 Myc-like protein [Arabidopsis thaliana]
Length = 240
Score = 134 bits (336), Expect = 3e-30
Identities = 70/170 (41%), Positives = 116/170 (68%), Gaps = 2/170 (1%)
Query: 63 LMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQ 122
++ KKLNHNASERDRR+K+N+L SSLR+LLP DQ +K+SIP+T++RV+KYIP+ ++++Q
Sbjct: 63 VLEKKLNHNASERDRRRKLNALYSSLRALLPLSDQKRKLSIPMTVARVVKYIPEQKQELQ 122
Query: 123 GLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANK 182
L+++KEELL RIS + + + +S+ ++ + L DTE+ + I++ +
Sbjct: 123 RLSRRKEELLKRISRKTHQEQLRNKAMMDSIDSSSSQRIAANWLTDTEIAVQIATSKWTS 182
Query: 183 IPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLI 232
+ S++L+ LE NGL +++ SSS + R+FY LH Q+ + ++LI
Sbjct: 183 V--SDMLLRLEENGLNVISVSSSVSSTARIFYTLHLQMRGDCKVRLEELI 230
>UniRef100_Q9STJ7 Hypothetical protein T30C3.70 [Arabidopsis thaliana]
Length = 163
Score = 56.2 bits (134), Expect = 8e-07
Identities = 48/171 (28%), Positives = 83/171 (48%), Gaps = 12/171 (7%)
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
+KL H E+ RR+++ SL +SLRSLLP E K S + + YI LQ+ ++ +
Sbjct: 2 EKLVHKEIEKRRRQEMASLYASLRSLLPLEFIQGKRSTSDQVKGAVNYIDYLQRNIKDIN 61
Query: 126 KKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPL 185
K+++L+ + + + + E + +I N+ VV L E+V+ I + P
Sbjct: 62 SKRDDLV--LLSGRSFRSSNEQEWNEISNH----VVIRPCLVGIEIVLSIL-----QTPF 110
Query: 186 SEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLS 236
S +L L +GL +L S + RL + L +V+ + DL L+
Sbjct: 111 SSVLQVLREHGLYVLGYICS-SVNDRLIHTLQAEVNDLALIDLADLKDTLT 160
>UniRef100_Q9STJ6 Hypothetical protein T30C3.80 [Arabidopsis thaliana]
Length = 221
Score = 56.2 bits (134), Expect = 8e-07
Identities = 37/167 (22%), Positives = 85/167 (50%), Gaps = 4/167 (2%)
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
KKL H ER RR+++ +L ++LR+ LP + K ++ ++ + +I D + +++ L+
Sbjct: 43 KKLLHRDIERQRRQEMATLFATLRTHLPLKYIKGKRAVSDHVNGAVNFIKDTEARIKELS 102
Query: 126 KKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPL 185
+++EL + + + A V+ ++ E+V+ +S +PL
Sbjct: 103 ARRDELSRETGQGYKSNPDPGKTGSDVGKSEPATVMVQPHVSGLEVVVSSNSSGPEALPL 162
Query: 186 SEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLI 232
S++L ++ GL +++S +++ RL + + +V+ + C DL+
Sbjct: 163 SKVLETIQEKGLEVMSSFTTRV-NDRLMHTIQVEVNS---FGCIDLL 205
>UniRef100_Q5SMX5 Basic helix-loop-helix protein-like [Oryza sativa]
Length = 338
Score = 53.9 bits (128), Expect = 4e-06
Identities = 36/120 (30%), Positives = 59/120 (49%), Gaps = 10/120 (8%)
Query: 29 HKEKNYASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSL 88
H E Y +Y Y+Q ++ E P P +L H SER RR+K+N +L
Sbjct: 201 HMETTYRRRQY---YYQQAA----AAAEAAPPPPSGNNQLQHTMSERKRREKLNDSFVAL 253
Query: 89 RSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQ 148
+++LP + K SI + R +Y+ L+ ++ L +K EL +R+S R + N E +
Sbjct: 254 KAVLPPGSKKDKTSI---LIRAREYVKSLESKLSELEEKNRELEARLSTRPDDTKNDEEE 310
>UniRef100_Q6ESL3 DNA binding protein-like [Oryza sativa]
Length = 363
Score = 52.4 bits (124), Expect = 1e-05
Identities = 43/182 (23%), Positives = 77/182 (41%), Gaps = 23/182 (12%)
Query: 62 KLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQV 121
K +A +H+ +ER RR++IN ++ LRSLLP +T K S+ ++ V++++ +L++Q
Sbjct: 102 KALAASRSHSEAERRRRQRINGHLARLRSLLPNTTKTDKASL---LAEVIEHVKELKRQT 158
Query: 122 QGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEAN 181
++ + +Y N E + SA EL + +
Sbjct: 159 TA--------IAAAAAAGDYHGNDEDDDDAVVGRRSAAAQQLLPTEADELAVDAAVDAEG 210
Query: 182 KIPLSEILMCLENNGL------------LLLNSSSSKTFGGRLFYNLHFQVDKTQRYECD 229
K+ + L C + L L + T GGR+ L D+ Q+ CD
Sbjct: 211 KLVVRASLCCEDRPDLIPDIARALAALRLRARRAEITTLGGRVRSVLLITADEQQQQHCD 270
Query: 230 DL 231
D+
Sbjct: 271 DV 272
>UniRef100_Q9FLI1 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MIO24
[Arabidopsis thaliana]
Length = 174
Score = 52.0 bits (123), Expect = 2e-05
Identities = 49/158 (31%), Positives = 79/158 (49%), Gaps = 6/158 (3%)
Query: 66 KKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
+K+ H +ER RR+++ SL +SLRSLLP K S ++ + YI LQ++++ L+
Sbjct: 2 EKMMHRETERQRRQEMASLYASLRSLLPLHFIKGKRSTSDQVNEAVNYIKYLQRKIKELS 61
Query: 126 KKKEEL--LSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKI 183
++++L LSR S + + + I N VV L E+++ S +
Sbjct: 62 VRRDDLMVLSRGSLLGSSNGDFKEDVEMISGKN--HVVVRQCLVGVEIMLS-SRCCGGQP 118
Query: 184 PLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVD 221
S +L L GL LLNS SS RL Y + +V+
Sbjct: 119 RFSSVLQVLSEYGLCLLNSISS-IVDDRLVYTIQAEVN 155
>UniRef100_Q9FLI0 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MIO24
[Arabidopsis thaliana]
Length = 209
Score = 52.0 bits (123), Expect = 2e-05
Identities = 48/197 (24%), Positives = 88/197 (44%), Gaps = 15/197 (7%)
Query: 44 HQFSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLP-----GEDQT 98
HQ P+E E ++ KKL H ER RR+++ L +SLRS LP
Sbjct: 12 HQSHMPQERDETKKE-------KKLLHRNIERQRRQEMAILFASLRSQLPLKYIKALSSQ 64
Query: 99 KKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYAVNKESQRKKIPNYNSA 158
K ++ ++ + +I D Q +++ L+ +++EL I ++ A
Sbjct: 65 GKRAMSDHVNGAVSFIKDTQTRIKDLSARRDELKREIG--DPTSLTGSGSGSGSSRSEPA 122
Query: 159 FVVSTSRLNDTELVIHISSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHF 218
V+ ++ E+V+ + PLS +L L GL +++S +++ RL Y +
Sbjct: 123 SVMVQPCVSGFEVVVSSLASGLEAWPLSRVLEVLHGQGLEVISSLTARV-NERLMYTIQV 181
Query: 219 QVDKTQRYECDDLIQKL 235
+V+ ++ L QKL
Sbjct: 182 EVNSFDCFDLAWLQQKL 198
>UniRef100_Q41101 Phaseolin G-box binding protein PG1 [Phaseolus vulgaris]
Length = 642
Score = 51.2 bits (121), Expect = 3e-05
Identities = 35/116 (30%), Positives = 57/116 (48%), Gaps = 6/116 (5%)
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LR+++P + K S+ + + YI +L+ ++ L +
Sbjct: 461 LNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASL---LGDAISYINELKSKLSELESE 517
Query: 128 KEEL---LSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEA 180
K EL L + E A S P N +TS+L D EL + I ++A
Sbjct: 518 KGELEKQLELVKKELELATKSPSPPPGPPPSNKEAKETTSKLIDLELEVKIIGWDA 573
>UniRef100_O49687 BHLH protein-like [Arabidopsis thaliana]
Length = 589
Score = 50.8 bits (120), Expect = 3e-05
Identities = 40/141 (28%), Positives = 68/141 (47%), Gaps = 16/141 (11%)
Query: 31 EKNYASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKK--------LNHNASERDRRKKIN 82
+ N++ LE S+ S+ V +E P+ +K LNH +ER RR+K+N
Sbjct: 373 DSNHSDLEASVAKEAESN---RVVVEPEKKPRKRGRKPANGREEPLNHVEAERQRREKLN 429
Query: 83 SLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYA 142
SLR+++P + K S+ + + YI +L+ ++Q KEEL +I + A
Sbjct: 430 QRFYSLRAVVPNVSKMDKASL---LGDAISYISELKSKLQKAESDKEELQKQIDVMNKEA 486
Query: 143 VNKESQRK--KIPNYNSAFVV 161
N +S K K N S+ ++
Sbjct: 487 GNAKSSVKDRKCLNQESSVLI 507
>UniRef100_Q8W2F6 Putative transcription factor BHLH4 [Arabidopsis thaliana]
Length = 589
Score = 50.8 bits (120), Expect = 3e-05
Identities = 40/141 (28%), Positives = 68/141 (47%), Gaps = 16/141 (11%)
Query: 31 EKNYASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKK--------LNHNASERDRRKKIN 82
+ N++ LE S+ S+ V +E P+ +K LNH +ER RR+K+N
Sbjct: 373 DSNHSDLEASVAKEAESN---RVVVEPEKKPRKRGRKPANGREEPLNHVEAERQRREKLN 429
Query: 83 SLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISHRQEYA 142
SLR+++P + K S+ + + YI +L+ ++Q KEEL +I + A
Sbjct: 430 QRFYSLRAVVPNVSKMDKASL---LGDAISYISELKSKLQKAESDKEELQKQIDVMNKEA 486
Query: 143 VNKESQRK--KIPNYNSAFVV 161
N +S K K N S+ ++
Sbjct: 487 GNAKSSVKDRKCLNQESSVLI 507
>UniRef100_Q6JAH5 B1-1 [Sorghum bicolor]
Length = 509
Score = 50.4 bits (119), Expect = 4e-05
Identities = 34/88 (38%), Positives = 49/88 (55%), Gaps = 8/88 (9%)
Query: 69 NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKK 128
NH SER RR+K+N + L+SL+P + K SI ++ + Y+ +LQ++VQ L +
Sbjct: 314 NHVMSERKRREKLNEMFLILKSLVPSIHKVDKASI---LAETIAYLKELQRRVQEL-ESS 369
Query: 129 EELLSRISHRQEYAV----NKESQRKKI 152
EL SR S NKES RKK+
Sbjct: 370 RELTSRPSETTRPITRQHGNKESVRKKL 397
>UniRef100_Q41286 Myc-like regulatory R gene product [Sorghum bicolor]
Length = 145
Score = 49.7 bits (117), Expect = 7e-05
Identities = 32/89 (35%), Positives = 50/89 (55%), Gaps = 9/89 (10%)
Query: 69 NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKK 128
NH SER RR+K+N + L+ L+P + K+S+ ++ + Y+ +LQ++VQ L K
Sbjct: 2 NHVMSERKRREKLNEMFLILKLLVPSIQKVAKVSL---LAETIAYLKELQRKVQEL-KSS 57
Query: 129 EELLSRISH-----RQEYAVNKESQRKKI 152
ELLSR S + + ES RKK+
Sbjct: 58 RELLSRPSETTARPTKPCGIGSESVRKKL 86
>UniRef100_Q700B9 MYC transcription factor [Solanum tuberosum]
Length = 646
Score = 49.7 bits (117), Expect = 7e-05
Identities = 31/91 (34%), Positives = 50/91 (54%), Gaps = 8/91 (8%)
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LR+++P + K S+ + + YI +L+ +VQ
Sbjct: 472 LNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASL---LGDAIAYINELKSKVQNSDLD 528
Query: 128 KEELLSRISHRQEYAVNKESQRKKIPNYNSA 158
KEEL S+I ++ KE K NY+S+
Sbjct: 529 KEELRSQIE-----SLRKELANKGSSNYSSS 554
>UniRef100_P13027 Anthocyanin regulatory R-S protein [Zea mays]
Length = 612
Score = 49.7 bits (117), Expect = 7e-05
Identities = 33/95 (34%), Positives = 51/95 (52%), Gaps = 10/95 (10%)
Query: 64 MAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQG 123
M+ NH SER RR+K+N + L+SLLP + K SI ++ + Y+ +LQ++VQ
Sbjct: 413 MSATKNHVMSERKRREKLNEMFLVLKSLLPSIHRVNKASI---LAETIAYLKELQRRVQE 469
Query: 124 LTKKKE------ELLSRISHRQEYAVNKESQRKKI 152
L +E E +R+ R N ES RK++
Sbjct: 470 LESSREPASRPSETTTRLITRPSRG-NNESVRKEV 503
>UniRef100_Q700C0 MYC transcription factor [Solanum tuberosum]
Length = 692
Score = 49.3 bits (116), Expect = 1e-04
Identities = 32/113 (28%), Positives = 59/113 (51%), Gaps = 9/113 (7%)
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LR+++P + K S+ + + YI +L+ ++Q
Sbjct: 516 LNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASL---LGDAISYINELKSKLQNTESD 572
Query: 128 KEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEA 180
KE+L S+I + KES+R P N + ++ D ++ + I ++A
Sbjct: 573 KEDLKSQIED-----LKKESRRPGPPPPNQDLKIG-GKIVDVDIDVKIIGWDA 619
>UniRef100_Q40836 Myc-like regulatory R gene product [Pennisetum americanum]
Length = 145
Score = 48.9 bits (115), Expect = 1e-04
Identities = 27/73 (36%), Positives = 44/73 (59%), Gaps = 3/73 (4%)
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKE 129
H SER RR+K+N + L+SL P + K+SI +++ + Y+ DLQ++VQ L +E
Sbjct: 3 HVMSERKRREKLNEMFLVLKSLAPSIHRMDKVSI---LAQTIAYLKDLQRRVQELEYSRE 59
Query: 130 ELLSRISHRQEYA 142
++SR S + A
Sbjct: 60 PIISRPSETTKVA 72
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 420,661,418
Number of Sequences: 2790947
Number of extensions: 16725096
Number of successful extensions: 50948
Number of sequences better than 10.0: 1092
Number of HSP's better than 10.0 without gapping: 467
Number of HSP's successfully gapped in prelim test: 625
Number of HSP's that attempted gapping in prelim test: 50228
Number of HSP's gapped (non-prelim): 1215
length of query: 260
length of database: 848,049,833
effective HSP length: 125
effective length of query: 135
effective length of database: 499,181,458
effective search space: 67389496830
effective search space used: 67389496830
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC125478.4


