

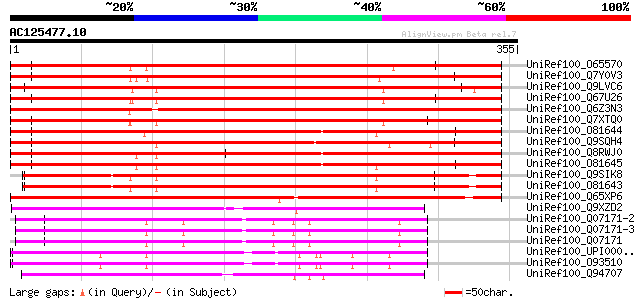
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125477.10 - phase: 0 /pseudo
(355 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O65570 Villin 4 [Arabidopsis thaliana] 570 e-161
UniRef100_Q7Y0V3 Actin filament bundling protein P-115-ABP [Lili... 543 e-153
UniRef100_Q9LVC6 Villin [Arabidopsis thaliana] 528 e-149
UniRef100_Q67U26 Putative villin 2 [Oryza sativa] 524 e-147
UniRef100_Q6Z3N3 Putative villin [Oryza sativa] 511 e-144
UniRef100_Q7XTQ0 OSJNBa0041A02.24 protein [Oryza sativa] 503 e-141
UniRef100_O81644 Villin 2 [Arabidopsis thaliana] 452 e-126
UniRef100_Q9SQH4 Actin bundling protein ABP135 [Lilium longiflorum] 443 e-123
UniRef100_Q8RWJ0 Hypothetical protein At3g57410 [Arabidopsis tha... 435 e-120
UniRef100_O81645 Villin 3 [Arabidopsis thaliana] 435 e-120
UniRef100_Q9SIK8 Putative villin [Arabidopsis thaliana] 339 6e-92
UniRef100_O81643 Villin 1 [Arabidopsis thaliana] 330 4e-89
UniRef100_Q65XP6 Putative villin [Oryza sativa] 310 4e-83
UniRef100_Q9XZD2 Fragmin A [Physarum polycephalum] 201 3e-50
UniRef100_Q07171-2 Splice isoform 2 of Q07171 [Drosophila melano... 201 3e-50
UniRef100_Q07171-3 Splice isoform 3 of Q07171 [Drosophila melano... 201 3e-50
UniRef100_Q07171 Gelsolin precursor [Drosophila melanogaster] 201 3e-50
UniRef100_UPI00003AB298 UPI00003AB298 UniRef100 entry 191 2e-47
UniRef100_O93510 Gelsolin precursor [Gallus gallus] 191 3e-47
UniRef100_Q94707 Actin-binding protein fragmin P [Physarum polyc... 190 4e-47
>UniRef100_O65570 Villin 4 [Arabidopsis thaliana]
Length = 974
Score = 570 bits (1468), Expect = e-161
Identities = 270/344 (78%), Positives = 311/344 (89%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTA K+GALRHDIHYWLGKDTSQDEAG AA+KTVELDA LGGRAVQYREVQGHET+KFLS
Sbjct: 53 TTALKTGALRHDIHYWLGKDTSQDEAGTAAVKTVELDAALGGRAVQYREVQGHETEKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIPQEGG ASGFKHV AEEH TRLFVC+GKHVV+VKEVPFARSSLNHDDI+ILDT
Sbjct: 113 YFKPCIIPQEGGVASGFKHVVAEEHITRLFVCRGKHVVHVKEVPFARSSLNHDDIYILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
+SKIFQFNGSNSSIQERAKALEVVQYIKDTYHDG CEVA++EDG+LMAD++SGEFWG FG
Sbjct: 173 KSKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGTCEVATVEDGKLMADADSGEFWGFFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAPLPRKT +D+DKT +S +L CVEKG+A P E D+L +E+LDTNKCYILDCG+EVF
Sbjct: 233 GFAPLPRKTANDEDKTYNSDITRLFCVEKGQANPVEGDTLKREMLDTNKCYILDCGIEVF 292
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNAAM 300
VW+GR TSLD+RK AS + +E++ S+ RPKSQ+IR++EGFETV FRSKF+SW Q TN +
Sbjct: 293 VWMGRTTSLDDRKIASKAAEEMIRSSERPKSQMIRIIEGFETVPFRSKFESWTQETNTTV 352
Query: 301 PEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
EDGRG+VAALL+RQG++V+GL+KA P KEEPQ +IDCTG+LQV
Sbjct: 353 SEDGRGRVAALLQRQGVNVRGLMKAAPPKEEPQVFIDCTGNLQV 396
Score = 75.9 bits (185), Expect = 2e-12
Identities = 75/301 (24%), Positives = 133/301 (43%), Gaps = 33/301 (10%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK + ++E G+A ++ + Q R +G E +F + I+ +GG +S
Sbjct: 440 WFGKQSVEEERGSAVSMASKMVESMKFVPAQARIYEGKEPIQFFVIMQSFIV-FKGGISS 498
Query: 76 GFKHVEAE---------EHKTRLFVCKG---KHVVYVKEVPFARSSLNHDDIFILDTESK 123
G+K AE E+ LF +G +++ ++ P A +SLN +IL +S
Sbjct: 499 GYKKYIAEKEVDDDTYNENGVALFRIQGSGPENMQAIQVDPVA-ASLNSSYYYILHNDSS 557
Query: 124 IFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFA 183
+F + G+ S+ ++ A + IK + ++G SES +FW L GG A
Sbjct: 558 VFTWAGNLSTATDQELAERQLDLIKPNQQS-----RAQKEG-----SESEQFWELLGGKA 607
Query: 184 PLPRKTVSDDDKTIDSHPPKLLCV-EKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVW 242
+ ++ K + P C K + E + T++ L T +I+DC E+FVW
Sbjct: 608 EYSSQKLT---KEPERDPHLFSCTFTKEVLKVTEIYNFTQDDLMTEDIFIIDCHSEIFVW 664
Query: 243 IGRNTSLDERKSASGSTDELVSSTN-----RPKSQIIRVMEGFETVMFRSKFDSWPQTTN 297
+G+ + A ++ + + P++ I +MEG E F F SW + +
Sbjct: 665 VGQEVVPKNKLLALTIGEKFIEKDSLLEKLSPEAPIYVIMEGGEPSFFTRFFTSWDSSKS 724
Query: 298 A 298
A
Sbjct: 725 A 725
>UniRef100_Q7Y0V3 Actin filament bundling protein P-115-ABP [Lilium longiflorum]
Length = 958
Score = 543 bits (1399), Expect = e-153
Identities = 264/346 (76%), Positives = 294/346 (84%), Gaps = 2/346 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TT KSG LRHDIHYWLGKDTSQDEAG AAIKTVELD LGGRAVQYREVQGHET+ FLS
Sbjct: 53 TTILKSGGLRHDIHYWLGKDTSQDEAGTAAIKTVELDVTLGGRAVQYREVQGHETEIFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAE--EHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFIL 118
YFKPCIIPQEGG ASGFKH E EH TRLFVCKGKHVV+VKEVPF RSSLNHDDIFIL
Sbjct: 113 YFKPCIIPQEGGVASGFKHSEINQHEHHTRLFVCKGKHVVHVKEVPFTRSSLNHDDIFIL 172
Query: 119 DTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGL 178
DTESKIFQFNGSNSSIQER KALEVVQ+IKDTYH+GKCE+A +EDG+LMAD E+GEFWG
Sbjct: 173 DTESKIFQFNGSNSSIQERGKALEVVQHIKDTYHNGKCEIAVVEDGKLMADVEAGEFWGF 232
Query: 179 FGGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLE 238
FGGFAPLPRK D D+ ++ KLLCVEKG+ + DSL +ELL+T+KCY+LDCG+E
Sbjct: 233 FGGFAPLPRKAAFDHDRKTETLATKLLCVEKGQPSSVQADSLIRELLNTDKCYLLDCGVE 292
Query: 239 VFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNA 298
VFVWIGRNTSL+ERKSAS + +EL+ + +R K +IRVMEG+ETV FRSKFD+WP
Sbjct: 293 VFVWIGRNTSLEERKSASSAAEELLRAHDRTKVHVIRVMEGYETVKFRSKFDAWPHAAVV 352
Query: 299 AMPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
E+GRGKVAALLKRQGLDVKGLVKA P KEEPQP+IDCTG+LQV
Sbjct: 353 TATEEGRGKVAALLKRQGLDVKGLVKAAPAKEEPQPFIDCTGNLQV 398
Score = 59.3 bits (142), Expect = 2e-07
Identities = 74/318 (23%), Positives = 133/318 (41%), Gaps = 42/318 (13%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W G + +D A ++ +AVQ + +G E + S F+ I+ +GG +S
Sbjct: 442 WFGNKSIEDGRTTAVSLARKMVESFKSQAVQAQVYEGMEPIQLFSIFQSLIV-FKGGVSS 500
Query: 76 GFKHVEAEEHKT---------RLFVCKGK---HVVYVKEVPFARSSLNHDDIFILDTESK 123
+K+ +E + T LF +G ++ ++ P A +SLN +IL
Sbjct: 501 VYKNFISENNLTDDTYTEDGLALFRVQGSGPDNMQAIQVEPVA-TSLNSSYCYILHNGDT 559
Query: 124 IFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFA 183
+F ++GS ++ + + IK +++ E+ +FW L GG
Sbjct: 560 VFTWSGSLTTSDDHDLVERQLDLIKP----------NVQSKPQKEGLETQQFWDLLGGKR 609
Query: 184 PL-PRKTVSDDDKTIDSHPPKLLCV-EKGKAEPFETDSLTKELLDTNKCYILDCGLEVFV 241
+K V + +K P C K + E + +++ L T +ILDC +FV
Sbjct: 610 EHGSQKIVKEPEKD----PHLFSCTFSKDDLKVTEVYNFSQDDLTTEDIFILDCHSNIFV 665
Query: 242 WIGRNTSLDERKSASG--------STDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWP 293
W+G+ +D + A D L+ +R ++ I VMEG E F ++F +W
Sbjct: 666 WVGQ--QVDSKSKAQALIIGEKFLEYDFLMEKISR-ETPIFIVMEGSEPQFF-TRFFTWD 721
Query: 294 QTTNAAMPEDGRGKVAAL 311
+A + K+A L
Sbjct: 722 SAKSAMHGNSFQRKLAIL 739
>UniRef100_Q9LVC6 Villin [Arabidopsis thaliana]
Length = 962
Score = 528 bits (1361), Expect = e-149
Identities = 251/346 (72%), Positives = 300/346 (86%), Gaps = 2/346 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTAS+SG+L HDIHYWLGKD+SQDEAGA A+ TVELD+ LGGRAVQYREVQGHET+KFLS
Sbjct: 53 TTASRSGSLHHDIHYWLGKDSSQDEAGAVAVMTVELDSALGGRAVQYREVQGHETEKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIPQEGG ASGF HV+ EEH+TRL++CKGKHVV VKEVPF RS+LNH+D+FILDT
Sbjct: 113 YFKPCIIPQEGGVASGFNHVKPEEHQTRLYICKGKHVVRVKEVPFVRSTLNHEDVFILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
ESKIFQF+GS SSIQERAKALEVVQYIKDTYHDGKC++A++EDGR+MAD+E+GEFWGLFG
Sbjct: 173 ESKIFQFSGSKSSIQERAKALEVVQYIKDTYHDGKCDIAAVEDGRMMADAEAGEFWGLFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAPLP+K +DD+T S KL VEKG+ + E + LTKELLDTNKCYILDCGLE+F
Sbjct: 233 GFAPLPKKPAVNDDETAASDGIKLFSVEKGQTDAVEAECLTKELLDTNKCYILDCGLELF 292
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNAAM 300
VW GR+TS+D+RKSA+ + +E S+ PKS ++ VMEG+ETVMFRSKFDSWP ++ A
Sbjct: 293 VWKGRSTSIDQRKSATEAAEEFFRSSEPPKSNLVSVMEGYETVMFRSKFDSWPASSTIAE 352
Query: 301 PEDGRGKVAALLKRQGLDVKGLVK--ADPVKEEPQPYIDCTGHLQV 344
P+ GRGKVAALL+RQG++V+GLVK + K+EP+PYID TG+LQV
Sbjct: 353 PQQGRGKVAALLQRQGVNVQGLVKTSSSSSKDEPKPYIDGTGNLQV 398
Score = 66.2 bits (160), Expect = 1e-09
Identities = 74/323 (22%), Positives = 138/323 (41%), Gaps = 33/323 (10%)
Query: 11 HDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQE 70
H + W GK + +++ +A ++ + Q R +G E +F + I +
Sbjct: 437 HLVGTWFGKQSVEEDRASAISLANKMVESMKFVPAQARINEGKEPIQFFVIMQS-FITFK 495
Query: 71 GGAASGFKHVEAEE---------HKTRLFVCKGKHVVYVK--EVPFARSSLNHDDIFILD 119
GG + FK AE LF +G ++ ++ A + LN +IL
Sbjct: 496 GGVSDAFKKYIAENDIPDTTYEAEGVALFRVQGSGPENMQAIQIEAASAGLNSSHCYILH 555
Query: 120 TESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLF 179
+S +F + G+ +S +++ E+++ + D + A E SES +FW L
Sbjct: 556 GDSTVFTWCGNLTSSEDQ----ELMERMLDLIKPNEPTKAQKEG------SESEQFWELL 605
Query: 180 GGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKA-EPFETDSLTKELLDTNKCYILDCGLE 238
GG + P + + D +S P C ++ + E + T++ L T +ILDC E
Sbjct: 606 GGKSEYPSQKIKRDG---ESDPHLFSCTYTNESLKATEIFNFTQDDLMTEDIFILDCHTE 662
Query: 239 VFVWIGRNTSLDERKSASGSTD-----ELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWP 293
VFVW+G+ ++ A + + + ++ I V EG E F ++F +W
Sbjct: 663 VFVWVGQQVDPKKKPQALDIGENFLKHDFLLENLASETPIYIVTEGNEPPFF-TRFFTW- 720
Query: 294 QTTNAAMPEDGRGKVAALLKRQG 316
++ + M D + A+L +G
Sbjct: 721 DSSKSGMHGDSFQRKLAILTNKG 743
>UniRef100_Q67U26 Putative villin 2 [Oryza sativa]
Length = 1016
Score = 524 bits (1350), Expect = e-147
Identities = 249/346 (71%), Positives = 294/346 (84%), Gaps = 2/346 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTA K+G+ RHDIHYWLGKDTSQDEAG AAIKTVELDA LGGRAVQYREVQG+ET++FLS
Sbjct: 99 TTALKNGSFRHDIHYWLGKDTSQDEAGTAAIKTVELDAALGGRAVQYREVQGNETERFLS 158
Query: 61 YFKPCIIPQEGGAASGFKHVEAEE--HKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFIL 118
YFKPCIIP+EGG ASGF+H E E H TRLFVC+GKH V+VKEVPFARSSLNHDDIFIL
Sbjct: 159 YFKPCIIPEEGGIASGFRHTEINEREHVTRLFVCRGKHTVHVKEVPFARSSLNHDDIFIL 218
Query: 119 DTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGL 178
DT+SKIFQFNGSNSSIQERAKALEVVQY+KD+ H+GKC+V S+EDG+LMAD+++GEFWGL
Sbjct: 219 DTKSKIFQFNGSNSSIQERAKALEVVQYLKDSNHEGKCDVGSVEDGKLMADADAGEFWGL 278
Query: 179 FGGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLE 238
FGGFAPLPRKT SD + + KL+C+ KG+ P + D LT+ELLD+ KCY+LDCG E
Sbjct: 279 FGGFAPLPRKTFSDLNGKDSAFSSKLICLNKGQTVPVDFDVLTRELLDSTKCYLLDCGSE 338
Query: 239 VFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNA 298
++VW+GR T L+ERK A + +EL+ NRPKS I+R+MEGFETV+FRSKF WP+ +A
Sbjct: 339 IYVWMGRETPLEERKRAGSAAEELLREVNRPKSHIVRLMEGFETVIFRSKFSKWPKKADA 398
Query: 299 AMPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
+ ++ RGKVAALLKRQG +VKGL KA PVKEEPQP IDCTG+LQV
Sbjct: 399 VVSDESRGKVAALLKRQGFNVKGLAKAAPVKEEPQPQIDCTGNLQV 444
Score = 80.1 bits (196), Expect = 9e-14
Identities = 75/300 (25%), Positives = 131/300 (43%), Gaps = 32/300 (10%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK + QDE A ++ L +AV R +G E +F S F+ +I +GG ++
Sbjct: 488 WFGKKSVQDEKTTAISVASKMVESLKFQAVMVRLYEGKEPAEFFSIFQNLVI-FKGGVST 546
Query: 76 GFKHVEAE---------EHKTRLFVCKGKHVVYVK--EVPFARSSLNHDDIFILDTESKI 124
G+K +E E+ LF +G ++ +V A +SLN ++L +
Sbjct: 547 GYKKFVSENGIEDDTYSENGVALFRVQGSGPENMQAIQVDTAATSLNSSYCYVLHDGDTL 606
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAP 184
F + G+ SS ++ A + IK +++ L SE +FW L G +
Sbjct: 607 FTWIGNLSSSMDQELAERQLDVIKP----------NLQSRMLKEGSEYDQFWKLLGVKSE 656
Query: 185 LPRKTVSDDDKTIDSHPPKLLCV-EKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWI 243
P + ++ D +S P C KG + E + T++ L T +ILDC VFVW+
Sbjct: 657 YPSQKIAKDQ---ESDPHLFSCTFSKGVLKVREIFNFTQDDLMTEDVFILDCHSCVFVWV 713
Query: 244 GRNTSLDERKSASGSTD-----ELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNA 298
G+ R A + +++ + ++ + + EG E F ++F +W +A
Sbjct: 714 GQRVDTKMRAQALSVGEKFLELDILMENSSQETPVYVITEGSEPQFF-TRFFTWDSAKSA 772
>UniRef100_Q6Z3N3 Putative villin [Oryza sativa]
Length = 911
Score = 511 bits (1317), Expect = e-144
Identities = 250/347 (72%), Positives = 293/347 (84%), Gaps = 6/347 (1%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTA K+G+LRHDIHYW+GKDTSQDE+G AAI TVELDA LGGRAVQYRE+QG+ET KFLS
Sbjct: 65 TTALKNGSLRHDIHYWIGKDTSQDESGTAAILTVELDAALGGRAVQYREIQGNETDKFLS 124
Query: 61 YFKPCIIPQEGGAASGFKHVEA--EEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFIL 118
YF+PCI+PQ GG ASGFKHVE +EH+TRL+VC G VV+V PFARSSLNHDDIFIL
Sbjct: 125 YFRPCIMPQPGGVASGFKHVEVNEQEHETRLYVCTGNRVVHV---PFARSSLNHDDIFIL 181
Query: 119 DTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGL 178
DT+SKIFQFNGSNSSIQERAKALEVVQYIKDT+H+GKCEVA++EDGRLMAD+E+GEFWG
Sbjct: 182 DTKSKIFQFNGSNSSIQERAKALEVVQYIKDTFHEGKCEVAAVEDGRLMADAEAGEFWGF 241
Query: 179 FGGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLE 238
FGGFAPLPR+ +D++ + KLLC +GK EP +SL ELL TNKCY+LDCG+E
Sbjct: 242 FGGFAPLPRRAPVEDNEKYEETVFKLLCFNQGKLEPINYESLLHELLKTNKCYLLDCGVE 301
Query: 239 VFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNA 298
+FVW+GR TSL ERKSAS + ++L+S NR K+ +I+V+EGFETVMF+SKF WPQT +
Sbjct: 302 LFVWMGRTTSLQERKSASEAAEKLLSDDNRTKTHVIKVIEGFETVMFKSKFKEWPQTPDL 361
Query: 299 AM-PEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
+ EDGRGKVAALLKRQGL+VKGL+KA P KEEPQ YIDCTG LQV
Sbjct: 362 KLSSEDGRGKVAALLKRQGLNVKGLMKAAPAKEEPQAYIDCTGSLQV 408
Score = 38.5 bits (88), Expect = 0.29
Identities = 62/285 (21%), Positives = 107/285 (36%), Gaps = 67/285 (23%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK + +++ A ++ +AVQ R +G E +F F+ + +GG +S
Sbjct: 452 WFGKKSIEEDRVTAISLASKMVESAKFQAVQTRLYEGKEPIQFFVIFQSFQV-FKGGLSS 510
Query: 76 GFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESKIFQFNGSNSSIQ 135
G+K AE DD L+ +F+ GS
Sbjct: 511 GYKKFIAEN--------------------------GIDDDTYLEDGLALFRIQGSGP--- 541
Query: 136 ERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPLPRKTVSDDDK 195
E +A++V D + ++ D + W T S D +
Sbjct: 542 ENMQAIQV---------DAAASSLNSSYSYILHDGNTVFTW--------TGNLTTSLDQE 584
Query: 196 TIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTSLDERKSA 255
++ + L + K K E T++ L T +ILDC ++FVW+G+ + R A
Sbjct: 585 VVE----RQLDIIKIK----EIYHFTQDDLMTEDVFILDCHSDIFVWVGQQVDVKVRLQA 636
Query: 256 SGSTDELVS--------STNRPKSQIIRVMEGFETVMFRSKFDSW 292
++ V S++ P I +MEG E F ++F +W
Sbjct: 637 LDIGEKFVKLDFLMENLSSDTP---IFVIMEGSEPTFF-TRFFTW 677
>UniRef100_Q7XTQ0 OSJNBa0041A02.24 protein [Oryza sativa]
Length = 946
Score = 503 bits (1295), Expect = e-141
Identities = 243/347 (70%), Positives = 288/347 (82%), Gaps = 3/347 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTA K+G+ RHD+HYWLGKDTSQDEAG AAI TVELDA LGGRAVQYREVQG ET+K LS
Sbjct: 53 TTALKNGSFRHDLHYWLGKDTSQDEAGTAAILTVELDAALGGRAVQYREVQGGETEKLLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEA--EEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFIL 118
YF+PCI+PQ GG ASGF HVE ++H TRL+VC+GKHVV+VKEVPF RSSLNH+DIFIL
Sbjct: 113 YFRPCIMPQPGGVASGFNHVEVNQQDHVTRLYVCQGKHVVHVKEVPFVRSSLNHEDIFIL 172
Query: 119 DTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGL 178
DT +KIFQFNGSNS IQERAKALEVVQYIKDT+H+GKCEVA++EDG+LMAD+E+GEFWGL
Sbjct: 173 DTANKIFQFNGSNSCIQERAKALEVVQYIKDTFHEGKCEVAAVEDGKLMADTEAGEFWGL 232
Query: 179 FGGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLE 238
FGGFAPLP+KT S+D+ KLLC +G E +SL ELL+TNKCY+LDCG E
Sbjct: 233 FGGFAPLPKKTSSEDNGDDKETVTKLLCFNQGTLEHISFESLEHELLETNKCYLLDCGAE 292
Query: 239 VFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNA 298
++VW+GR TSL RK AS + ++L+ NR S +I+V+EGFET+MF+SKF+ WP T +
Sbjct: 293 MYVWMGRGTSLQVRKGASEAAEKLLIDENRKGSNVIKVIEGFETIMFKSKFNKWPPTPDL 352
Query: 299 AM-PEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
+ EDGRGKVAALL+ QGLDVKGL+KA P +EEPQPYIDCTGHLQV
Sbjct: 353 KLSSEDGRGKVAALLRSQGLDVKGLMKAAPEEEEPQPYIDCTGHLQV 399
Score = 61.6 bits (148), Expect = 3e-08
Identities = 67/294 (22%), Positives = 121/294 (40%), Gaps = 32/294 (10%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK + +++ +A ++ +A Q R +G E +F F+ + +GG +S
Sbjct: 443 WFGKKSVEEDRTSAISLASKMFQAAKFQAAQARLYEGKEPIQFFVIFQSLQV-FKGGLSS 501
Query: 76 GFKHVEAE---------EHKTRLFVCKGKHVVYVK--EVPFARSSLNHDDIFILDTESKI 124
G+K+ A E LF +G ++ +V SSLN +IL + +
Sbjct: 502 GYKNFIAVNGTDDDTYVEGGLALFRIQGSGSENMQAIQVDAVSSSLNSSYCYILHNGNTV 561
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAP 184
F + G+ ++ + + IK S ++GR E+ +FW L GG
Sbjct: 562 FTWTGNLTTSLDNDLVERQLDVIKPDLPS-----RSQKEGR-----ETDQFWELLGGKCK 611
Query: 185 LPRKTVSDDDKTIDSHPPKLLCV-EKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWI 243
K + ++ +S P C+ KG + E T++ L ++LDC ++FVW+
Sbjct: 612 YSNKKIGKEN---ESDPHLFSCILSKGNQKVKEIHHFTQDDLMAEDIFVLDCRTDLFVWV 668
Query: 244 GRNTSLDERKSASGSTD-----ELVSSTNRPKSQIIRVMEGFETVMFRSKFDSW 292
G+ R A + + + + I V EG E F ++F +W
Sbjct: 669 GQEVDAKLRSQAMDIGEKFLLHDFLMENLSQDTPIFIVTEGSEPQFF-TRFFTW 721
>UniRef100_O81644 Villin 2 [Arabidopsis thaliana]
Length = 976
Score = 452 bits (1163), Expect = e-126
Identities = 213/345 (61%), Positives = 273/345 (78%), Gaps = 2/345 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TT +K GA DIH+W+GKDTSQDEAG AA+KTVELDAVLGGRAVQ+RE+QGHE+ KFLS
Sbjct: 51 TTQNKGGAYLFDIHFWIGKDTSQDEAGTAAVKTVELDAVLGGRAVQHREIQGHESDKFLS 110
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIP EGG ASGFK VE E +TRL+ CKGK + +K+VPFARSSLNHDD+FILDT
Sbjct: 111 YFKPCIIPLEGGVASGFKTVEEEVFETRLYTCKGKRAIRLKQVPFARSSLNHDDVFILDT 170
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
E KI+QFNG+NS+IQERAKALEVVQY+KD YH+G C+VA ++DG+L +S+SG FW LFG
Sbjct: 171 EEKIYQFNGANSNIQERAKALEVVQYLKDKYHEGTCDVAIVDDGKLDTESDSGAFWVLFG 230
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAP+ RK +DDD +S PPKL C+ GK EP + D L+K +L+ KCY+LDCG E++
Sbjct: 231 GFAPIGRKVANDDDIVPESTPPKLYCITDGKMEPIDGD-LSKSMLENTKCYLLDCGAEIY 289
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSWPQTTNAA 299
+W+GR T +DERK+AS S +E ++S NRPK + + RV++G+E+ F+S FDSWP +
Sbjct: 290 IWVGRVTQVDERKAASQSAEEFLASENRPKATHVTRVIQGYESHSFKSNFDSWPSGSATP 349
Query: 300 MPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
E+GRGKVAALLK+QG+ +KG+ K+ PV E+ P ++ G L+V
Sbjct: 350 GNEEGRGKVAALLKQQGVGLKGIAKSAPVNEDIPPLLESGGKLEV 394
Score = 81.3 bits (199), Expect = 4e-14
Identities = 81/318 (25%), Positives = 132/318 (41%), Gaps = 41/318 (12%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK + ++ A + L GR VQ R +G E +F++ F+P ++ +GG +S
Sbjct: 439 WFGKKSIPEDQDTAIRLANTMSNSLKGRPVQGRIYEGKEPPQFVALFQPMVV-LKGGLSS 497
Query: 76 GFKHVEAEEHKTRLFVCK-----------GKHVVYVKEVPFARSSLNHDDIFILDTESKI 124
G+K E T G H +V +SLN + F+L + + +
Sbjct: 498 GYKSSMGESESTDETYTPESIALVQVSGTGVHNNKAVQVETVATSLNSYECFLLQSGTSM 557
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAP 184
F ++G+ S+ ++ A +V +++K I +ES FW GG
Sbjct: 558 FLWHGNQSTHEQLELATKVAEFLKP----------GITLKHAKEGTESSTFWFALGGKQN 607
Query: 185 LPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIG 244
K S + TI +GK + E + ++ L T Y LD EVFVW+G
Sbjct: 608 FTSKKASSE--TIRDPHLFSFAFNRGKFQVEEIYNFAQDDLLTEDIYFLDTHAEVFVWVG 665
Query: 245 RNTSLDERKSA----------SGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQ 294
+ E+++ +GS + L PK I ++ EG E F + F SW
Sbjct: 666 QCVEPKEKQTVFEIGQKYIDLAGSLEGL-----HPKVPIYKINEGNEPCFFTTYF-SW-D 718
Query: 295 TTNAAMPEDGRGKVAALL 312
T A + + K A+LL
Sbjct: 719 ATKAIVQGNSFQKKASLL 736
>UniRef100_Q9SQH4 Actin bundling protein ABP135 [Lilium longiflorum]
Length = 965
Score = 443 bits (1139), Expect = e-123
Identities = 214/347 (61%), Positives = 274/347 (78%), Gaps = 4/347 (1%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTA K GA +DIH+W+GKDTSQDEAG AAIKTVELDAVLGGRAVQ+RE+QGHE+ KFLS
Sbjct: 53 TTAGKGGAHLYDIHFWIGKDTSQDEAGTAAIKTVELDAVLGGRAVQHRELQGHESDKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YF+PCIIP EGG SGFK E E +TRL+VC+GK VV +K+VPFAR+SLNHDD+FILDT
Sbjct: 113 YFRPCIIPLEGGVVSGFKTPEEETFETRLYVCRGKRVVRLKQVPFARTSLNHDDVFILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
E KI+QFNG+NS+IQERAKALEV+Q++KD YH+G C+VA I+DGRL A+S SGEFW LFG
Sbjct: 173 EKKIYQFNGANSNIQERAKALEVIQFLKDKYHEGTCDVAIIDDGRLAAESGSGEFWVLFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAP+ ++ V DDD T+++ P KL + G+ + E +L+K +L+ NKCY+LDCG E+F
Sbjct: 233 GFAPIGKRVVGDDDVTLETTPGKLYSINDGQLK-LEEGTLSKAMLENNKCYLLDCGAEIF 291
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSWP--QTTN 297
VW+GR T +++RK+AS S +E + + NRPK ++I RV++GFET F+S F+SWP T
Sbjct: 292 VWVGRVTQVEDRKAASKSAEEFIINENRPKVTRITRVIQGFETRTFKSNFESWPLGSATG 351
Query: 298 AAMPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
+ E+GRGKVAALLK+QG+ VKG+ K P EE P I+ TG +V
Sbjct: 352 TSGGEEGRGKVAALLKQQGVGVKGMSKGSPANEEVPPLIEGTGKTEV 398
Score = 90.9 bits (224), Expect = 5e-17
Identities = 79/313 (25%), Positives = 142/313 (45%), Gaps = 32/313 (10%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W+GK++++D+ A + L G+ VQ R VQG E +F++ F+P ++ +GG +
Sbjct: 443 WIGKNSAKDDQLMATKLASSMCNSLKGKPVQGRIVQGREPPQFIALFQPMVV-LKGGISP 501
Query: 76 GFKHVEAEEH-KTRLFVCKGKHVVYVK----------EVPFARSSLNHDDIFILDTESKI 124
G+K + A+++ +V G ++ + +V +SL+ D F+L + + +
Sbjct: 502 GYKKLIADKNLNDDTYVSDGIALIRISKTSVHNNKVIQVDAVATSLSSTDSFLLQSGNSM 561
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAP 184
F ++G+ S+ +++ A +V +++K + +ES FW GG
Sbjct: 562 FLWHGNASTFEQQQWAAKVAEFLK----------PGVVLKHAKEGTESSAFWFALGGKQS 611
Query: 185 LPRKTVSDDDKTIDSHPPKLLC-VEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWI 243
K D + I P +C KGK E E + +++ L T ILD E+FVW+
Sbjct: 612 YSPK---KDAQEIVRDPHLYVCSFNKGKLEVTEVYNFSQDDLLTEDILILDTHEEIFVWV 668
Query: 244 GRNTSLDERKSASGSTDELVS-----STNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNA 298
G++ E+++A + + P + +V EG E F + F SW T A
Sbjct: 669 GQSVDSKEKQNAFDIGQKYIDLAITLEGLSPDVPLYKVTEGNEPCFFTAYF-SWDGTKAA 727
Query: 299 AMPEDGRGKVAAL 311
KVA L
Sbjct: 728 VQGNSFEKKVAML 740
>UniRef100_Q8RWJ0 Hypothetical protein At3g57410 [Arabidopsis thaliana]
Length = 618
Score = 435 bits (1118), Expect = e-120
Identities = 206/345 (59%), Positives = 270/345 (77%), Gaps = 2/345 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TT +K GA DIH+W+GKDTSQDEAG AA+KTVELDA LGGRAVQYRE+QGHE+ KFLS
Sbjct: 53 TTQNKGGAYLFDIHFWIGKDTSQDEAGTAAVKTVELDAALGGRAVQYREIQGHESDKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIP EGG ASGFK E EE +TRL+ CKGK V++K+VPFARSSLNHDD+FILDT
Sbjct: 113 YFKPCIIPLEGGVASGFKKPEEEEFETRLYTCKGKRAVHLKQVPFARSSLNHDDVFILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
+ KI+QFNG+NS+IQERAKAL V+QY+KD +H+G +VA ++DG+L +S+SGEFW LFG
Sbjct: 173 KEKIYQFNGANSNIQERAKALVVIQYLKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAP+ RK S+D+ ++ PPKL + G+ E + D L+K +L+ NKCY+LDCG E+F
Sbjct: 233 GFAPIARKVASEDEIIPETTPPKLYSIADGQVESIDGD-LSKSMLENNKCYLLDCGSEIF 291
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSWPQTTNAA 299
+W+GR T ++ERK+A + ++ V+S NRPK ++I RV++G+E F+S FDSWP +
Sbjct: 292 IWVGRVTQVEERKTAIQAAEDFVASENRPKATRITRVIQGYEPHSFKSNFDSWPSGSATP 351
Query: 300 MPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
E+GRGKVAALLK+QG+ +KGL K+ PV E+ P ++ G L+V
Sbjct: 352 ANEEGRGKVAALLKQQGVGLKGLSKSTPVNEDIPPLLEGGGKLEV 396
Score = 54.3 bits (129), Expect = 5e-06
Identities = 38/147 (25%), Positives = 75/147 (50%), Gaps = 12/147 (8%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK+++Q++ A + L GR VQ R +G E +F++ F+ ++ +GG +S
Sbjct: 441 WFGKNSNQEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVV-LKGGLSS 499
Query: 76 GFKHVEAEEHKT---------RLFVCKGKHVVYVK--EVPFARSSLNHDDIFILDTESKI 124
G+K+ E+ + L G V K +V +SLN D F+L + + +
Sbjct: 500 GYKNSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSM 559
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTY 151
F + G++S+ +++ A +V +++K +
Sbjct: 560 FLWVGNHSTHEQQELAAKVAEFLKSAW 586
>UniRef100_O81645 Villin 3 [Arabidopsis thaliana]
Length = 966
Score = 435 bits (1118), Expect = e-120
Identities = 206/345 (59%), Positives = 270/345 (77%), Gaps = 2/345 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TT +K GA DIH+W+GKDTSQDEAG AA+KTVELDA LGGRAVQYRE+QGHE+ KFLS
Sbjct: 53 TTQNKGGAYLFDIHFWIGKDTSQDEAGTAAVKTVELDAALGGRAVQYREIQGHESDKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIP EGG ASGFK E EE +TRL+ CKGK V++K+VPFARSSLNHDD+FILDT
Sbjct: 113 YFKPCIIPLEGGVASGFKKPEEEEFETRLYTCKGKRAVHLKQVPFARSSLNHDDVFILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
+ KI+QFNG+NS+IQERAKAL V+QY+KD +H+G +VA ++DG+L +S+SGEFW LFG
Sbjct: 173 KEKIYQFNGANSNIQERAKALVVIQYLKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAP+ RK S+D+ ++ PPKL + G+ E + D L+K +L+ NKCY+LDCG E+F
Sbjct: 233 GFAPIARKVASEDEIIPETTPPKLYSIADGQVESIDGD-LSKSMLENNKCYLLDCGSEIF 291
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSWPQTTNAA 299
+W+GR T ++ERK+A + ++ V+S NRPK ++I RV++G+E F+S FDSWP +
Sbjct: 292 IWVGRVTQVEERKTAIQAAEDFVASENRPKATRITRVIQGYEPHSFKSNFDSWPSGSATP 351
Query: 300 MPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
E+GRGKVAALLK+QG+ +KGL K+ PV E+ P ++ G L+V
Sbjct: 352 ANEEGRGKVAALLKQQGVGLKGLSKSTPVNEDIPPLLEGGGKLEV 396
Score = 87.8 bits (216), Expect = 4e-16
Identities = 83/318 (26%), Positives = 147/318 (46%), Gaps = 41/318 (12%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK+++Q++ A + L GR VQ R +G E +F++ F+ ++ +GG +S
Sbjct: 441 WFGKNSNQEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVV-LKGGLSS 499
Query: 76 GFKHVEAEEHKT---------RLFVCKGKHVVYVK--EVPFARSSLNHDDIFILDTESKI 124
G+K+ E+ + L G V K +V +SLN D F+L + + +
Sbjct: 500 GYKNSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSM 559
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAP 184
F + G++S+ +++ A +V +++K + ++G +ES FW GG
Sbjct: 560 FLWVGNHSTHEQQELAAKVAEFLKPG-----TTIKHAKEG-----TESSSFWFALGGKQN 609
Query: 185 LPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIG 244
K VS + T+ +GK + E + ++ L T + ++LD EVFVW+G
Sbjct: 610 FTSKKVSSE--TVRDPHLFSFSFNRGKFQVEEIHNFDQDDLLTEEMHLLDTHAEVFVWVG 667
Query: 245 RNTSLDERKSA----------SGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQ 294
+ E+++A +GS + L PK + ++ EG E F + F SW
Sbjct: 668 QCVDPKEKQTAFEIGQRYINLAGSLEGL-----SPKVPLYKITEGNEPCFFTTYF-SW-D 720
Query: 295 TTNAAMPEDGRGKVAALL 312
+T A + + K AALL
Sbjct: 721 STKATVQGNSYQKKAALL 738
>UniRef100_Q9SIK8 Putative villin [Arabidopsis thaliana]
Length = 909
Score = 339 bits (870), Expect = 6e-92
Identities = 173/337 (51%), Positives = 232/337 (68%), Gaps = 7/337 (2%)
Query: 10 RHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQ 69
++DIHYWLG D ++ ++ A+ K ++LDA LG VQYREVQG ET+KFLSYFKPCIIP
Sbjct: 62 QYDIHYWLGIDANEVDSILASDKALDLDAALGCCTVQYREVQGQETEKFLSYFKPCIIPV 121
Query: 70 EGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESKIFQFNG 129
EG S + E ++ L CKG HVV VKEVPF RSSLNHDD+FILDT SK+F F G
Sbjct: 122 EG-KYSPKTGIAGETYQVTLLRCKGDHVVRVKEVPFLRSSLNHDDVFILDTASKVFLFAG 180
Query: 130 SNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPLPRKT 189
NSS QE+AKA+EVV+YIKD HDG+CEVA+IEDG+ DS++GEFW FGG+AP+P+ +
Sbjct: 181 CNSSTQEKAKAMEVVEYIKDNKHDGRCEVATIEDGKFSGDSDAGEFWSFFGGYAPIPKLS 240
Query: 190 VSDDDKTIDSHPPKLLCVE-KGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTS 248
S + + +L ++ KG P T SL K++L+ NKCY+LDC EVFVW+GRNTS
Sbjct: 241 SSTTQEQTQTPCAELFWIDTKGNLHPTGTSSLDKDMLEKNKCYMLDCHSEVFVWMGRNTS 300
Query: 249 LDERKSASGSTDELVSSTNR-PKSQIIRVMEGFETVMFRSKFDSWPQTTNAAMPEDGRGK 307
L ERK++ S++E + R + ++ + EG E FRS F+ WPQT +++ +GR K
Sbjct: 301 LTERKTSISSSEEFLRKEGRSTTTSLVLLTEGLENARFRSFFNKWPQTVESSLYNEGREK 360
Query: 308 VAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
VAAL K++G DV+ L P +E+ Y +C +L+V
Sbjct: 361 VAALFKQKGYDVEEL----PDEEDDPLYTNCRDNLKV 393
Score = 70.9 bits (172), Expect = 5e-11
Identities = 70/303 (23%), Positives = 128/303 (42%), Gaps = 29/303 (9%)
Query: 11 HDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQE 70
H ++ W+G ++ Q + A + G +V QG+E +F F+ ++ +
Sbjct: 432 HLLYVWIGCESIQQDRADAITNASAIVGTTKGESVLCHIYQGNEPSRFFPMFQSLVV-FK 490
Query: 71 GGAASGFKHVEAE---------EHKTRLFVCKGKHVVYVK--EVPFARSSLNHDDIFILD 119
GG + +K + AE E+K LF G ++ +V +SLN +IL
Sbjct: 491 GGLSRRYKVLLAEKEKIGEEYNENKASLFRVVGTSPRNMQAIQVNLVATSLNSSYSYILQ 550
Query: 120 TESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLF 179
+ F + G SS + + L+ + Y DT C+ I +G +E+ FW L
Sbjct: 551 YGASAFTWIGKLSSDSDH-EVLDRMLYFLDT----SCQPIYIREG-----NETDTFWNLL 600
Query: 180 GGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEV 239
GG + P++ + K I+ + E + ++ L T ++LDC EV
Sbjct: 601 GGKSEYPKE--KEMRKQIEEPHLFTCSCSSDVLKVKEIYNFVQDDLTTEDVFLLDCQSEV 658
Query: 240 FVWIGRNTSLDERKSA-----SGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQ 294
+VWIG N+++ ++ A +++ ++ + V EG E F F+ P+
Sbjct: 659 YVWIGSNSNIKSKEEALTLGLKFLEMDILEEGLTMRTPVYVVTEGHEPPFFTRFFEWVPE 718
Query: 295 TTN 297
N
Sbjct: 719 KAN 721
>UniRef100_O81643 Villin 1 [Arabidopsis thaliana]
Length = 910
Score = 330 bits (846), Expect = 4e-89
Identities = 172/338 (50%), Positives = 229/338 (66%), Gaps = 8/338 (2%)
Query: 10 RHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQ 69
++DIHYWLG D ++ ++ A+ K ++LDA LG VQYREVQG ET+KFLSYFKPCIIP
Sbjct: 62 QYDIHYWLGIDANEVDSILASDKALDLDAALGCCTVQYREVQGQETEKFLSYFKPCIIPV 121
Query: 70 EGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESKIFQFNG 129
EG S + E ++ L CKG HVV VKEVPF RSSLNHDD+FILDT SK+F F G
Sbjct: 122 EG-KYSPKTGIAGETYQVTLLRCKGDHVVRVKEVPFLRSSLNHDDVFILDTASKVFLFAG 180
Query: 130 SNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPLPRKT 189
NSS QE+AKA+EVV+YIKD HDG+CEVA+IEDG+ DS++GEFW FGG+AP+P+ +
Sbjct: 181 CNSSTQEKAKAMEVVEYIKDNKHDGRCEVATIEDGKFSGDSDAGEFWSFFGGYAPIPKLS 240
Query: 190 VSDDDKTIDSHPPKLLCVE-KGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTS 248
S + + +L ++ KG P T SL K++L+ NKCY+LDC EVFVW+GRNTS
Sbjct: 241 SSTTQEQTQTPCAELFWIDTKGNLHPTGTSSLDKDMLEKNKCYMLDCHSEVFVWMGRNTS 300
Query: 249 LDERKSASGSTDELVSSTNR-PKSQIIRVMEGFETVMFRSKFDSWPQTT-NAAMPEDGRG 306
L ERK++ S++E + R + ++ + EG E FRS F+ WP + A +GR
Sbjct: 301 LTERKTSISSSEEFLRKEGRSTTTSLVLLTEGLENARFRSFFNKWPSDRWSLAFYNEGRE 360
Query: 307 KVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
KVAAL K++G DV+ L P +E+ Y +C +L+V
Sbjct: 361 KVAALFKQKGYDVEEL----PDEEDDPLYTNCRDNLKV 394
Score = 71.2 bits (173), Expect = 4e-11
Identities = 70/303 (23%), Positives = 128/303 (42%), Gaps = 29/303 (9%)
Query: 11 HDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQE 70
H ++ W+G ++ Q + A + G +V QG+E +F F+ ++ +
Sbjct: 433 HLLYVWIGCESIQQDRADAITNASAIVGTTKGESVLCHIYQGNEPSRFFPMFQSLVV-FK 491
Query: 71 GGAASGFKHVEAE---------EHKTRLFVCKGKHVVYVK--EVPFARSSLNHDDIFILD 119
GG + +K + AE E+K LF G ++ +V +SLN +IL
Sbjct: 492 GGLSRRYKVLLAEKEKIGEEYNENKASLFRVVGTSPRNMQAIQVNLVATSLNSSYSYILQ 551
Query: 120 TESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLF 179
+ F + G SS + + L+ + Y DT C+ I +G +E+ FW L
Sbjct: 552 YGASAFTWIGKLSSDSDH-EVLDRMLYFLDT----SCQPTYIREG-----NETDTFWNLL 601
Query: 180 GGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEV 239
GG + P++ + K I+ + E + ++ L T ++LDC EV
Sbjct: 602 GGKSEYPKE--KEMRKQIEEPHLFTCSCSSDVLKVKEIYNFVQDDLTTEDVFLLDCQSEV 659
Query: 240 FVWIGRNTSLDERKSA-----SGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQ 294
+VWIG N+++ ++ A +++ ++ + V EG E F F+ P+
Sbjct: 660 YVWIGSNSNIKSKEEALTLGLKFLEMDILEEGLTMRTPVYVVTEGHEPPFFTRFFEWVPE 719
Query: 295 TTN 297
N
Sbjct: 720 KAN 722
>UniRef100_Q65XP6 Putative villin [Oryza sativa]
Length = 634
Score = 310 bits (794), Expect = 4e-83
Identities = 155/348 (44%), Positives = 229/348 (65%), Gaps = 11/348 (3%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
T KSG +H++HYW+G++ +++ A+ K +ELD LG VQYRE QG E+ KFLS
Sbjct: 49 TVELKSGVRQHNVHYWVGEEAKEEDCLTASDKAIELDVALGSNTVQYRETQGEESDKFLS 108
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIP +G +S + + T +F C+G+HV V EVPF+RSSL+H +F++DT
Sbjct: 109 YFKPCIIPIQGSLSSHMRIYGDKSKDTTMFRCEGEHVARVTEVPFSRSSLDHKAVFVVDT 168
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
ESKIF F+G NSS+Q RAKAL+VV+++K+ H G+CE+A+IEDG+L+ DS++G+FW LFG
Sbjct: 169 ESKIFLFSGCNSSMQTRAKALDVVKHLKENRHCGRCEIATIEDGKLVGDSDAGDFWNLFG 228
Query: 181 GFAPLPR---KTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGL 237
G+AP+PR TV + T S KL + K P ET+ L +E+L++++ YILDCG
Sbjct: 229 GYAPIPRDVQDTVMTELMTTSS--KKLFWINKRNLVPVETNLLEREMLNSDRNYILDCGT 286
Query: 238 EVFVWIGRNTSLDERKSASGSTDELVSSTNR-PKSQIIRVMEGFETVMFRSKFDSWPQTT 296
EVF+W+G T + ER+++ + ++ V R ++ + + EG ETV F+ F WP+
Sbjct: 287 EVFLWMGMTTLVSERRTSVTALEDYVRCEGRQSNARSVILTEGHETVEFKMHFQHWPKNA 346
Query: 297 NAAMPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
+ E GR KVAA+ K QG DV + +++P+ +I C G L+V
Sbjct: 347 VPKLYEAGREKVAAIFKHQGYDV-----TEIPEDKPRHFISCNGSLKV 389
Score = 42.4 bits (98), Expect = 0.020
Identities = 52/195 (26%), Positives = 83/195 (41%), Gaps = 25/195 (12%)
Query: 11 HDIHYWLGKDT-SQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQ 69
H W G ++ ++D AA++ + +D+V G AV + +G E + F FK II
Sbjct: 428 HLFFAWSGLNSINEDRVAAASLMSGMIDSVKG-HAVVAQVFEGREPEMFFLVFKSLII-F 485
Query: 70 EGGAASGFKHVEA---------EEHKTRLFVCKG-KH-VVYVKEVPFARSSLNHDDIFIL 118
+GG + +K+ + +++ LF +G KH + +V A SSLN +IL
Sbjct: 486 KGGRSMAYKNFVSQRSDANGWYQKNGVALFRVQGLKHDCIRAIQVDLAASSLNSSHCYIL 545
Query: 119 DTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGL 178
F + GS SS + + D D C + + + SE FW
Sbjct: 546 QAGGSFFTWLGSLSSPSD--------HNLLDRMMDKLCPLK--QSLLVREGSEPDRFWEA 595
Query: 179 FGGFAP-LPRKTVSD 192
GG + L K V D
Sbjct: 596 LGGRSEYLREKQVKD 610
>UniRef100_Q9XZD2 Fragmin A [Physarum polycephalum]
Length = 368
Score = 201 bits (511), Expect = 3e-50
Identities = 122/299 (40%), Positives = 166/299 (54%), Gaps = 17/299 (5%)
Query: 2 TASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSY 61
T ++ L D+H+WLG T+QDEAG AA KTVELD VLGG VQ+REVQG+E+Q+FLSY
Sbjct: 77 TYKQNDRLAWDVHFWLGTYTTQDEAGTAAYKTVELDDVLGGAPVQHREVQGYESQRFLSY 136
Query: 62 FKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTE 121
F I EGG +GF HV+ EE++ RL KGK + V EVP + SLN D+FI+D
Sbjct: 137 FPNGIRILEGGFDTGFHHVKPEEYRPRLLHLKGKKFIRVSEVPLSHKSLNSGDVFIVDLG 196
Query: 122 SKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSE-SGEFWGLFG 180
+++ QFNGS S + ERAKA +VQ I+ +GK + GR++ +SE FW G
Sbjct: 197 AELIQFNGSKSGVAERAKAAALVQAIEGE-RNGKSK------GRVVEESEDDAAFWKALG 249
Query: 181 GFAPLPRKTVSDDDKTIDS--------HPPKLLCVEKGKAEPFETDSLTKELLDTNKCYI 232
G + D DS H AE + + K LLD+ +I
Sbjct: 250 GKGAIASAEAGGSDVEADSIANVEKTLHRLSDATGNMKLAEVAKGKKIKKSLLDSTDVFI 309
Query: 233 LDCGLEVFVWIGRNTSLDERKSASGSTDELVSSTNR-PKSQIIRVMEGFETVMFRSKFD 290
+D G EV W+G S+ ERK A E V+ N+ P + + RV+EG E ++ S F+
Sbjct: 310 IDAGQEVIAWVGAKASVGERKYALRYAQEFVTQHNKNPATPVSRVLEGGENEVWNSLFE 368
>UniRef100_Q07171-2 Splice isoform 2 of Q07171 [Drosophila melanogaster]
Length = 740
Score = 201 bits (510), Expect = 3e-50
Identities = 117/299 (39%), Positives = 172/299 (57%), Gaps = 13/299 (4%)
Query: 5 KSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKP 64
K L D+H+WLG +TS DEAGAAAI TV+LD +L G VQ+REVQ HE+Q FLSYFK
Sbjct: 51 KDKKLSWDVHFWLGLETSTDEAGAAAILTVQLDDLLNGGPVQHREVQDHESQLFLSYFKN 110
Query: 65 CIIPQEGGAASGFKHVEAE-EHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESK 123
I ++GG +GFKHVE + +TRLF KGK V V++V + SS+N D FILD S
Sbjct: 111 GIRYEQGGVGTGFKHVETNAQGETRLFQVKGKRNVRVRQVNLSVSSMNTGDCFILDAGSD 170
Query: 124 IFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFA 183
I+ + GS + E+ KA+ I+D H+G+ V ++D D++ F+ + G +
Sbjct: 171 IYVYVGSQAKRVEKLKAISAANQIRDQDHNGRARVQIVDD--FSTDADKQHFFDVLGSGS 228
Query: 184 --PLPRKTVSDDDKTI---DSHPPKLLCVE----KGKAEPFETDSLTKELLDTNKCYILD 234
+P ++ +D+D D+ L V K K + LT+ +LDT +C+ILD
Sbjct: 229 ADQVPDESTADEDSAFERTDAAAVSLYKVSDASGKLKVDIIGQKPLTQAMLDTRECFILD 288
Query: 235 CGLEVFVWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSW 292
G +FVW+G+ + E+ A E + + P +QI R++EG E+ F+ FD+W
Sbjct: 289 TGSGIFVWVGKGATQKEKTDAMAKAQEFLRTKKYPAWTQIHRIVEGSESAPFKQYFDTW 347
Score = 63.9 bits (154), Expect = 6e-09
Identities = 71/277 (25%), Positives = 115/277 (40%), Gaps = 29/277 (10%)
Query: 25 EAGAAAIKTVELDAVLGGR-AVQYREVQGHETQKFLSYFKPCIIPQEGGAASGFKHVEAE 83
+A AAA K + ++G + + + QGHE + F FK G + F +
Sbjct: 459 KASAAARKRAFEEGLVGSKDGLLVQTNQGHEPRHFYKIFK-------GKLLTSFTALPVT 511
Query: 84 EHKTRLFVCKG--KHVVYVKEVPFARSSLNHDDIFILDT--ESKIFQFNGSNSSIQERAK 139
+LF +G + V+ EV SSL D F+L + KI+ +NG +S E+
Sbjct: 512 ---AQLFRIRGTVESDVHASEVAADSSSLASSDAFVLHSGKSHKIYIWNGLGASAFEKQA 568
Query: 140 ALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPLPRKTVSDDDKTIDS 199
A V D + D E+ +E+G +E EFW G R D ++S
Sbjct: 569 A---VDRFSDYWDD--VELEQVEEG-----AEPDEFWEELNGEGQYDRSLGDDGAPLLES 618
Query: 200 HPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTSLDERKSASGST 259
G + E +E LD++ +LD G E+++W+G S +E +
Sbjct: 619 RLFHCHLSSGGFLKVEEVAQYEQEDLDSDDIMLLDAGDEIYLWVGYGVSEEENGKLLDTA 678
Query: 260 DELVSSTNRPKS----QIIRVMEGFETVMFRSKFDSW 292
+ +S IIRV +G E +F+ F +W
Sbjct: 679 KLYFNLEPTARSFDTVSIIRVPQGKEPRVFKRMFPNW 715
>UniRef100_Q07171-3 Splice isoform 3 of Q07171 [Drosophila melanogaster]
Length = 883
Score = 201 bits (510), Expect = 3e-50
Identities = 117/299 (39%), Positives = 172/299 (57%), Gaps = 13/299 (4%)
Query: 5 KSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKP 64
K L D+H+WLG +TS DEAGAAAI TV+LD +L G VQ+REVQ HE+Q FLSYFK
Sbjct: 194 KDKKLSWDVHFWLGLETSTDEAGAAAILTVQLDDLLNGGPVQHREVQDHESQLFLSYFKN 253
Query: 65 CIIPQEGGAASGFKHVEAE-EHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESK 123
I ++GG +GFKHVE + +TRLF KGK V V++V + SS+N D FILD S
Sbjct: 254 GIRYEQGGVGTGFKHVETNAQGETRLFQVKGKRNVRVRQVNLSVSSMNTGDCFILDAGSD 313
Query: 124 IFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFA 183
I+ + GS + E+ KA+ I+D H+G+ V ++D D++ F+ + G +
Sbjct: 314 IYVYVGSQAKRVEKLKAISAANQIRDQDHNGRARVQIVDD--FSTDADKQHFFDVLGSGS 371
Query: 184 --PLPRKTVSDDDKTI---DSHPPKLLCVE----KGKAEPFETDSLTKELLDTNKCYILD 234
+P ++ +D+D D+ L V K K + LT+ +LDT +C+ILD
Sbjct: 372 ADQVPDESTADEDSAFERTDAAAVSLYKVSDASGKLKVDIIGQKPLTQAMLDTRECFILD 431
Query: 235 CGLEVFVWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSW 292
G +FVW+G+ + E+ A E + + P +QI R++EG E+ F+ FD+W
Sbjct: 432 TGSGIFVWVGKGATQKEKTDAMAKAQEFLRTKKYPAWTQIHRIVEGSESAPFKQYFDTW 490
Score = 63.9 bits (154), Expect = 6e-09
Identities = 71/277 (25%), Positives = 115/277 (40%), Gaps = 29/277 (10%)
Query: 25 EAGAAAIKTVELDAVLGGR-AVQYREVQGHETQKFLSYFKPCIIPQEGGAASGFKHVEAE 83
+A AAA K + ++G + + + QGHE + F FK G + F +
Sbjct: 602 KASAAARKRAFEEGLVGSKDGLLVQTNQGHEPRHFYKIFK-------GKLLTSFTALPVT 654
Query: 84 EHKTRLFVCKG--KHVVYVKEVPFARSSLNHDDIFILDT--ESKIFQFNGSNSSIQERAK 139
+LF +G + V+ EV SSL D F+L + KI+ +NG +S E+
Sbjct: 655 ---AQLFRIRGTVESDVHASEVAADSSSLASSDAFVLHSGKSHKIYIWNGLGASAFEKQA 711
Query: 140 ALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPLPRKTVSDDDKTIDS 199
A V D + D E+ +E+G +E EFW G R D ++S
Sbjct: 712 A---VDRFSDYWDD--VELEQVEEG-----AEPDEFWEELNGEGQYDRSLGDDGAPLLES 761
Query: 200 HPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTSLDERKSASGST 259
G + E +E LD++ +LD G E+++W+G S +E +
Sbjct: 762 RLFHCHLSSGGFLKVEEVAQYEQEDLDSDDIMLLDAGDEIYLWVGYGVSEEENGKLLDTA 821
Query: 260 DELVSSTNRPKS----QIIRVMEGFETVMFRSKFDSW 292
+ +S IIRV +G E +F+ F +W
Sbjct: 822 KLYFNLEPTARSFDTVSIIRVPQGKEPRVFKRMFPNW 858
>UniRef100_Q07171 Gelsolin precursor [Drosophila melanogaster]
Length = 798
Score = 201 bits (510), Expect = 3e-50
Identities = 117/299 (39%), Positives = 172/299 (57%), Gaps = 13/299 (4%)
Query: 5 KSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKP 64
K L D+H+WLG +TS DEAGAAAI TV+LD +L G VQ+REVQ HE+Q FLSYFK
Sbjct: 109 KDKKLSWDVHFWLGLETSTDEAGAAAILTVQLDDLLNGGPVQHREVQDHESQLFLSYFKN 168
Query: 65 CIIPQEGGAASGFKHVEAE-EHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESK 123
I ++GG +GFKHVE + +TRLF KGK V V++V + SS+N D FILD S
Sbjct: 169 GIRYEQGGVGTGFKHVETNAQGETRLFQVKGKRNVRVRQVNLSVSSMNTGDCFILDAGSD 228
Query: 124 IFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFA 183
I+ + GS + E+ KA+ I+D H+G+ V ++D D++ F+ + G +
Sbjct: 229 IYVYVGSQAKRVEKLKAISAANQIRDQDHNGRARVQIVDD--FSTDADKQHFFDVLGSGS 286
Query: 184 --PLPRKTVSDDDKTI---DSHPPKLLCVE----KGKAEPFETDSLTKELLDTNKCYILD 234
+P ++ +D+D D+ L V K K + LT+ +LDT +C+ILD
Sbjct: 287 ADQVPDESTADEDSAFERTDAAAVSLYKVSDASGKLKVDIIGQKPLTQAMLDTRECFILD 346
Query: 235 CGLEVFVWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSW 292
G +FVW+G+ + E+ A E + + P +QI R++EG E+ F+ FD+W
Sbjct: 347 TGSGIFVWVGKGATQKEKTDAMAKAQEFLRTKKYPAWTQIHRIVEGSESAPFKQYFDTW 405
Score = 63.9 bits (154), Expect = 6e-09
Identities = 71/277 (25%), Positives = 115/277 (40%), Gaps = 29/277 (10%)
Query: 25 EAGAAAIKTVELDAVLGGR-AVQYREVQGHETQKFLSYFKPCIIPQEGGAASGFKHVEAE 83
+A AAA K + ++G + + + QGHE + F FK G + F +
Sbjct: 517 KASAAARKRAFEEGLVGSKDGLLVQTNQGHEPRHFYKIFK-------GKLLTSFTALPVT 569
Query: 84 EHKTRLFVCKG--KHVVYVKEVPFARSSLNHDDIFILDT--ESKIFQFNGSNSSIQERAK 139
+LF +G + V+ EV SSL D F+L + KI+ +NG +S E+
Sbjct: 570 ---AQLFRIRGTVESDVHASEVAADSSSLASSDAFVLHSGKSHKIYIWNGLGASAFEKQA 626
Query: 140 ALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPLPRKTVSDDDKTIDS 199
A V D + D E+ +E+G +E EFW G R D ++S
Sbjct: 627 A---VDRFSDYWDD--VELEQVEEG-----AEPDEFWEELNGEGQYDRSLGDDGAPLLES 676
Query: 200 HPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTSLDERKSASGST 259
G + E +E LD++ +LD G E+++W+G S +E +
Sbjct: 677 RLFHCHLSSGGFLKVEEVAQYEQEDLDSDDIMLLDAGDEIYLWVGYGVSEEENGKLLDTA 736
Query: 260 DELVSSTNRPKS----QIIRVMEGFETVMFRSKFDSW 292
+ +S IIRV +G E +F+ F +W
Sbjct: 737 KLYFNLEPTARSFDTVSIIRVPQGKEPRVFKRMFPNW 773
>UniRef100_UPI00003AB298 UPI00003AB298 UniRef100 entry
Length = 778
Score = 191 bits (486), Expect = 2e-47
Identities = 121/303 (39%), Positives = 172/303 (55%), Gaps = 17/303 (5%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
T +SG L++D+H+WLG ++SQDE GAAAI TV++D L G+AVQ+REVQGHE+ FL
Sbjct: 96 TIRQRSGNLQYDLHFWLGDESSQDERGAAAIFTVQMDDYLQGKAVQHREVQGHESSTFLG 155
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKT-RLFVCKGKHVVYVKEVPFARSSLNHDDIFILD 119
YFK I + GG ASGF+HV E RL KG+ V EVP + S N D FILD
Sbjct: 156 YFKSGIKYKAGGVASGFRHVVPNEVTVQRLLQVKGRRTVRATEVPVSWESFNTGDCFILD 215
Query: 120 TESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLF 179
S I+Q+ GSNS+ QER KA + + I+D +G+ +V E+G +E E +
Sbjct: 216 LGSNIYQWCGSNSNRQERLKATVLAKGIRDNERNGRAKVFVSEEG-----AEREEMLQVL 270
Query: 180 GGFAPLPRKTVSDDDKTIDSHP--PKLLCVEKGKAE-----PFETDSLTKELLDTNKCYI 232
G LP + SDD KT ++ KL V G + + ++ L+T C+I
Sbjct: 271 GPKPSLP-QGASDDTKTDTANRKLAKLYKVSNGAGNMAVSLVADENPFSQAALNTEDCFI 329
Query: 233 LDCGLE--VFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVM-EGFETVMFRSKF 289
LD G + +FVW GR+ + DERK+A + + + PK ++V+ E ET +F+ F
Sbjct: 330 LDHGTDGKIFVWKGRSANSDERKAALKTATDFIEKMGYPKHTQVQVLPESGETPLFKQFF 389
Query: 290 DSW 292
+W
Sbjct: 390 KNW 392
Score = 109 bits (273), Expect = 1e-22
Identities = 92/300 (30%), Positives = 137/300 (45%), Gaps = 29/300 (9%)
Query: 3 ASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYF 62
A K G + I+ W G ++QDE +A TV+LD LGG VQ R VQG E +S F
Sbjct: 479 AGKQGQI---IYTWQGAHSTQDEIATSAFLTVQLDEELGGSPVQKRVVQGKEPPHLMSMF 535
Query: 63 --KPCIIPQEGGAASGFKHVEAEEHKTRLFVCKG--KHVVYVKEVPFARSSLNHDDIFIL 118
KP I+ + G + G + A+ TRLF + E+ S LN +D F+L
Sbjct: 536 GGKPLIVYKGGTSREGGQTTPAQ---TRLFQVRSSTSGATRAVELDPVASQLNSNDAFVL 592
Query: 119 DTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGL 178
T S + + G S+ E + A E+++ + + +GR E FW
Sbjct: 593 KTPSAAYLWVGRGSNSAELSGAQELLKVL-------GARPVQVSEGR-----EPDNFWVA 640
Query: 179 FGGFAPLPRKTVSDDDKTIDSHPPKLL-CVEKGKAEPFE--TDSLTKELLDTNKCYILDC 235
GG AP R + DK +D+HPP+L C K E LT++ L T+ ILD
Sbjct: 641 LGGKAPY-RTSPRLKDKKMDAHPPRLFACSNKSGRFTIEEVPGDLTQDDLATDDVMILDT 699
Query: 236 GLEVFVWIGRNTSLDERKSASGSTDELVS---STNRPKSQIIRVMEGFETVMFRSKFDSW 292
+VFVWIG++ +E+ A S + ++ ++ + V +G E F F W
Sbjct: 700 WDQVFVWIGKDAQEEEKTEALKSAKRYIETDPASRDKRTPVTLVKQGLEPPTFSGWFLGW 759
>UniRef100_O93510 Gelsolin precursor [Gallus gallus]
Length = 778
Score = 191 bits (485), Expect = 3e-47
Identities = 121/303 (39%), Positives = 172/303 (55%), Gaps = 17/303 (5%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
T +SG L++D+H+WLG ++SQDE GAAAI TV++D L G+AVQ+REVQGHE+ FL
Sbjct: 96 TIRQRSGNLQYDLHFWLGDESSQDERGAAAIFTVQMDDYLQGKAVQHREVQGHESSTFLG 155
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKT-RLFVCKGKHVVYVKEVPFARSSLNHDDIFILD 119
YFK I + GG ASGF+HV E RL KG+ V EVP + S N D FILD
Sbjct: 156 YFKSGIKYKAGGVASGFRHVVPNEVTVQRLLQVKGRRTVRATEVPVSWESFNTGDCFILD 215
Query: 120 TESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLF 179
S I+Q+ GSNS+ QER KA + + I+D +G+ +V E+G +E E +
Sbjct: 216 LGSNIYQWCGSNSNRQERLKATVLAKGIRDNEKNGRAKVFVSEEG-----AEREEMLQVL 270
Query: 180 GGFAPLPRKTVSDDDKTIDSHP--PKLLCVEKGKAE-----PFETDSLTKELLDTNKCYI 232
G LP + SDD KT ++ KL V G + + ++ L+T C+I
Sbjct: 271 GPKPSLP-QGASDDTKTDTANRKLAKLYKVSNGAGNMAVSLVADENPFSQAALNTEDCFI 329
Query: 233 LDCGLE--VFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVM-EGFETVMFRSKF 289
LD G + +FVW GR+ + DERK+A + + + PK ++V+ E ET +F+ F
Sbjct: 330 LDHGTDGKIFVWKGRSANSDERKAALKTATDFIDKMGYPKHTQVQVLPESGETPLFKQFF 389
Query: 290 DSW 292
+W
Sbjct: 390 KNW 392
Score = 108 bits (271), Expect = 2e-22
Identities = 92/300 (30%), Positives = 138/300 (45%), Gaps = 29/300 (9%)
Query: 3 ASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYF 62
A K G + I+ W G ++QDE +A TV+LD LGG VQ R VQG E +S F
Sbjct: 479 AGKQGQI---IYTWQGAHSTQDEIATSAFLTVQLDEELGGSPVQKRVVQGKEPPHLMSMF 535
Query: 63 --KPCIIPQEGGAASGFKHVEAEEHKTRLFVCKG--KHVVYVKEVPFARSSLNHDDIFIL 118
KP I+ + G + G + A+ TRLF + E+ A S LN +D F+L
Sbjct: 536 GGKPLIVYKGGTSREGGQTTPAQ---TRLFQVRSSTSGATRAVELDPAASQLNSNDAFVL 592
Query: 119 DTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGL 178
T S + + G S+ E + A E+++ + + +GR E FW
Sbjct: 593 KTPSAAYLWVGRGSNSAELSGAQELLKVL-------GARPVQVSEGR-----EPDNFWVA 640
Query: 179 FGGFAPLPRKTVSDDDKTIDSHPPKLL-CVEKGKAEPFE--TDSLTKELLDTNKCYILDC 235
GG AP R + DK +D++PP+L C K E LT++ L T+ ILD
Sbjct: 641 LGGKAPY-RTSPRLKDKKMDAYPPRLFACSNKSGRFTIEEVPGDLTQDDLATDDVMILDT 699
Query: 236 GLEVFVWIGRNTSLDERKSASGSTDELVS---STNRPKSQIIRVMEGFETVMFRSKFDSW 292
+VFVWIG++ +E+ A S + ++ ++ + V +G E F F W
Sbjct: 700 WDQVFVWIGKDAQEEEKTEALKSAKRYIETDPASRDKRTPVTLVKQGLEPPTFSGWFLGW 759
>UniRef100_Q94707 Actin-binding protein fragmin P [Physarum polycephalum]
Length = 371
Score = 190 bits (483), Expect = 4e-47
Identities = 115/292 (39%), Positives = 166/292 (56%), Gaps = 17/292 (5%)
Query: 9 LRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYF-KPCII 67
L +D+H+WLG T+QDEAG AA KTVELD LGG VQYREVQG+E+++FLS F K +
Sbjct: 87 LAYDVHFWLGAFTTQDEAGTAAYKTVELDDYLGGLPVQYREVQGYESERFLSLFPKGGLR 146
Query: 68 PQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESKIFQF 127
+GG +GF HVEA++++TRL KGK + V EVP SLN D+F+LD + Q+
Sbjct: 147 ILDGGVETGFHHVEADKYRTRLLHLKGKKHIRVHEVPKTYKSLNSGDVFVLDAGKTVIQW 206
Query: 128 NGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESG-EFWGLFGGFAPLP 186
NG+ + + E+ KA E++Q I+ E I GR++A++++ EF+ L G P+
Sbjct: 207 NGAKAGLLEKVKAAELLQAIEG-------EREGIASGRVVAEADNDTEFFTLLGDKGPIA 259
Query: 187 RKTVSDDDKTID--SHPPKLLCVE--KGKAEPFETD---SLTKELLDTNKCYILDCGLEV 239
D D P LL + GK E E + + LLD+N ++L G EV
Sbjct: 260 DAAAGGSDLEADKKDQPAVLLRLSDASGKFEFTEVARGLKVKRNLLDSNDVFVLYTGAEV 319
Query: 240 FVWIGRNTSLDERKSASGSTDELVSSTNRP-KSQIIRVMEGFETVMFRSKFD 290
F W+G++ S+ E+K A E V P + + R++EG E +F FD
Sbjct: 320 FAWVGKHASVGEKKKALSFAQEYVQKAGLPIHTPVARILEGGENEVFEDFFD 371
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 587,981,542
Number of Sequences: 2790947
Number of extensions: 24345207
Number of successful extensions: 54404
Number of sequences better than 10.0: 289
Number of HSP's better than 10.0 without gapping: 241
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 52252
Number of HSP's gapped (non-prelim): 753
length of query: 355
length of database: 848,049,833
effective HSP length: 128
effective length of query: 227
effective length of database: 490,808,617
effective search space: 111413556059
effective search space used: 111413556059
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC125477.10


