

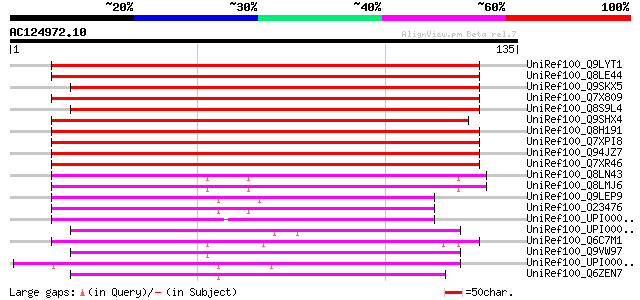
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124972.10 - phase: 1 /pseudo
(135 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LYT1 Hypothetical protein F17J16_100 [Arabidopsis th... 164 4e-40
UniRef100_Q8LE44 Putative amine oxidase [Arabidopsis thaliana] 164 5e-40
UniRef100_Q9SKX5 Putative amine oxidase 1 [Arabidopsis thaliana] 162 2e-39
UniRef100_Q7X809 OSJNBb0085C12.17 protein [Oryza sativa] 159 1e-38
UniRef100_Q8S9L4 At2g43020/MFL8.12 [Arabidopsis thaliana] 159 1e-38
UniRef100_Q9SHX4 F1E22.18 [Arabidopsis thaliana] 135 2e-31
UniRef100_Q8H191 Hypothetical protein At1g65840 [Arabidopsis tha... 133 8e-31
UniRef100_Q7XPI8 OSJNBb0004A17.1 protein [Oryza sativa] 133 8e-31
UniRef100_Q94JZ7 Hypothetical protein F1E22.18 [Arabidopsis thal... 133 8e-31
UniRef100_Q7XR46 OSJNBa0043A12.39 protein [Oryza sativa] 130 6e-30
UniRef100_Q8LN43 Putative polyamine oxidase [Oryza sativa] 71 5e-12
UniRef100_Q8LMJ6 Putative polyamine oxidase, 3'-partial [Oryza s... 71 5e-12
UniRef100_Q9LEP9 Putative corticosteroid binding protein [Brassi... 66 2e-10
UniRef100_O23476 Hypothetical protein dl4185w [Arabidopsis thali... 65 3e-10
UniRef100_UPI000030F65C UPI000030F65C UniRef100 entry 54 6e-07
UniRef100_UPI00002357EF UPI00002357EF UniRef100 entry 54 8e-07
UniRef100_Q6C7M1 Similar to tr|Q9Y802 Schizosaccharomyces pombe ... 53 1e-06
UniRef100_Q9VW97 CG17149-PA, isoform A [Drosophila melanogaster] 51 5e-06
UniRef100_UPI0000235AB2 UPI0000235AB2 UniRef100 entry 50 8e-06
UniRef100_Q6ZEN7 Slr5093 protein [Synechocystis sp.] 50 8e-06
>UniRef100_Q9LYT1 Hypothetical protein F17J16_100 [Arabidopsis thaliana]
Length = 488
Score = 164 bits (415), Expect = 4e-40
Identities = 80/114 (70%), Positives = 92/114 (80%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L + +I FEPKL WK+ AI D+ VG+ENKIIL+F NVFWPN +FL VVAE S GCSYFL
Sbjct: 281 LKSGMITFEPKLPQWKQEAINDLGVGIENKIILNFDNVFWPNVEFLGVVAETSYGCSYFL 340
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA H VLVYMP G+ A+DIEK SDEAAANFAF QL+KILPDASSP+ +V
Sbjct: 341 NLHKATSHPVLVYMPAGQLARDIEKKSDEAAANFAFSQLQKILPDASSPINYLV 394
>UniRef100_Q8LE44 Putative amine oxidase [Arabidopsis thaliana]
Length = 488
Score = 164 bits (414), Expect = 5e-40
Identities = 80/114 (70%), Positives = 92/114 (80%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L + +I FEPKL WK+ AI D+ VG+ENKIIL+F NVFWPN +FL VVAE S GCSYFL
Sbjct: 281 LKSGMITFEPKLPQWKQEAINDLGVGIENKIILNFDNVFWPNVEFLGVVAETSYGCSYFL 340
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA H VLVYMP G+ A+DIEK SDEAAANFAF QL+KILPDASSP+ +V
Sbjct: 341 NLHKATSHPVLVYMPAGQLARDIEKNSDEAAANFAFSQLQKILPDASSPINYLV 394
>UniRef100_Q9SKX5 Putative amine oxidase 1 [Arabidopsis thaliana]
Length = 490
Score = 162 bits (409), Expect = 2e-39
Identities = 79/109 (72%), Positives = 89/109 (81%)
Query: 17 IKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFLNLHKA 76
IKFEPKL +WK+ AI D+ VG+ENKIILHF+ VFWP +FL VVAE S GCSYFLNLHKA
Sbjct: 285 IKFEPKLPEWKQEAINDLGVGIENKIILHFEKVFWPKVEFLGVVAETSYGCSYFLNLHKA 344
Query: 77 AGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
GH VLVYMP G+ AKDIEKMSDEAAANFA QL++ILPDA PV +V
Sbjct: 345 TGHPVLVYMPAGQLAKDIEKMSDEAAANFAVLQLQRILPDALPPVQYLV 393
>UniRef100_Q7X809 OSJNBb0085C12.17 protein [Oryza sativa]
Length = 484
Score = 159 bits (403), Expect = 1e-38
Identities = 77/114 (67%), Positives = 89/114 (77%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L IKFEP+L +WKE AI ++ VGVENKIILHF VFWPN +FL VV+ + GCSYFL
Sbjct: 270 LKANTIKFEPRLPEWKEEAIRELSVGVENKIILHFSEVFWPNVEFLGVVSSTTYGCSYFL 329
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA GH VLVYMP GR A DIEK+SDEAAA FAF QLKKILP+A+ P+ +V
Sbjct: 330 NLHKATGHPVLVYMPAGRLACDIEKLSDEAAAQFAFSQLKKILPNAAEPIHYLV 383
>UniRef100_Q8S9L4 At2g43020/MFL8.12 [Arabidopsis thaliana]
Length = 490
Score = 159 bits (402), Expect = 1e-38
Identities = 78/109 (71%), Positives = 88/109 (80%)
Query: 17 IKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFLNLHKA 76
IKF PKL +WK+ AI D+ VG+ENKIILHF+ VFWP +FL VVAE S GCSYFLNLHKA
Sbjct: 285 IKFGPKLPEWKQEAINDLGVGIENKIILHFEKVFWPKVEFLGVVAETSYGCSYFLNLHKA 344
Query: 77 AGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
GH VLVYMP G+ AKDIEKMSDEAAANFA QL++ILPDA PV +V
Sbjct: 345 TGHPVLVYMPAGQLAKDIEKMSDEAAANFAVLQLQRILPDALPPVQYLV 393
>UniRef100_Q9SHX4 F1E22.18 [Arabidopsis thaliana]
Length = 516
Score = 135 bits (340), Expect = 2e-31
Identities = 66/111 (59%), Positives = 77/111 (68%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L LI+FEP+L WK +AI+ + VG ENKI L F FWPN +FL +VA S C YFL
Sbjct: 282 LKANLIQFEPELPQWKTSAISGLGVGNENKIALRFDRAFWPNVEFLGMVAPTSYACGYFL 341
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVT 122
NLHKA GH VLVYM G A+D+EK+SDEA ANF QLKK+ PDA PVT
Sbjct: 342 NLHKATGHPVLVYMAAGNLAQDLEKLSDEATANFVMLQLKKMFPDAPDPVT 392
>UniRef100_Q8H191 Hypothetical protein At1g65840 [Arabidopsis thaliana]
Length = 497
Score = 133 bits (335), Expect = 8e-31
Identities = 65/114 (57%), Positives = 77/114 (67%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L LI+FEP+L WK +AI+ + VG ENKI L F FWPN +FL +VA S C YFL
Sbjct: 282 LKANLIQFEPELPQWKTSAISGLGVGNENKIALRFDRAFWPNVEFLGMVAPTSYACGYFL 341
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA GH VLVYM G A+D+EK+SDEA ANF QLKK+ PDA P +V
Sbjct: 342 NLHKATGHPVLVYMAAGNLAQDLEKLSDEATANFVMLQLKKMFPDAPDPAQYLV 395
>UniRef100_Q7XPI8 OSJNBb0004A17.1 protein [Oryza sativa]
Length = 492
Score = 133 bits (335), Expect = 8e-31
Identities = 64/114 (56%), Positives = 80/114 (70%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L +IKFEP+L DWK ++I+D+ +G+ENKI L F +VFWPN + L VA SN C YFL
Sbjct: 278 LKANIIKFEPELPDWKLSSISDLGIGIENKIALRFNSVFWPNVEVLGRVAPTSNACGYFL 337
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA GH VLV M GR A + EK+SDE + NF QLKK+LP A+ PV +V
Sbjct: 338 NLHKATGHPVLVCMVAGRFAYEFEKLSDEESVNFVMSQLKKMLPGATEPVQYLV 391
>UniRef100_Q94JZ7 Hypothetical protein F1E22.18 [Arabidopsis thaliana]
Length = 497
Score = 133 bits (335), Expect = 8e-31
Identities = 65/114 (57%), Positives = 77/114 (67%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L LI+FEP+L WK +AI+ + VG ENKI L F FWPN +FL +VA S C YFL
Sbjct: 282 LRANLIQFEPELPQWKTSAISGLGVGNENKIALRFDRAFWPNVEFLGMVAPTSYACGYFL 341
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA GH VLVYM G A+D+EK+SDEA ANF QLKK+ PDA P +V
Sbjct: 342 NLHKATGHPVLVYMAAGNLAQDLEKLSDEATANFVMLQLKKMFPDAPDPAQYLV 395
>UniRef100_Q7XR46 OSJNBa0043A12.39 protein [Oryza sativa]
Length = 487
Score = 130 bits (327), Expect = 6e-30
Identities = 60/114 (52%), Positives = 78/114 (67%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L +IKFEP+L WK +AIAD+ VG+ENKI +HF VFWPN + L +V C YFL
Sbjct: 276 LKANIIKFEPELPSWKSSAIADLGVGIENKIAMHFDTVFWPNVEVLGMVGPTPKACGYFL 335
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA G+ VLVYM GR A+++EK+SD+ A + LKK+LPDA+ P +V
Sbjct: 336 NLHKATGNPVLVYMAAGRFAQEVEKLSDKEAVDLVMSHLKKMLPDATEPTKYLV 389
>UniRef100_Q8LN43 Putative polyamine oxidase [Oryza sativa]
Length = 1862
Score = 71.2 bits (173), Expect = 5e-12
Identities = 46/122 (37%), Positives = 63/122 (50%), Gaps = 6/122 (4%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFW-PNEDFLEVVAE---ISNGC 67
L + IKF P L DWK ++I + G+ NKI+L F VFW N D+ AE + C
Sbjct: 1045 LKAQTIKFSPSLPDWKLSSIDRLGFGLLNKIVLEFPEVFWDDNVDYFGATAEQTDLRGQC 1104
Query: 68 SYFLNLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDAS--SPVTIIV 125
F NL K G VL+ + VG+ A D + +S + A L+K+ DAS PV +V
Sbjct: 1105 FMFWNLKKTVGVPVLIALLVGKAAIDGQSISSDDHVKNAIVVLRKLFKDASVPDPVASVV 1164
Query: 126 LN 127
N
Sbjct: 1165 TN 1166
>UniRef100_Q8LMJ6 Putative polyamine oxidase, 3'-partial [Oryza sativa]
Length = 1348
Score = 71.2 bits (173), Expect = 5e-12
Identities = 46/122 (37%), Positives = 63/122 (50%), Gaps = 6/122 (4%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFW-PNEDFLEVVAE---ISNGC 67
L + IKF P L DWK ++I + G+ NKI+L F VFW N D+ AE + C
Sbjct: 1045 LKAQTIKFSPSLPDWKLSSIDRLGFGLLNKIVLEFPEVFWDDNVDYFGATAEQTDLRGQC 1104
Query: 68 SYFLNLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDAS--SPVTIIV 125
F NL K G VL+ + VG+ A D + +S + A L+K+ DAS PV +V
Sbjct: 1105 FMFWNLKKTVGVPVLIALLVGKAAIDGQSISSDDHVKNAIVVLRKLFKDASVPDPVASVV 1164
Query: 126 LN 127
N
Sbjct: 1165 TN 1166
>UniRef100_Q9LEP9 Putative corticosteroid binding protein [Brassica napus]
Length = 1238
Score = 65.9 bits (159), Expect = 2e-10
Identities = 38/106 (35%), Positives = 55/106 (51%), Gaps = 4/106 (3%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNE-DFLEVVAEISN---GC 67
L + IKF P L DWK ++I + GV NK++L F VFW + D+ AE ++ C
Sbjct: 864 LKAETIKFSPPLPDWKYSSIKQLGFGVLNKVVLEFSKVFWDDSLDYFGATAEETDQRGEC 923
Query: 68 SYFLNLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKI 113
F N+ K G VL+ + VG+ A D + S N A L+K+
Sbjct: 924 FMFWNVKKTVGAPVLIALVVGKAAVDYKDKSKSEHVNHAMMVLRKL 969
>UniRef100_O23476 Hypothetical protein dl4185w [Arabidopsis thaliana]
Length = 1265
Score = 65.1 bits (157), Expect = 3e-10
Identities = 38/106 (35%), Positives = 53/106 (49%), Gaps = 4/106 (3%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNE-DFLEVVAE---ISNGC 67
L + IKF P L DWK A+I + GV NK++L F VFW + D+ AE + C
Sbjct: 899 LKAETIKFSPPLPDWKYASIKQLGFGVLNKVVLEFPTVFWDDSVDYFGATAEETDLRGEC 958
Query: 68 SYFLNLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKI 113
F N+ K G VL+ + VG+ A + S N A L+K+
Sbjct: 959 FMFWNVKKTVGAPVLIALVVGKAAFEYTNKSKSEHVNHAMMVLRKL 1004
>UniRef100_UPI000030F65C UPI000030F65C UniRef100 entry
Length = 200
Score = 54.3 bits (129), Expect = 6e-07
Identities = 34/102 (33%), Positives = 52/102 (50%), Gaps = 1/102 (0%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L ++IKF P+L K+ +I + G NK++L F + FW + DF+ VVA + ++
Sbjct: 15 LKKEVIKFIPELPIKKQLSIKKLGWGTLNKVMLKFTHKFWGDTDFI-VVANKESKFHAWI 73
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKI 113
N V+V G AK +E SDE A A +LK +
Sbjct: 74 NEEPICKEPVIVATITGENAKKLENKSDEEVAQIALNELKAV 115
>UniRef100_UPI00002357EF UPI00002357EF UniRef100 entry
Length = 536
Score = 53.9 bits (128), Expect = 8e-07
Identities = 33/109 (30%), Positives = 50/109 (45%), Gaps = 5/109 (4%)
Query: 17 IKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSY--FLNLH 74
+ F P L DWK+ AIA +G KI + F FWP++ + A+ + Y F +L
Sbjct: 307 VNFTPSLPDWKQTAIAKFNMGTYTKIFMQFNETFWPDDTQFFLYADPTTRGYYPVFQSLS 366
Query: 75 K---AAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSP 120
G +++ V QA E+ SDE + L+K+ PD P
Sbjct: 367 TDGFLPGSNIIFVTVVQDQAYRAERQSDEQTKREVLEVLQKMFPDKHIP 415
>UniRef100_Q6C7M1 Similar to tr|Q9Y802 Schizosaccharomyces pombe [Yarrowia
lipolytica]
Length = 1293
Score = 53.1 bits (126), Expect = 1e-06
Identities = 45/130 (34%), Positives = 59/130 (44%), Gaps = 16/130 (12%)
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFW-PNEDFLEVVAEISNG---- 66
L + I+F P L WK +I + GV NKI L F FW ++D L VV + +NG
Sbjct: 835 LKARAIQFIPDLPQWKTDSIERLAFGVVNKICLVFDECFWDDSKDVLCVVKDAANGSADD 894
Query: 67 ---------CSYFLNLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKIL-PD 116
C+ F N G L+ G AK + SDE + A K L+ I D
Sbjct: 895 AGFKQARGFCNMFWNNSAVVGKPCLIGTVSGEAAKIMADKSDEEIVDAALKSLQVITGKD 954
Query: 117 AS-SPVTIIV 125
A+ SPV IV
Sbjct: 955 ATPSPVESIV 964
>UniRef100_Q9VW97 CG17149-PA, isoform A [Drosophila melanogaster]
Length = 890
Score = 51.2 bits (121), Expect = 5e-06
Identities = 30/105 (28%), Positives = 47/105 (44%), Gaps = 1/105 (0%)
Query: 17 IKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFW-PNEDFLEVVAEISNGCSYFLNLHK 75
+KF+P L DWK+ AI + G NK++L F +FW PN + V +
Sbjct: 638 VKFDPPLPDWKQQAIKRLGFGNLNKVVLCFDRIFWDPNANLFGHVGSTTASRGEMFLFWS 697
Query: 76 AAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSP 120
+ VL+ + G A +E ++D+ LK I + S P
Sbjct: 698 ISSSPVLLALVAGMAANLVESVTDDIIIGRCMSVLKNIFGNTSVP 742
>UniRef100_UPI0000235AB2 UPI0000235AB2 UniRef100 entry
Length = 1274
Score = 50.4 bits (119), Expect = 8e-06
Identities = 38/136 (27%), Positives = 57/136 (40%), Gaps = 17/136 (12%)
Query: 2 AQVLIHTHA--RLLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNE-DFLE 58
A ++++T + L + ++F P L DWK AI + GV NK+IL F FW E D
Sbjct: 641 ADMVVYTGSLGTLQHRTVQFSPPLPDWKVGAIDRLGFGVMNKVILAFDQPFWDTERDMFG 700
Query: 59 VVAEISNGCS--------------YFLNLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAAN 104
++ E +N S F N K G VL+ + G A E+ D
Sbjct: 701 LLREPTNRDSMAQEDYASNRGRFYLFWNCMKTTGLPVLIALMAGDAAHQAERTPDAEIVA 760
Query: 105 FAFKQLKKILPDASSP 120
QL+ + + P
Sbjct: 761 EVMSQLRNVFKQVAVP 776
>UniRef100_Q6ZEN7 Slr5093 protein [Synechocystis sp.]
Length = 458
Score = 50.4 bits (119), Expect = 8e-06
Identities = 31/101 (30%), Positives = 50/101 (48%), Gaps = 1/101 (0%)
Query: 17 IKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNE-DFLEVVAEISNGCSYFLNLHK 75
+KF P+L K AI + +G+ NK L F VFWP + D++E V S ++N+ +
Sbjct: 280 VKFIPELPSPKRKAIKALGMGILNKCYLRFPKVFWPKKVDWIEQVPTERGLWSEWVNIFR 339
Query: 76 AAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPD 116
+L+ + K+IE +DE A K L+ + D
Sbjct: 340 VNQLPILLGFNAADEGKEIETWTDEEIIKSAMKTLRHLFGD 380
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 209,155,472
Number of Sequences: 2790947
Number of extensions: 7622118
Number of successful extensions: 23219
Number of sequences better than 10.0: 186
Number of HSP's better than 10.0 without gapping: 92
Number of HSP's successfully gapped in prelim test: 94
Number of HSP's that attempted gapping in prelim test: 23084
Number of HSP's gapped (non-prelim): 190
length of query: 135
length of database: 848,049,833
effective HSP length: 111
effective length of query: 24
effective length of database: 538,254,716
effective search space: 12918113184
effective search space used: 12918113184
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC124972.10


