

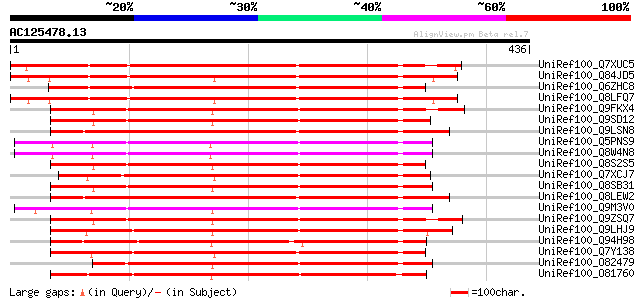
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.13 - phase: 0
(436 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XUC5 OSJNBa0088A01.23 protein [Oryza sativa] 338 2e-91
UniRef100_Q84JD5 Hypothetical protein At5g06750 [Arabidopsis tha... 329 1e-88
UniRef100_Q6ZHC8 Hypothetical protein OJ1717_A09.24 [Oryza sativa] 327 4e-88
UniRef100_Q8LFQ7 Protein phosphatase 2C-like [Arabidopsis thaliana] 327 5e-88
UniRef100_Q9FKX4 Protein phosphatase 2C-like protein [Arabidopsi... 288 2e-76
UniRef100_Q9SD12 Protein phosphatase 2C-like protein [Arabidopsi... 286 1e-75
UniRef100_Q9LSN8 Protein phosphatase 2C-like protein [Arabidopsi... 282 1e-74
UniRef100_Q5PNS9 At4g38520 [Arabidopsis thaliana] 280 6e-74
UniRef100_Q8W4N8 Hypothetical protein At4g38520; F20M13.80 [Arab... 280 6e-74
UniRef100_Q8S2S5 Protein phosphatase 2c-like protein [Thellungie... 280 6e-74
UniRef100_Q7XCJ7 Hypothetical protein [Oryza sativa] 278 3e-73
UniRef100_Q8SB31 Hypothetical protein OJ1540_H01.4 [Oryza sativa] 277 4e-73
UniRef100_Q8LEW2 Protein phosphatase-2c, putative [Arabidopsis t... 276 6e-73
UniRef100_Q9M3V0 Protein phosphatase 2C [Fagus sylvatica] 275 1e-72
UniRef100_Q9ZSQ7 Protein phosphatase 2C homolog [Mesembryanthemu... 274 4e-72
UniRef100_Q9LHJ9 Protein phosphatase 2C [Arabidopsis thaliana] 273 5e-72
UniRef100_Q94H98 Hypothetical protein OSJNBb0048A17.8 [Oryza sat... 272 1e-71
UniRef100_Q7Y138 Hypothetical protein OSJNBa0078D06.30 [Oryza sa... 271 3e-71
UniRef100_O82479 Protein phosphatase-2c [Mesembryanthemum crysta... 270 5e-71
UniRef100_O81760 Hypothetical protein F17I5.110 [Arabidopsis tha... 269 1e-70
>UniRef100_Q7XUC5 OSJNBa0088A01.23 protein [Oryza sativa]
Length = 388
Score = 338 bits (866), Expect = 2e-91
Identities = 193/392 (49%), Positives = 243/392 (61%), Gaps = 29/392 (7%)
Query: 1 MHPWLENIVNLC----------RKRKNDNINMALFDQDPHASSIDLKRHCYGQFSSAFVQ 50
M PWLE I + C R+ + D D S DL RH G+FS A VQ
Sbjct: 1 MWPWLERIASACWDRVRRYALTRRDEEDGSGSGGDADDLLLWSRDLVRHAAGEFSFAVVQ 60
Query: 51 ANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITEPTL 110
AN+ +EDHSQVE + A F+GVYDGH G EAS FI+ HL HL+R + I+E +
Sbjct: 61 ANDVLEDHSQVETGAA-ATFIGVYDGHGGAEASRFISNHLAAHLVRLAQ-ERGTISEDIV 118
Query: 111 RDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTMV-N 169
R+A SATE GFL V + + K ++ +GSCCL GIIWK TL++ANLGDSRAV+G + +
Sbjct: 119 RNAFSATEEGFLSLVRRTHLIKPSIASIGSCCLVGIIWKGTLYLANLGDSRAVVGCLTGS 178
Query: 170 NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKR 229
NKI AEQLTRDHN E +R+EL S HPDD+ IV+ + VWR+KGII VSRSIGD YLK+
Sbjct: 179 NKIVAEQLTRDHNASMEEVRQELRSLHPDDSQIVVLKNGVWRIKGIIQVSRSIGDAYLKK 238
Query: 230 PEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAVE 289
EF+LD S +F + EP R VL++EP + +R L D F IFASDGLW+ L+N+QAVE
Sbjct: 239 QEFALDPSMTRF-HLSEPLRRPVLTSEPSIYTRVLHSQDSFFIFASDGLWEHLTNQQAVE 297
Query: 290 IVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFLD 349
IV NN R GIA+RLV L +AA R YN +K G RR+FHDDI+V+VVF+D
Sbjct: 298 IVHNNPREGIARRLVKAALKEAARKREMKYNDIKKLE----KGVRRFFHDDITVVVVFID 353
Query: 350 KKSILRMPLHNLSYKSSSARPTTS--AFAESG 379
H L + P S F +SG
Sbjct: 354 ---------HELLQDGDESTPEISVRGFVDSG 376
>UniRef100_Q84JD5 Hypothetical protein At5g06750 [Arabidopsis thaliana]
Length = 393
Score = 329 bits (843), Expect = 1e-88
Identities = 193/396 (48%), Positives = 249/396 (62%), Gaps = 27/396 (6%)
Query: 1 MHPWLENIVNLCRK--RKNDNINMALFDQDPHAS-----------SIDLKRHCYGQFSSA 47
M WL + C + R+ +N D D H S +L+RH +G FS A
Sbjct: 1 MFSWLARMALFCLRPMRRYGRMNRDDDDDDDHDGDSSSSGDSLLWSRELERHSFGDFSIA 60
Query: 48 FVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITE 107
VQANE +EDHSQVE + A+F+GVYDGH G EAS +I+ HLF HL+R R + I+E
Sbjct: 61 VVQANEVIEDHSQVETGNG-AVFVGVYDGHGGPEASRYISDHLFSHLMRVSR-ERSCISE 118
Query: 108 PTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM 167
LR A SATE GFL V + K + VGSCCL G+IWK TL +AN+GDSRAV+G+M
Sbjct: 119 EALRAAFSATEEGFLTLVRRTCGLKPLIAAVGSCCLVGVIWKGTLLIANVGDSRAVLGSM 178
Query: 168 VNN-----KIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSI 222
+N KI AEQLT DHN E +R+EL S HPDD+ IV+ + VWR+KGII VSRSI
Sbjct: 179 GSNNNRSNKIVAEQLTSDHNAALEEVRQELRSLHPDDSHIVVLKHGVWRIKGIIQVSRSI 238
Query: 223 GDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFL 282
GD YLKRPEFSLD SFP+F + E R VLSAEP + +R L +DKF+IFASDGLW+ +
Sbjct: 239 GDAYLKRPEFSLDPSFPRF-HLAEELQRPVLSAEPCVYTRVLQTSDKFVIFASDGLWEQM 297
Query: 283 SNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDIS 342
+N+QAVEIV + R GIA+RLV + AA R Y+ +K G RR+FHDDI+
Sbjct: 298 TNQQAVEIVNKHPRPGIARRLVRRAITIAAKKREMNYDDLKKVE----RGVRRFFHDDIT 353
Query: 343 VIVVFLDKKSIL--RMPLHNLSYKSSSARPTTSAFA 376
V+V+F+D + ++ + + LS K S S F+
Sbjct: 354 VVVIFIDNELLMVEKATVPELSIKGFSHTVGPSKFS 389
>UniRef100_Q6ZHC8 Hypothetical protein OJ1717_A09.24 [Oryza sativa]
Length = 387
Score = 327 bits (838), Expect = 4e-88
Identities = 176/318 (55%), Positives = 224/318 (70%), Gaps = 8/318 (2%)
Query: 33 SIDLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFD 92
S DL RH G+FS A VQANEA+EDHSQVE S A F+GVYDGH G +A+ FI+ HLF
Sbjct: 42 SRDLGRHAAGEFSFAVVQANEALEDHSQVETGSA-ATFVGVYDGHGGADAARFISDHLFA 100
Query: 93 HLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTL 152
HL+R R +E ++E +R A SATE GFL V + K + VGSCCL GIIW+ L
Sbjct: 101 HLIRLARESET-VSEEVVRGAFSATEEGFLTLVRRTQFLKPMIAAVGSCCLVGIIWRGVL 159
Query: 153 HVANLGDSRAVIGTMVN-NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWR 211
+VANLGDSRAV+G + NKI AEQ+TRDHN E +R+EL+S HPDD+ IV+ + VWR
Sbjct: 160 YVANLGDSRAVVGYLGRTNKITAEQITRDHNACKEEVRQELISRHPDDSQIVVLKHGVWR 219
Query: 212 VKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFL 271
+KGII VSR+IGD YLKR EF+LD S +F + EP R VL+AEP + +R L+ D+F+
Sbjct: 220 IKGIIQVSRTIGDAYLKRREFALDPSITRF-RLSEPLRRPVLTAEPSICTRVLSLQDQFV 278
Query: 272 IFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGD 331
IFASDGLW+ L+N+QAV+IV N R GIAKRLV+T L +AA R + +K
Sbjct: 279 IFASDGLWEHLTNQQAVDIVYKNPRAGIAKRLVNTALKEAARKREMRFVDLKKVE----K 334
Query: 332 GNRRYFHDDISVIVVFLD 349
G RR+FHDDI+V+VV++D
Sbjct: 335 GVRRFFHDDITVVVVYID 352
>UniRef100_Q8LFQ7 Protein phosphatase 2C-like [Arabidopsis thaliana]
Length = 393
Score = 327 bits (837), Expect = 5e-88
Identities = 192/396 (48%), Positives = 248/396 (62%), Gaps = 27/396 (6%)
Query: 1 MHPWLENIVNLCRK--RKNDNINMALFDQDPHAS-----------SIDLKRHCYGQFSSA 47
M WL + C + R+ +N D D H S +L+RH +G FS A
Sbjct: 1 MFSWLARMALFCLRPMRRYGRMNRDDDDDDDHDGDSSSSGDSLLWSRELERHSFGDFSIA 60
Query: 48 FVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITE 107
VQANE +EDHSQVE + A+F+GVYDGH G EAS +I+ HLF HL+R R + I+E
Sbjct: 61 VVQANEVIEDHSQVETGNG-AVFVGVYDGHGGPEASRYISDHLFSHLMRVSR-ERSCISE 118
Query: 108 PTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM 167
LR A SATE GFL V + K + VGSCCL G+IWK TL +AN+GDSRAV+G+M
Sbjct: 119 EALRAAFSATEEGFLTLVRRTCGLKPLIAAVGSCCLVGVIWKGTLLIANVGDSRAVLGSM 178
Query: 168 VNN-----KIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSI 222
+N KI AEQLT DHN E +R+EL S HPDD+ IV+ + VWR+KGII VSRSI
Sbjct: 179 GSNNNRSNKIVAEQLTSDHNAALEEVRQELRSLHPDDSHIVVLKHGVWRIKGIIQVSRSI 238
Query: 223 GDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFL 282
GD YLKRPEFSLD SFP+F + E R V SAEP + +R L +DKF+IFASDGLW+ +
Sbjct: 239 GDAYLKRPEFSLDPSFPRF-HLAEELQRPVSSAEPCVYTRVLQTSDKFVIFASDGLWEQM 297
Query: 283 SNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDIS 342
+N+QAVEIV + R GIA+RLV + AA R Y+ +K G RR+FHDDI+
Sbjct: 298 TNQQAVEIVNKHPRPGIARRLVRRAITIAAKKREMNYDDLKKVE----RGVRRFFHDDIT 353
Query: 343 VIVVFLDKKSIL--RMPLHNLSYKSSSARPTTSAFA 376
V+V+F+D + ++ + + LS K S S F+
Sbjct: 354 VVVIFIDNELLMVEKATVPELSIKGFSHTVGPSKFS 389
>UniRef100_Q9FKX4 Protein phosphatase 2C-like protein [Arabidopsis thaliana]
Length = 385
Score = 288 bits (737), Expect = 2e-76
Identities = 162/358 (45%), Positives = 221/358 (61%), Gaps = 25/358 (6%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKAL--------FLGVYDGHAGFEASVFI 86
D H +G FS A VQAN +ED SQVE L F+GVYDGH G E S F+
Sbjct: 39 DSAHHLFGDFSMAVVQANNLLEDQSQVESGPLTTLSSSGPYGTFVGVYDGHGGPETSRFV 98
Query: 87 TQHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGI 146
HLF HL R A ++ ++ +R A ATE GFL V K + K ++ VGSCCL G+
Sbjct: 99 NDHLFHHLKRFA-AEQDSMSVDVIRKAYEATEEGFLGVVAKQWAVKPHIAAVGSCCLIGV 157
Query: 147 IWKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVM 204
+ L+VAN+GDSRAV+G ++ ++ A QL+ +HN E++R+E+ S HPDD+ IV+
Sbjct: 158 VCDGKLYVANVGDSRAVLGKVIKATGEVNALQLSAEHNVSIESVRQEMHSLHPDDSHIVV 217
Query: 205 YEREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDL 264
+ VWRVKGII VSRSIGD YLK+ EF+ + + K+ + EP R +LS EP + DL
Sbjct: 218 LKHNVWRVKGIIQVSRSIGDVYLKKSEFNKEPLYTKYR-LREPMKRPILSWEPSITVHDL 276
Query: 265 TENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKN 324
+D+FLIFASDGLW+ LSN++AVEIVQN+ RNGIA+RLV L +AA R Y+ +
Sbjct: 277 QPDDQFLIFASDGLWEQLSNQEAVEIVQNHPRNGIARRLVKAALQEAAKKREMRYSDLNK 336
Query: 325 ANLGRGDGNRRYFHDDISVIVVFLDKKSILRMPLHNLSYKSSSARPTTSAFAESGLTM 382
G RR+FHDDI+V+V+FLD NL ++SS + + + G+T+
Sbjct: 337 IE----RGVRRHFHDDITVVVLFLDT---------NLLSRASSLKTPSVSIRGGGITL 381
>UniRef100_Q9SD12 Protein phosphatase 2C-like protein [Arabidopsis thaliana]
Length = 379
Score = 286 bits (731), Expect = 1e-75
Identities = 151/328 (46%), Positives = 208/328 (63%), Gaps = 15/328 (4%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKAL-------FLGVYDGHAGFEASVFIT 87
D +H G+FS A VQAN +ED SQVE L F+G+YDGH G E S F+
Sbjct: 37 DFGQHLVGEFSMAVVQANNLLEDQSQVESGPLSTLDSGPYGTFIGIYDGHGGPETSRFVN 96
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
HLF HL R A + ++ ++ A ATE GFL V K + K + VGSCCL G+I
Sbjct: 97 DHLFQHLKRFA-AEQASMSVDVIKKAYEATEEGFLGVVTKQWPTKPQIAAVGSCCLVGVI 155
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L++AN+GDSRAV+G + ++ A QL+ +HN E++R+E+ S HPDD+ IVM
Sbjct: 156 CGGMLYIANVGDSRAVLGRAMKATGEVIALQLSAEHNVSIESVRQEMHSLHPDDSHIVML 215
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ VWRVKG+I +SRSIGD YLK+ EF+ + + K+ + EPF R +LS EP + ++
Sbjct: 216 KHNVWRVKGLIQISRSIGDVYLKKAEFNKEPLYTKYR-IREPFKRPILSGEPTITEHEIQ 274
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
DKFLIFASDGLW+ +SN++AV+IVQN+ RNGIA+RLV L +AA R Y+ +K
Sbjct: 275 PQDKFLIFASDGLWEQMSNQEAVDIVQNHPRNGIARRLVKMALQEAAKKREMRYSDLKKI 334
Query: 326 NLGRGDGNRRYFHDDISVIVVFLDKKSI 353
G RR+FHDDI+V+++FLD +
Sbjct: 335 E----RGVRRHFHDDITVVIIFLDTNQV 358
>UniRef100_Q9LSN8 Protein phosphatase 2C-like protein [Arabidopsis thaliana]
Length = 379
Score = 282 bits (722), Expect = 1e-74
Identities = 156/336 (46%), Positives = 210/336 (62%), Gaps = 7/336 (2%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHL 94
DL ++C G FS A +QAN+ +ED SQVE + F+GVYDGH G EA+ ++ HLF+H
Sbjct: 45 DLGKYCGGDFSMAVIQANQVLEDQSQVE-SGNFGTFVGVYDGHGGPEAARYVCDHLFNHF 103
Query: 95 LRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHV 154
+ +T T+ A ATE GF V + +++ NL VG+CCL G+I++ TL V
Sbjct: 104 REISAETQGVVTRETIERAFHATEEGFASIVSELWQEIPNLATVGTCCLVGVIYQNTLFV 163
Query: 155 ANLGDSRAVIGTMVN-NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVK 213
A+LGDSR V+G N + A QL+ +HN +E IR EL HPDD IV++ VWRVK
Sbjct: 164 ASLGDSRVVLGKKGNCGGLSAIQLSTEHNANNEDIRWELKDLHPDDPQIVVFRHGVWRVK 223
Query: 214 GIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIF 273
GII VSRSIGD Y+KRPEF+ + KF + EP R ++SA P + S L ND FLIF
Sbjct: 224 GIIQVSRSIGDMYMKRPEFNKEPISQKF-RIAEPMKRPLMSATPTILSHPLHPNDSFLIF 282
Query: 274 ASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGN 333
ASDGLW+ L+NE+AVEIV N+ R G AKRL+ L +AA R Y+ ++ +
Sbjct: 283 ASDGLWEHLTNEKAVEIVHNHPRAGSAKRLIKAALHEAARKREMRYSDLRKID----KKV 338
Query: 334 RRYFHDDISVIVVFLDKKSILRMPLHNLSYKSSSAR 369
RR+FHDDI+VIVVFL+ I R +++ + S R
Sbjct: 339 RRHFHDDITVIVVFLNHDLISRGHINSTQDTTVSIR 374
>UniRef100_Q5PNS9 At4g38520 [Arabidopsis thaliana]
Length = 400
Score = 280 bits (716), Expect = 6e-74
Identities = 159/364 (43%), Positives = 216/364 (58%), Gaps = 19/364 (5%)
Query: 5 LENIVNLCRKRKNDNINMALFDQDPHASSI----DLKRHCYGQFSSAFVQANEAMEDHSQ 60
L N +N C ++D + D + D +H +G FS A VQAN +ED SQ
Sbjct: 5 LMNFLNACLWPRSDQQARSASDSGGRQEGLLWFRDSGQHVFGDFSMAVVQANSLLEDQSQ 64
Query: 61 VEVASRKA-------LFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITEPTLRDA 113
+E S + F+GVYDGH G E S FI H+F HL R A + ++ ++ A
Sbjct: 65 LESGSLSSHDSGPFGTFVGVYDGHGGPETSRFINDHMFHHLKRFT-AEQQCMSSEVIKKA 123
Query: 114 VSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM--VNNK 171
ATE GFL V ++ + + VGSCCL +I L+VAN GDSRAV+G + V +
Sbjct: 124 FQATEEGFLSIVTNQFQTRPQIATVGSCCLVSVICDGKLYVANAGDSRAVLGQVMRVTGE 183
Query: 172 IQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPE 231
A QL+ +HN E++R+EL + HPD IV+ + VWRVKGII VSRSIGD YLKR E
Sbjct: 184 AHATQLSAEHNASIESVRRELQALHPDHPDIVVLKHNVWRVKGIIQVSRSIGDVYLKRSE 243
Query: 232 FSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAVEIV 291
F+ + + KF + PF + +LSAEP + L +D+F+I ASDGLW+ +SN++AV+IV
Sbjct: 244 FNREPLYAKFR-LRSPFSKPLLSAEPAITVHTLEPHDQFIICASDGLWEHMSNQEAVDIV 302
Query: 292 QNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFLDKK 351
QN+ RNGIAKRLV L +AA R Y+ +K + G RR+FHDDI+VIVVF D
Sbjct: 303 QNHPRNGIAKRLVKVALQEAAKKREMRYSDLKKID----RGVRRHFHDDITVIVVFFDTN 358
Query: 352 SILR 355
+ R
Sbjct: 359 LVSR 362
>UniRef100_Q8W4N8 Hypothetical protein At4g38520; F20M13.80 [Arabidopsis thaliana]
Length = 400
Score = 280 bits (716), Expect = 6e-74
Identities = 159/364 (43%), Positives = 216/364 (58%), Gaps = 19/364 (5%)
Query: 5 LENIVNLCRKRKNDNINMALFDQDPHASSI----DLKRHCYGQFSSAFVQANEAMEDHSQ 60
L N +N C ++D + D + D +H +G FS A VQAN +ED SQ
Sbjct: 5 LMNFLNACLWPRSDQQARSASDSGGRQEGLLWFSDSGQHVFGDFSMAVVQANSLLEDQSQ 64
Query: 61 VEVASRKA-------LFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITEPTLRDA 113
+E S + F+GVYDGH G E S FI H+F HL R A + ++ ++ A
Sbjct: 65 LESGSLSSHDSGPFGTFVGVYDGHGGPETSRFINDHMFHHLKRFT-AEQQCMSSEVIKKA 123
Query: 114 VSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM--VNNK 171
ATE GFL V ++ + + VGSCCL +I L+VAN GDSRAV+G + V +
Sbjct: 124 FQATEEGFLSIVTNQFQTRPQIATVGSCCLVSVICDGKLYVANAGDSRAVLGQVMRVTGE 183
Query: 172 IQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPE 231
A QL+ +HN E++R+EL + HPD IV+ + VWRVKGII VSRSIGD YLKR E
Sbjct: 184 AHATQLSAEHNASIESVRRELQALHPDHPDIVVLKHNVWRVKGIIQVSRSIGDVYLKRSE 243
Query: 232 FSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAVEIV 291
F+ + + KF + PF + +LSAEP + L +D+F+I ASDGLW+ +SN++AV+IV
Sbjct: 244 FNREPLYAKFR-LRSPFSKPLLSAEPAITVHTLEPHDQFIICASDGLWEHMSNQEAVDIV 302
Query: 292 QNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFLDKK 351
QN+ RNGIAKRLV L +AA R Y+ +K + G RR+FHDDI+VIVVF D
Sbjct: 303 QNHPRNGIAKRLVKVALQEAAKKREMRYSDLKKID----RGVRRHFHDDITVIVVFFDTN 358
Query: 352 SILR 355
+ R
Sbjct: 359 LVSR 362
>UniRef100_Q8S2S5 Protein phosphatase 2c-like protein [Thellungiella halophila]
Length = 378
Score = 280 bits (716), Expect = 6e-74
Identities = 151/324 (46%), Positives = 205/324 (62%), Gaps = 15/324 (4%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKAL-------FLGVYDGHAGFEASVFIT 87
D +H G+FS A VQAN +ED SQVE L F+G+YDGH G E S F+
Sbjct: 36 DSGQHLLGEFSMAVVQANNLLEDQSQVESGPLSTLDSGPFGTFIGIYDGHGGPETSRFVN 95
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
HLF HL R A ++ +R A ATE GFL V K + K + VGSCCL G+I
Sbjct: 96 DHLFQHLKRFA-AEHASMSVDVIRKAYEATEEGFLGVVTKQWPVKPQIAAVGSCCLVGVI 154
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L++AN+GDSRAV+G +N ++ A QL+ +HN E++R+E+ S HPDD+ IV+
Sbjct: 155 CGGRLYIANVGDSRAVLGRAMNATGEVIALQLSAEHNVSIESVRQEMRSLHPDDSHIVVL 214
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ VWRVKG+I +SRSIGD YLK+ EF+ + + K+ + EP R +LS EP + ++
Sbjct: 215 KHNVWRVKGLIQISRSIGDIYLKKAEFNKEPLYTKYR-LREPIKRPILSGEPSITEHEIQ 273
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
D+FLIFASDGLW+ +SN++AV+IVQN+ RNGIA+RLV L AA R Y+ +K
Sbjct: 274 PQDQFLIFASDGLWEQMSNQEAVDIVQNHPRNGIARRLVKMALQAAAKKREMRYSDLKKI 333
Query: 326 NLGRGDGNRRYFHDDISVIVVFLD 349
G RR+FHDDI+V+V+FLD
Sbjct: 334 E----RGVRRHFHDDITVVVIFLD 353
>UniRef100_Q7XCJ7 Hypothetical protein [Oryza sativa]
Length = 393
Score = 278 bits (710), Expect = 3e-73
Identities = 148/321 (46%), Positives = 203/321 (63%), Gaps = 15/321 (4%)
Query: 42 GQFSSAFVQANEAMEDHSQVEVA-------SRKALFLGVYDGHAGFEASVFITQHLFDHL 94
G+FS A VQAN +EDHSQVE + + +GVYDGH G E + +I HLF+HL
Sbjct: 48 GEFSMAVVQANNLLEDHSQVESGPLSTTDPNLQGTLVGVYDGHGGPETARYINDHLFNHL 107
Query: 95 LRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHV 154
R + ++ +R A ATE GF V + + L VGSCCL G+I L++
Sbjct: 108 -RGFASEHKCMSADVIRKAFRATEEGFFSVVSSQWSMRPQLAAVGSCCLVGVICAGNLYI 166
Query: 155 ANLGDSRAVIGTMVNN--KIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRV 212
ANLGDSRAV+G +V ++ A QL+ +HN E +R+EL + HPDD IV+ + VWRV
Sbjct: 167 ANLGDSRAVLGRLVKGTGEVLAMQLSAEHNASFEEVRRELQAAHPDDPHIVVLKHNVWRV 226
Query: 213 KGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLI 272
KGII ++RSIGD YLK+PEF+ + KF + E F R +LS+EP + L D+F+I
Sbjct: 227 KGIIQITRSIGDVYLKKPEFNREPLHSKFR-LQETFRRPLLSSEPAIVVHQLQTTDQFII 285
Query: 273 FASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDG 332
FASDGLW+ +SN++AV++VQ+N RNGIA+RLV + QAA R Y+ +K + G
Sbjct: 286 FASDGLWEHISNQEAVDLVQHNPRNGIARRLVKAAMQQAAKKREMRYSDLKKID----RG 341
Query: 333 NRRYFHDDISVIVVFLDKKSI 353
RR+FHDDI+V+VVF D +I
Sbjct: 342 VRRHFHDDITVVVVFFDSNAI 362
>UniRef100_Q8SB31 Hypothetical protein OJ1540_H01.4 [Oryza sativa]
Length = 392
Score = 277 bits (709), Expect = 4e-73
Identities = 152/330 (46%), Positives = 206/330 (62%), Gaps = 15/330 (4%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKAL-------FLGVYDGHAGFEASVFIT 87
D +H G+FS A VQAN +ED Q+E L F+GVYDGH G E + +I
Sbjct: 39 DTGQHVNGEFSMAVVQANNLLEDQCQIESGPLSFLDSGPYGTFVGVYDGHGGPETACYIN 98
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
HLF HL R + +N I+ L+ A ATE GF V K + K + VGSCCL G+I
Sbjct: 99 DHLFHHLKRFA-SEQNSISADVLKKAYEATEDGFFSVVTKQWPVKPQIAAVGSCCLVGVI 157
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L+VAN+GDSR V+G V ++ A QL+ +HN E++RKEL S HP+D IV+
Sbjct: 158 CGGILYVANVGDSRVVLGRHVKATGEVLAVQLSAEHNVSIESVRKELQSMHPEDRHIVVL 217
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ VWRVKG+I V RSIGD YLKR EF+ + + KF + EPF + +LS+EP + + L
Sbjct: 218 KHNVWRVKGLIQVCRSIGDAYLKRSEFNREPLYAKFR-LREPFHKPILSSEPSISVQPLQ 276
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
+D+FLIFASDGLW+ L+N++AV+IV ++ RNG A+RL+ L +AA R Y+ +K
Sbjct: 277 PHDQFLIFASDGLWEHLTNQEAVDIVHSSPRNGSARRLIKAALQEAAKKREMRYSDLKKI 336
Query: 326 NLGRGDGNRRYFHDDISVIVVFLDKKSILR 355
+ G RR+FHDDI+VIVVFLD + R
Sbjct: 337 D----RGVRRHFHDDITVIVVFLDSSLVSR 362
>UniRef100_Q8LEW2 Protein phosphatase-2c, putative [Arabidopsis thaliana]
Length = 384
Score = 276 bits (707), Expect = 6e-73
Identities = 155/336 (46%), Positives = 208/336 (61%), Gaps = 7/336 (2%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHL 94
DL ++C G FS A +QAN+ +ED SQVE + F+GVYDGH G EA+ ++ HLF H
Sbjct: 50 DLGKYCGGDFSMAVIQANQVLEDQSQVE-SGNFGTFVGVYDGHGGPEAARYVCDHLFXHF 108
Query: 95 LRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHV 154
+ +T T+ A ATE GF V + +++ NL VG+CCL G+I++ TL V
Sbjct: 109 REISAETQGVVTRETIERAFHATEEGFASIVSELWQEIPNLATVGTCCLVGVIYQNTLFV 168
Query: 155 ANLGDSRAVIGTMVN-NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVK 213
A+LGDSR V+G N + A QL+ +HN +E IR EL HPDD IV++ V RVK
Sbjct: 169 ASLGDSRVVLGKKGNCGGLSAIQLSTEHNANNEDIRWELKDLHPDDPQIVVFRHGVXRVK 228
Query: 214 GIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIF 273
GII VSRSIGD Y+KRPEF+ + KF + EP R ++SA P + S L ND FLIF
Sbjct: 229 GIIQVSRSIGDMYMKRPEFNKEPISQKF-RIAEPMKRPLMSATPTILSHPLHPNDSFLIF 287
Query: 274 ASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGN 333
ASDGLW+ L+NE+AVEIV N+ R G AKRL+ L +AA R Y+ ++ +
Sbjct: 288 ASDGLWEHLTNEKAVEIVHNHPRAGSAKRLIKAALHEAARKREMRYSDLRKID----KKV 343
Query: 334 RRYFHDDISVIVVFLDKKSILRMPLHNLSYKSSSAR 369
RR+FHDDI+VIVVFL+ I R +++ + S R
Sbjct: 344 RRHFHDDITVIVVFLNHDLISRGHINSTQDTTVSIR 379
>UniRef100_Q9M3V0 Protein phosphatase 2C [Fagus sylvatica]
Length = 397
Score = 275 bits (704), Expect = 1e-72
Identities = 156/364 (42%), Positives = 221/364 (59%), Gaps = 19/364 (5%)
Query: 5 LENIVNLCRKRKNDNI----NMALFDQDPHASSIDLKRHCYGQFSSAFVQANEAMEDHSQ 60
L N + C + K+D + A QD D +H G+FS QAN +ED SQ
Sbjct: 5 LMNFLRACFRAKSDRHVHTGSNARGRQDGLLWYKDSGQHFNGEFSYGCGQANNLLEDQSQ 64
Query: 61 VEVASRK-------ALFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITEPTLRDA 113
+E S F+GVYDGH G E S ++ HLF HL R + ++ ++ +R A
Sbjct: 65 IESGSLSLNETGPYGTFIGVYDGHGGPETSRYVNNHLFQHLKRFT-SEQHSMSVEVIRKA 123
Query: 114 VSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTMVN--NK 171
ATE GFL V K + K + VGSCCL G+I TL++ANLGDSRAV+G ++ +
Sbjct: 124 YQATEEGFLSQVTKQWPLKPQIAAVGSCCLVGVICGGTLYIANLGDSRAVLGRVMKATGE 183
Query: 172 IQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPE 231
+ + QL+ +HN E++R+EL S HP+D IV+ + VWRVKG+I +SRSIGD YLK+ E
Sbjct: 184 VLSIQLSAEHNVAIESVRQELHSLHPEDPQIVVLKHNVWRVKGLIQISRSIGDVYLKKAE 243
Query: 232 FSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAVEIV 291
F+ + + KF + EPF + +LSA+P + L +D+F+IFASDGLW+ LSN++AV+IV
Sbjct: 244 FNREPLYAKFR-LREPFKKPILSADPAISVHQLQPHDQFVIFASDGLWEHLSNQEAVDIV 302
Query: 292 QNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFLDKK 351
QN+ R+G +RL+ L +AA R Y+ +K + G RR+FHDDI+VIVVFLD
Sbjct: 303 QNHPRSGSVRRLIKVALQEAAKKREMRYSDLKKID----RGVRRHFHDDITVIVVFLDSN 358
Query: 352 SILR 355
+ R
Sbjct: 359 LVSR 362
>UniRef100_Q9ZSQ7 Protein phosphatase 2C homolog [Mesembryanthemum crystallinum]
Length = 396
Score = 274 bits (700), Expect = 4e-72
Identities = 150/355 (42%), Positives = 214/355 (60%), Gaps = 24/355 (6%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKAL-------FLGVYDGHAGFEASVFIT 87
D +H G+FS A VQAN +ED SQ+E L F+GVYDGH G E S +I
Sbjct: 38 DTGQHINGEFSMAVVQANNLLEDQSQIESGCLSLLDSGPYGTFVGVYDGHGGPETSRYIN 97
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
HLF HL R A + ++ ++ A ATE GF V K + K + VGSCCL G++
Sbjct: 98 DHLFHHLKRFA-AEQQSMSVDVIKKAFQATEEGFFSVVAKQWPMKPQIAAVGSCCLVGVV 156
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L++ANLGDSRAV+G V ++ A QL+ +HN E++R+E+ + HP+D IV+
Sbjct: 157 CNGILYIANLGDSRAVLGRAVKATGEVLAIQLSAEHNASIESVRQEMQATHPEDKDIVVL 216
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ VWRVKG+I ++RSIGD YLK+ E++ + + KF + EPF + +LS++P + +L
Sbjct: 217 KHNVWRVKGLIQITRSIGDVYLKKTEYNREPLYSKFR-LREPFKKPILSSDPAISVHELQ 275
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
+D+ IFASDGLW+ L+N++AV++VQ + RNG AKRLV L +AA R Y+ +K
Sbjct: 276 PHDQVCIFASDGLWEHLTNQEAVDLVQKSPRNGSAKRLVKVALQEAAKKREMRYSDLKKI 335
Query: 326 NLGRGDGNRRYFHDDISVIVVFLDKKSILRMPLHNLSYKSSSARPTTSAFAESGL 380
+ G RR+FHDDI+V+VVFLD NL + SS R T + G+
Sbjct: 336 D----RGVRRHFHDDITVVVVFLDS---------NLISRGSSVRGPTLSLRGGGI 377
>UniRef100_Q9LHJ9 Protein phosphatase 2C [Arabidopsis thaliana]
Length = 385
Score = 273 bits (699), Expect = 5e-72
Identities = 154/351 (43%), Positives = 217/351 (60%), Gaps = 19/351 (5%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEV-------ASRKALFLGVYDGHAGFEASVFIT 87
D H G+FS + +QAN +EDHS++E + +A F+GVYDGH G EA+ F+
Sbjct: 41 DSGNHVAGEFSMSVIQANNLLEDHSKLESGPVSMFDSGPQATFVGVYDGHGGPEAARFVN 100
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
+HLFD++ + N ++ + A ATE FL V + ++ K + VG+CCL GII
Sbjct: 101 KHLFDNIRKFTSENHG-MSANVITKAFLATEEDFLSLVRRQWQIKPQIASVGACCLVGII 159
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L++AN GDSR V+G + ++A QL+ +HN E++R+EL S HP+D IV+
Sbjct: 160 CSGLLYIANAGDSRVVLGRLEKAFKIVKAVQLSSEHNASLESVREELRSLHPNDPQIVVL 219
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ +VWRVKGII VSRSIGD YLK+ EF+ + KF VPE F + +L AEP + +
Sbjct: 220 KHKVWRVKGIIQVSRSIGDAYLKKAEFNREPLLAKFR-VPEVFHKPILRAEPAITVHKIH 278
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
D+FLIFASDGLW+ LSN++AV+IV RNGIA++L+ T L +AA R Y+ +K
Sbjct: 279 PEDQFLIFASDGLWEHLSNQEAVDIVNTCPRNGIARKLIKTALREAAKKREMRYSDLKKI 338
Query: 326 NLGRGDGNRRYFHDDISVIVVFLD----KKSILRMPLHNLSYKSSSARPTT 372
+ G RR+FHDDI+VIVVFLD +S R PL ++S A P+T
Sbjct: 339 D----RGVRRHFHDDITVIVVFLDSHLVSRSTSRRPLLSISGGGDLAGPST 385
>UniRef100_Q94H98 Hypothetical protein OSJNBb0048A17.8 [Oryza sativa]
Length = 380
Score = 272 bits (696), Expect = 1e-71
Identities = 158/320 (49%), Positives = 201/320 (62%), Gaps = 13/320 (4%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHL 94
+L+ H G+FS A QAN AMED +QV +AS A +GVYDGH G +AS F+ LF H+
Sbjct: 27 ELRPHAAGEFSMAAAQANLAMEDQAQV-LASPAATLVGVYDGHGGADASRFLRSRLFPHV 85
Query: 95 LRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHV 154
R + TE +R A A E FL+ V + +RQ+ + VGSCCL G I TL+V
Sbjct: 86 QRFEKEQGGMSTE-VIRRAFGAAEEEFLQQVRQAWRQRPKMAAVGSCCLLGAISGDTLYV 144
Query: 155 ANLGDSRAVIGTMV--NNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRV 212
ANLGDSRAV+G V AE+LT +HN E +R+EL + +PDD IV++ R WRV
Sbjct: 145 ANLGDSRAVLGRRVVGGGVAVAERLTDEHNAASEEVRRELTALNPDDAQIVVHARGAWRV 204
Query: 213 KGIITVSRSIGDTYLKRPEFSLDESFPKFEEV--PEPFIRGVLSAEPEMRSRDLTENDKF 270
KGII VSR+IGD YLK+ E+S+D P F V P P R LSAEP ++ R L ND F
Sbjct: 205 KGIIQVSRTIGDVYLKKQEYSMD---PVFRNVGPPIPLKRPALSAEPSIQVRKLKPNDLF 261
Query: 271 LIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRG 330
LIFASDGLW+ LS++ AV+IV N R GIA RLV L +A R ++ +K
Sbjct: 262 LIFASDGLWEHLSDDAAVQIVFKNPRTGIANRLVKAALKEATRKREVSFRDLKTIE---- 317
Query: 331 DGNRRYFHDDISVIVVFLDK 350
G RR+FHDDISVIVV+LD+
Sbjct: 318 KGVRRHFHDDISVIVVYLDR 337
>UniRef100_Q7Y138 Hypothetical protein OSJNBa0078D06.30 [Oryza sativa]
Length = 386
Score = 271 bits (693), Expect = 3e-71
Identities = 149/319 (46%), Positives = 205/319 (63%), Gaps = 10/319 (3%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRK--ALFLGVYDGHAGFEASVFITQHLFD 92
DL R G+ S A VQ N +ED +VE A +GV+DGHAG +A+ F HL
Sbjct: 52 DLARCHAGELSVAVVQGNHVLEDQCRVESGPPPLAATCIGVFDGHAGPDAARFACDHLLP 111
Query: 93 HLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTL 152
+L A E +T +RDA ATE GFL V + + + ++ VG+CCL G++ ++TL
Sbjct: 112 NLREAASGPEG-VTADAIRDAFLATEEGFLAVVSRMWEAQPDMATVGTCCLVGVVHQRTL 170
Query: 153 HVANLGDSRAVIGTMVNN--KIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVW 210
VANLGDSRAV+G V +I AEQL+ +HN +E +R+ELM++HPDD IV + VW
Sbjct: 171 FVANLGDSRAVLGKKVGRAGQITAEQLSSEHNANEEDVRQELMAQHPDDPQIVALKHGVW 230
Query: 211 RVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKF 270
RVKGII VSRS+GD YLK +++ ++ PKF +PEPF R +LSA P + +R L +D F
Sbjct: 231 RVKGIIQVSRSLGDAYLKHSQYNTEQIKPKF-RLPEPFSRPILSANPSIIARCLQPSDCF 289
Query: 271 LIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRG 330
+IFASDGLW+ LSN+QAVEIV N+ R G A+RL+ L +AA R Y+ + +
Sbjct: 290 IIFASDGLWEHLSNQQAVEIVHNHQRAGSARRLIKAALHEAARKREMRYSDLMKID---- 345
Query: 331 DGNRRYFHDDISVIVVFLD 349
RR+FHDDI+VIV+F++
Sbjct: 346 KKVRRHFHDDITVIVLFIN 364
>UniRef100_O82479 Protein phosphatase-2c [Mesembryanthemum crystallinum]
Length = 309
Score = 270 bits (691), Expect = 5e-71
Identities = 148/289 (51%), Positives = 195/289 (67%), Gaps = 10/289 (3%)
Query: 70 FLGVYDGHAGFEASVFITQHLFDHLLRAVRANENK-ITEPTLRDAVSATEAGFLEYVEKN 128
F+GVYDGH G E S +I HLF HL R A+E++ ++ +R AV ATE GFL V K
Sbjct: 1 FVGVYDGHGGPETSRYINDHLFHHLRRF--ASEHQCMSADVIRKAVQATEEGFLSIVSKQ 58
Query: 129 YRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDE 186
+ K + VGSCCL G+I L+VANLGDSRAV+G +V ++ A QL+ +HN E
Sbjct: 59 WPVKPQIAAVGSCCLLGVICNGMLYVANLGDSRAVLGRLVKATGEVLAVQLSTEHNACLE 118
Query: 187 AIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPE 246
A+R+EL S HPDD+ IV+ + VWRVKG+I VSRSIGD YLK+ EF+ + + KF + E
Sbjct: 119 AVRQELRSTHPDDSHIVVLKHNVWRVKGLIQVSRSIGDVYLKKAEFNREPLYAKFR-LRE 177
Query: 247 PFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVST 306
PF R +LS+EP + +L +D+F+IFASDGLW+ LSN++AV+IVQNN NG AKRLV
Sbjct: 178 PFRRPILSSEPSISVHELQPHDQFVIFASDGLWEHLSNQKAVDIVQNNPHNGSAKRLVKI 237
Query: 307 VLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFLDKKSILR 355
L +AA R Y+ +K + G RR+FHDDI+VIVVFLD + R
Sbjct: 238 ALQEAAKKREMRYSDLKKID----RGVRRHFHDDITVIVVFLDSNLVSR 282
>UniRef100_O81760 Hypothetical protein F17I5.110 [Arabidopsis thaliana]
Length = 380
Score = 269 bits (687), Expect = 1e-70
Identities = 153/321 (47%), Positives = 202/321 (62%), Gaps = 12/321 (3%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHL 94
+L+ H G +S A VQAN +ED SQV +S A ++GVYDGH G EAS F+ +HLF ++
Sbjct: 27 ELRPHAGGDYSIAVVQANSRLEDQSQVFTSS-SATYVGVYDGHGGPEASRFVNRHLFPYM 85
Query: 95 LRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHV 154
+ R + ++ ++ A TE F V+++ K + VGSCCL G I TL+V
Sbjct: 86 HKFAREHGG-LSVDVIKKAFKETEEEFCGMVKRSLPMKPQMATVGSCCLVGAISNDTLYV 144
Query: 155 ANLGDSRAVIGTMV-----NNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREV 209
ANLGDSRAV+G++V N AE+L+ DHN E +RKE+ + +PDD+ IV+Y R V
Sbjct: 145 ANLGDSRAVLGSVVSGVDSNKGAVAERLSTDHNVAVEEVRKEVKALNPDDSQIVLYTRGV 204
Query: 210 WRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDK 269
WR+KGII VSRSIGD YLK+PE+ D F + P P R ++AEP + R L D
Sbjct: 205 WRIKGIIQVSRSIGDVYLKKPEYYRDPIFQRHGN-PIPLRRPAMTAEPSIIVRKLKPQDL 263
Query: 270 FLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGR 329
FLIFASDGLW+ LS+E AVEIV + R GIA+RLV L +AA R Y +K
Sbjct: 264 FLIFASDGLWEHLSDETAVEIVLKHPRTGIARRLVRAALEEAAKKREMRYGDIKKI---- 319
Query: 330 GDGNRRYFHDDISVIVVFLDK 350
G RR+FHDDISVIVV+LD+
Sbjct: 320 AKGIRRHFHDDISVIVVYLDQ 340
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.131 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 677,902,012
Number of Sequences: 2790947
Number of extensions: 26665854
Number of successful extensions: 78565
Number of sequences better than 10.0: 763
Number of HSP's better than 10.0 without gapping: 512
Number of HSP's successfully gapped in prelim test: 251
Number of HSP's that attempted gapping in prelim test: 76454
Number of HSP's gapped (non-prelim): 1081
length of query: 436
length of database: 848,049,833
effective HSP length: 130
effective length of query: 306
effective length of database: 485,226,723
effective search space: 148479377238
effective search space used: 148479377238
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC125478.13


