

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124968.5 - phase: 0
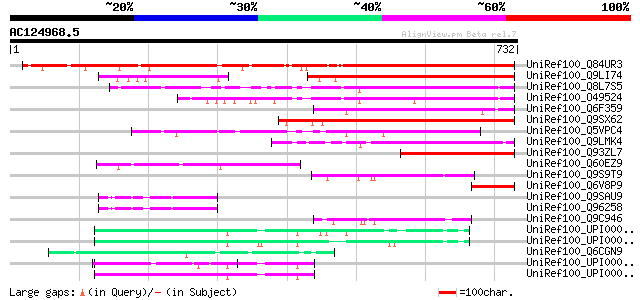
(732 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84UR3 Proline-rich protein family-like [Oryza sativa] 710 0.0
UniRef100_Q9LI74 Similarity to pherophorin [Arabidopsis thaliana] 329 2e-88
UniRef100_Q8L7S5 AT4g18560/F28J12_220 [Arabidopsis thaliana] 318 3e-85
UniRef100_O49524 Pherophorin - like protein [Arabidopsis thaliana] 307 8e-82
UniRef100_Q6F359 Hypothetical protein OJ1268_B08.2 [Oryza sativa] 293 1e-77
UniRef100_Q9SX62 F11A17.16 [Arabidopsis thaliana] 289 2e-76
UniRef100_Q5VPC4 Pherophorin-like protein [Oryza sativa] 248 5e-64
UniRef100_Q9LMK4 F10K1.18 protein [Arabidopsis thaliana] 223 2e-56
UniRef100_Q93ZL7 At1g48280/F11A17_25 [Arabidopsis thaliana] 192 2e-47
UniRef100_Q60EZ9 Hypothetical protein OJ1111_A10.14 [Oryza sativa] 83 3e-14
UniRef100_Q9S9T9 T32N4.10 protein [Arabidopsis thaliana] 81 1e-13
UniRef100_Q6V8P9 Putative actin-binding protein [Malus domestica] 79 4e-13
UniRef100_Q9SAU9 F5F19.14 protein [Arabidopsis thaliana] 72 5e-11
UniRef100_Q96258 AR791 [Arabidopsis thaliana] 72 5e-11
UniRef100_Q9C946 Hypothetical protein T7P1.21 [Arabidopsis thali... 70 3e-10
UniRef100_UPI000043298F UPI000043298F UniRef100 entry 62 8e-08
UniRef100_UPI000043298E UPI000043298E UniRef100 entry 60 2e-07
UniRef100_Q6CGN9 Similarity [Yarrowia lipolytica] 59 5e-07
UniRef100_UPI0000432993 UPI0000432993 UniRef100 entry 55 6e-06
UniRef100_UPI0000432990 UPI0000432990 UniRef100 entry 55 6e-06
>UniRef100_Q84UR3 Proline-rich protein family-like [Oryza sativa]
Length = 798
Score = 710 bits (1833), Expect = 0.0
Identities = 417/791 (52%), Positives = 518/791 (64%), Gaps = 107/791 (13%)
Query: 19 FSDQNQPPKLQTTKTTNPNNNNHSKPRL---WGAHIVKGFSADKKTKQLPTKKQQNTTTI 75
F +Q +P + ++ + +N N KP+ WG+ IVKGF+ADKKTK+ +
Sbjct: 7 FDNQTKPCR---SRVDSKSNINVLKPKFGSSWGSQIVKGFTADKKTKKTAAAASKKPPLA 63
Query: 76 TTSDVVTNQKNVNPFVPPHSRVKRSLMGDLSCS----QVHPHAFPTH--RRQSSTDLFTE 129
+ +V T+ N +P HSRVKRSLMGD CS QVHPH F H R +S DLF E
Sbjct: 64 SVENVNTS----NQQIPYHSRVKRSLMGDFPCSPAGAQVHPHVFDCHGIRSPASHDLFLE 119
Query: 130 LDHMRSLLQESKEREAKLNAELVECRKN-------------QSEVDELVKKVALLEEEKS 176
LDH+R L+ESKERE L +EL +CR+N ++E+D LV+ LE EK+
Sbjct: 120 LDHLREQLRESKERELALQSELRQCRENPRVSELEKDLDSRKNEIDRLVRLKTSLEVEKT 179
Query: 177 GLSEQLVALSRSCGLERQEEDK--DG--------------STQNLELEVVELRRLNKELH 220
LSEQL ALS C +E+ EE+ DG S++NLE+EVVELRRLNKEL
Sbjct: 180 SLSEQLSALS--CMVEQHEENARLDGHGNRVSSMNGGNASSSENLEIEVVELRRLNKELQ 237
Query: 221 MQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNE 280
QKRNL +LSS ES+L+ + ++ES+IVAK +AEASLLR TN +LSKQVEGLQ SRL E
Sbjct: 238 FQKRNLAIKLSSAESKLAVIEKNAESEIVAKVQAEASLLRHTNANLSKQVEGLQMSRLTE 297
Query: 281 VEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQC-GSA----- 334
VEELAYLRW+NSCLR EL N SD+ + + + D I+ D C G A
Sbjct: 298 VEELAYLRWINSCLRHELSN------SDQAARAMT-DADYNDEIACHVDDCDGDARLDQN 350
Query: 335 ----NSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSS 390
F++ ++ K+W S + + +++D+ WIE+ S RRHS+ G
Sbjct: 351 SSDHKKFSIAERIKQW---SQNDKNCEASKKEALLDRAWIEAAECRSPTRRHSLGGPKGC 407
Query: 391 EEEVSVLSKRRQSNCFDSFECLKEIEKESV----------PMPLFV-------------- 426
+E S++ KRRQS D+F CL E E++ L V
Sbjct: 408 AQEFSIV-KRRQS---DTFICLPEATDEAISCNKDETIREKRELLVDKYDFGRSESSRFL 463
Query: 427 ---------QQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFA 477
Q +EKRALRIPNPPPRPS S+ SA PP PPPPPPPP F+
Sbjct: 464 LGKSEVCKSQSMDVEKRALRIPNPPPRPSVSVPHSGPSNGSA-ANPPKPPPPPPPP-KFS 521
Query: 478 SRGNTAMVKRAPQVVELYHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLL 537
+R N ++KRAPQV ELYHSLM+RDS++D+S G+ + + A+VRSSMIGEIENRSSHL
Sbjct: 522 TR-NAGVMKRAPQVAELYHSLMRRDSKKDTSGSGICETANSANVRSSMIGEIENRSSHLQ 580
Query: 538 AIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEK 597
AIKAD+ETQGEFV SLI+EV +A Y++I+DVVAFVKWLDDELGFLVDERAVLKHFDWPE+
Sbjct: 581 AIKADVETQGEFVKSLIKEVTNAAYKDIEDVVAFVKWLDDELGFLVDERAVLKHFDWPER 640
Query: 598 KADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDS 657
KADTLREAAFGYQDLKKLESEVS+YKDDPRLPCDIALKKMV +SEK ER+VY LLRTRD+
Sbjct: 641 KADTLREAAFGYQDLKKLESEVSNYKDDPRLPCDIALKKMVTISEKTERSVYNLLRTRDA 700
Query: 658 LMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVL 717
MR CKEF IP +WMLDN +IGKIK SVKLAK YMKRVA+E+Q +KDPA++YM+L
Sbjct: 701 TMRQCKEFNIPTDWMLDNNLIGKIKFSSVKLAKMYMKRVAMELQYMGPLNKDPALEYMLL 760
Query: 718 QGVRFAFRIHQ 728
Q VRFAFR+HQ
Sbjct: 761 QAVRFAFRMHQ 771
>UniRef100_Q9LI74 Similarity to pherophorin [Arabidopsis thaliana]
Length = 1004
Score = 329 bits (844), Expect = 2e-88
Identities = 173/319 (54%), Positives = 220/319 (68%), Gaps = 21/319 (6%)
Query: 431 LEKRALRIPNPPPRP-----SCSISSKTKQECSAQVQPPPPPP-------------PPPP 472
+EKR R+P PPPR S ++ S PPPPPP PPPP
Sbjct: 644 IEKRPPRVPRPPPRSAGGGKSTNLPSARPPLPGGGPPPPPPPPGGGPPPPPGGGPPPPPP 703
Query: 473 PMSFASRGNTA--MVKRAPQVVELYHSLMKRDSRRDSSSGGLSDAP-DVADVRSSMIGEI 529
P RG V RAP++VE Y SLMKR+S+++ + +S + + R++MIGEI
Sbjct: 704 PPGALGRGAGGGNKVHRAPELVEFYQSLMKRESKKEGAPSLISSGTGNSSAARNNMIGEI 763
Query: 530 ENRSSHLLAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVL 589
ENRS+ LLA+KAD+ETQG+FV SL EV + + +I+D++AFV WLD+EL FLVDERAVL
Sbjct: 764 ENRSTFLLAVKADVETQGDFVQSLATEVRASSFTDIEDLLAFVSWLDEELSFLVDERAVL 823
Query: 590 KHFDWPEKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVY 649
KHFDWPE KAD LREAAF YQDL KLE +V+S+ DDP L C+ ALKKM L EK+E++VY
Sbjct: 824 KHFDWPEGKADALREAAFEYQDLMKLEKQVTSFVDDPNLSCEPALKKMYKLLEKVEQSVY 883
Query: 650 TLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKD 709
LLRTRD + KEF IPV+W+ D G++GKIKL SV+LAKKYMKRVA E+ + S DKD
Sbjct: 884 ALLRTRDMAISRYKEFGIPVDWLSDTGVVGKIKLSSVQLAKKYMKRVAYELDSVSGSDKD 943
Query: 710 PAMDYMVLQGVRFAFRIHQ 728
P ++++LQGVRFAFR+HQ
Sbjct: 944 PNREFLLLQGVRFAFRVHQ 962
Score = 120 bits (300), Expect = 2e-25
Identities = 87/256 (33%), Positives = 132/256 (50%), Gaps = 69/256 (26%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVE---CRKNQSEVDELVKKVAL--------------L 171
EL+ ++ L++E +ERE KL EL+E ++ +S++ EL +++ + L
Sbjct: 130 ELERLKQLVKELEEREVKLEGELLEYYGLKEQESDIVELQRQLKIKTVEIDMLNITINSL 189
Query: 172 EEEKSGLSEQL-----------VALSRSCGLERQ-------------------------- 194
+ E+ L E+L VA ++ L+RQ
Sbjct: 190 QAERKKLQEELSQNGIVRKELEVARNKIKELQRQIQLDANQTKGQLLLLKQHVSSLQMKE 249
Query: 195 ---------EEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSE 245
E K + Q+LE++V+EL+R N+EL +KR L+ +L S E++++ N +E
Sbjct: 250 EEAMNKDTEVERKLKAVQDLEVQVMELKRKNRELQHEKRELSIKLDSAEARIATLSNMTE 309
Query: 246 SDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKN----- 300
SD VAK + E + L+ NEDL KQVEGLQ +R +EVEEL YLRWVN+CLR EL+N
Sbjct: 310 SDKVAKVREEVNNLKHNNEDLLKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYQTPA 369
Query: 301 -TCSTLDSDKLSSPQS 315
S D K SP+S
Sbjct: 370 GKISARDLSKNLSPKS 385
>UniRef100_Q8L7S5 AT4g18560/F28J12_220 [Arabidopsis thaliana]
Length = 642
Score = 318 bits (816), Expect = 3e-85
Identities = 232/590 (39%), Positives = 307/590 (51%), Gaps = 77/590 (13%)
Query: 145 AKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQN 204
A N + E R+ +V+EL ++ ALL+ E + ++S LE Q DK+G
Sbjct: 81 ASHNGVVSELRR---QVEELREREALLKTENLEVKLLRESVSVIPLLESQIADKNGEIDE 137
Query: 205 LELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNE 264
L E L N+ L R R M + + E++IV K +S
Sbjct: 138 LRKETARLAEDNERL----RREFDRSEEMRRECETREKEMEAEIVELRKLVSSESDDHAL 193
Query: 265 DLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSI 324
+S++ +GL S L LK S + L P + ++ SI
Sbjct: 194 SVSQRFQGLMDVS------------AKSNLIRSLKRVGSLRN---LPEPITNQENTNKSI 238
Query: 325 SSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSI 384
SS D G I D++ ++NS E ++E S S+
Sbjct: 239 SSSGDADGD--------------IYRKDEIESYSRSSNS-------EELTESS-----SL 272
Query: 385 SGSNSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPR 444
S S V +R + DS E + P P IP PPP
Sbjct: 273 STVRSRVPRVPKPPPKRSISLGDS------TENRADPPP-----------QKSIPPPPPP 315
Query: 445 PSCSISSKTKQECSAQVQPPPPPPPPPPP-MSFASRGNTAMVKRAPQVVELYHSLMKRDS 503
P + + S PPPPPPPPPP +S AS A V+R P+VVE YHSLM+RDS
Sbjct: 316 PPPPLLQQPPPPPSVSKAPPPPPPPPPPKSLSIAS----AKVRRVPEVVEFYHSLMRRDS 371
Query: 504 ---RRDSSSGGLSDAPDVADVRSS--MIGEIENRSSHLLAIKADIETQGEFVNSLIREVN 558
RRDS+ GG + A + ++ MIGEIENRS +LLAIK D+ETQG+F+ LI+EV
Sbjct: 372 TNSRRDSTGGGNAAAEAILANSNARDMIGEIENRSVYLLAIKTDVETQGDFIRFLIKEVG 431
Query: 559 DAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTLREAAFGYQDLKKLESE 618
+A + +I+DVV FVKWLDDEL +LVDERAVLKHF+WPE+KAD LREAAF Y DLKKL SE
Sbjct: 432 NAAFSDIEDVVPFVKWLDDELSYLVDERAVLKHFEWPEQKADALREAAFCYFDLKKLISE 491
Query: 619 VSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWMLDNGII 678
S +++DPR ALKKM AL EK+E VY+L R R+S K FQIPV+WML+ GI
Sbjct: 492 ASRFREDPRQSSSSALKKMQALFEKLEHGVYSLSRMRESAATKFKSFQIPVDWMLETGIT 551
Query: 679 GKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
+IKL SVKLA KYMKRV+ E++ P + +++QGVRFAFR+HQ
Sbjct: 552 SQIKLASVKLAMKYMKRVSAELEAIEG--GGPEEEELIVQGVRFAFRVHQ 599
>UniRef100_O49524 Pherophorin - like protein [Arabidopsis thaliana]
Length = 637
Score = 307 bits (786), Expect = 8e-82
Identities = 218/539 (40%), Positives = 293/539 (53%), Gaps = 78/539 (14%)
Query: 243 SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLN-EVEEL----AYLRWVNSCLRTE 297
+S + +V++ + + LR + + E L+ + N E++EL A L N LR E
Sbjct: 81 ASHNGVVSELRRQVEELR--EREALLKTENLEIADKNGEIDELRKETARLAEDNERLRRE 138
Query: 298 ------LKNTCSTLDSD---KLSSPQSVVSSSGD----SISS-FSDQCGSANSFNLVKKP 343
++ C T + + ++ + +VSS D S+S F + NL++
Sbjct: 139 FDRSEEMRRECETREKEMEAEIVELRKLVSSESDDHALSVSQRFQGLMDVSAKSNLIRSL 198
Query: 344 KKW--------PITSSDQ----LSQVECTNNSIIDKNWIESISEGSNRRR----HSISGS 387
K+ PIT+ + +S + I K+ IES S SN S+S
Sbjct: 199 KRVGSLRNLPEPITNQENTNKSISSSGDADGDIYRKDEIESYSRSSNSEELTESSSLSTV 258
Query: 388 NSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPSC 447
S V +R + DS E + P P IP PPP P
Sbjct: 259 RSRVPRVPKPPPKRSISLGDS------TENRADPPP-----------QKSIPPPPPPPPP 301
Query: 448 SISSKTKQECSAQVQPPPPPPPPPPP-MSFASRGNTAMVKRAPQVVELYHSLMKRDS--- 503
+ + S PPPPPPPPPP +S AS A V+R P+VVE YHSLM+RDS
Sbjct: 302 PLLQQPPPPPSVSKAPPPPPPPPPPKSLSIAS----AKVRRVPEVVEFYHSLMRRDSTNS 357
Query: 504 RRDSSSGGLSDAPDVADVRSS--MIGEIENRSSHLLAIKADIETQGEFVNSLIREVNDAV 561
RRDS+ GG + A + ++ MIGEIENRS +LLAIK D+ETQG+F+ LI+EV +A
Sbjct: 358 RRDSTGGGNAAAEAILANSNARDMIGEIENRSVYLLAIKTDVETQGDFIRFLIKEVGNAA 417
Query: 562 YENIDDVVAFVKWLDDELGFL------------VDERAVLKHFDWPEKKADTLREAAFGY 609
+ +I+DVV FVKWLDDEL +L VDERAVLKHF+WPE+KAD LREAAF Y
Sbjct: 418 FSDIEDVVPFVKWLDDELSYLFKVCKFVVVSLKVDERAVLKHFEWPEQKADALREAAFCY 477
Query: 610 QDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPV 669
DLKKL SE S +++DPR ALKKM AL EK+E VY+L R R+S K FQIPV
Sbjct: 478 FDLKKLISEASRFREDPRQSSSSALKKMQALFEKLEHGVYSLSRMRESAATKFKSFQIPV 537
Query: 670 EWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
+WML+ GI +IKL SVKLA KYMKRV+ E++ P + +++QGVRFAFR+HQ
Sbjct: 538 DWMLETGITSQIKLASVKLAMKYMKRVSAELEAIEG--GGPEEEELIVQGVRFAFRVHQ 594
>UniRef100_Q6F359 Hypothetical protein OJ1268_B08.2 [Oryza sativa]
Length = 694
Score = 293 bits (750), Expect = 1e-77
Identities = 168/341 (49%), Positives = 202/341 (58%), Gaps = 54/341 (15%)
Query: 439 PNPPPRP-SCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAM------------V 485
P PPP P S S S S PP PPPPPPP SR +T V
Sbjct: 314 PPPPPMPRSRSASPSPSTSSSGSAGPPAPPPPPPPAAKRTSRTSTPATTSSSAPASGPCV 373
Query: 486 KRAPQVVELYHSLMKRDSRRDSSSGG--LSDAPDV-ADVRSSMIGEIENRSSHLLAIKAD 542
+R P+VVE YHSLM+RDS+RD GG P A MIGEIENRS+HLLAIK+D
Sbjct: 374 RRVPEVVEFYHSLMRRDSKRDGGGGGGGAEACPGGGAAAARDMIGEIENRSAHLLAIKSD 433
Query: 543 IETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTL 602
+E QG+F+ LI+EV A + +I+DVV FVKWLD EL LVDERAVLKHF+WPE+KAD L
Sbjct: 434 VERQGDFIRFLIKEVEGAAFVDIEDVVTFVKWLDVELSRLVDERAVLKHFEWPEQKADAL 493
Query: 603 REAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNC 662
REAAFGY+DLKK+E E SS+ DDPR PC ALKKM AL EK+E VY+L R RD M
Sbjct: 494 REAAFGYRDLKKIEEEASSFCDDPRQPCSSALKKMQALFEKLEHGVYSLARVRDGAMNRY 553
Query: 663 KEFQIPVEWMLDNGIIGK-----------------------------------IKLGSVK 687
+ + IP EWM D GI+ + IKL SVK
Sbjct: 554 RGYHIPWEWMQDTGIVSQNTHKDAKNINTYKTYTRDNNKKNLGCNVHVCQGALIKLQSVK 613
Query: 688 LAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
LA KY++RV+ E++ A P + ++LQGVRFAFR+HQ
Sbjct: 614 LAMKYLRRVSSELE---AIKDGPDEEELMLQGVRFAFRVHQ 651
>UniRef100_Q9SX62 F11A17.16 [Arabidopsis thaliana]
Length = 558
Score = 289 bits (739), Expect = 2e-76
Identities = 159/354 (44%), Positives = 226/354 (62%), Gaps = 14/354 (3%)
Query: 389 SSEEEVSVLS------KRRQSNCFDSFECL--KEIEKESVPMPLFVQQCALEKRA---LR 437
S+E ++S LS K Q++ F + L ++E+ V + V+ L + R
Sbjct: 169 SAEAKISSLSSNDKPAKEHQNSRFKDIQRLIASKLEQPKVKKEVAVESSRLSPPSPSPSR 228
Query: 438 IPNPPPRPSCSIS---SKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVEL 494
+P PP P +S S K++ ++ PP PPPPPPP A +++P V +L
Sbjct: 229 LPPTPPLPKFLVSPASSLGKRDENSSPFAPPTPPPPPPPPPPRPLAKAARAQKSPPVSQL 288
Query: 495 YHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLI 554
+ L K+D+ R+ S + V +S++GEI+NRS+HL+AIKADIET+GEF+N LI
Sbjct: 289 FQLLNKQDNSRNLSQSVNGNKSQVNSAHNSIVGEIQNRSAHLIAIKADIETKGEFINDLI 348
Query: 555 REVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTLREAAFGYQDLKK 614
++V + +++DV+ FV WLD EL L DERAVLKHF WPEKKADTL+EAA Y++LKK
Sbjct: 349 QKVLTTCFSDMEDVMKFVDWLDKELATLADERAVLKHFKWPEKKADTLQEAAVEYRELKK 408
Query: 615 LESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWMLD 674
LE E+SSY DDP + +ALKKM L +K E+ + L+R R S MR+ ++F+IPVEWMLD
Sbjct: 409 LEKELSSYSDDPNIHYGVALKKMANLLDKSEQRIRRLVRLRGSSMRSYQDFKIPVEWMLD 468
Query: 675 NGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
+G+I KIK S+KLAK YM RVA E+Q+ D++ + ++LQGVRFA+R HQ
Sbjct: 469 SGMICKIKRASIKLAKTYMNRVANELQSARNLDRESTKEALLLQGVRFAYRTHQ 522
>UniRef100_Q5VPC4 Pherophorin-like protein [Oryza sativa]
Length = 591
Score = 248 bits (633), Expect = 5e-64
Identities = 190/547 (34%), Positives = 264/547 (47%), Gaps = 87/547 (15%)
Query: 177 GLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQ 236
G + + V+ +RS G+ EV +L R ++L Q+R R+ +E +
Sbjct: 45 GSASKAVSFARSLGVHFPRSSAQVQPARAPPEVADLLRAIEQL--QERESRLRVELLEQK 102
Query: 237 LSC----------SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAY 286
+ ++ +++S + K K A+ L N L +++ + + + +
Sbjct: 103 ILKETVAIVPFLEAELAAKSSELEKCKDTAARLESENMRLCAELDAAVLEVTSRKQRIVH 162
Query: 287 LRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKK-PKK 345
+ + L+ + + + D+D SS SV +S SS +AN +LV++ P
Sbjct: 163 MEKEMAELKKQQE--AAAADADDCSSTASVSHEQPESASS------AANPASLVQRGPPI 214
Query: 346 WPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNC 405
P + + + S + + S S S S S + S +V + R
Sbjct: 215 PPPPPPVPPAAFKSKSYSASSRVSLPSTSAPSPSSSTSTSPTYSCSSSDTVTTPRN---- 270
Query: 406 FDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPP 465
++ E +P IP PPP P+ S+ C PPP
Sbjct: 271 -------RKPELSKLPP---------------IPPPPPMPALSV-------CGRAAAPPP 301
Query: 466 PPPPPPPPMSFASRGNTAM---VKRAPQVVELYHSLMKRDSR-RDSSSGG-LSDAPDVAD 520
PPPPPP + + A V R P+VVE YHSLM+RDSR RD S GG ++ VA
Sbjct: 302 PPPPPPARRTSGAASPAASGPRVTRVPEVVEFYHSLMRRDSRSRDGSGGGETANGGGVAA 361
Query: 521 VRSSMIGEIENRSSHLLA--------------------------IKADIETQGEFVNSLI 554
R MIGEIENRS+HLLA IK+D+E QG+F+ LI
Sbjct: 362 TRD-MIGEIENRSAHLLAAIIYLSAGREFGGGADRNSCMRGVRRIKSDVERQGDFIRFLI 420
Query: 555 REVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTLREAAFGYQDLKK 614
+EV A + +I+DVV FVKWLD+EL LVDERAVLKHF+WPE K D LREAAFGY DLKK
Sbjct: 421 KEVEGAAFVDIEDVVTFVKWLDNELSRLVDERAVLKHFEWPENKEDALREAAFGYCDLKK 480
Query: 615 LESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWML- 673
LE E SS++DD R PC ALKKM AL EK+E VY L R RD FQIP EWM
Sbjct: 481 LEVEASSFRDDARQPCSTALKKMQALFEKLEHGVYNLARFRDGATGRYSRFQIPCEWMQP 540
Query: 674 DNGIIGK 680
D GI+ +
Sbjct: 541 DTGIVSQ 547
>UniRef100_Q9LMK4 F10K1.18 protein [Arabidopsis thaliana]
Length = 392
Score = 223 bits (567), Expect = 2e-56
Identities = 140/355 (39%), Positives = 207/355 (57%), Gaps = 34/355 (9%)
Query: 379 RRRHSISGSNSSEEE-VSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALR 437
R R +S S E E S+L K+ QS+ S + ++ P V+ +
Sbjct: 41 RLRAQVSNLKSHENERKSMLWKKLQSSYDGS-----NTDGSNLKAPESVKS---NTKGQE 92
Query: 438 IPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHS 497
+ NP P+P+ S + PPPPPP P + R V+RAP+VVE Y +
Sbjct: 93 VRNPNPKPTIQGQSTATK--------PPPPPPLPSKRTLGKRS----VRRAPEVVEFYRA 140
Query: 498 LMKRDSR---RDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLI 554
L KR+S + + +G LS A +MIGEIENRS +L IK+D + + ++ LI
Sbjct: 141 LTKRESHMGNKINQNGVLSPA-----FNRNMIGEIENRSKYLSDIKSDTDRHRDHIHILI 195
Query: 555 REVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFD-WPEKKADTLREAAFGYQDLK 613
+V A + +I +V FVKW+D+EL LVDERAVLKHF WPE+K D+LREAA Y+ K
Sbjct: 196 SKVEAATFTDISEVETFVKWIDEELSSLVDERAVLKHFPKWPERKVDSLREAACNYKRPK 255
Query: 614 KLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWML 673
L +E+ S+KD+P+ AL+++ +L +++E +V + RDS + K+FQIP EWML
Sbjct: 256 NLGNEILSFKDNPKDSLTQALQRIQSLQDRLEESVNNTEKMRDSTGKRYKDFQIPWEWML 315
Query: 674 DNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
D G+IG++K S++LA++YMKR+A E+++ + + M LQGVRFA+ IHQ
Sbjct: 316 DTGLIGQLKYSSLRLAQEYMKRIAKELESNGSGKEGNLM----LQGVRFAYTIHQ 366
>UniRef100_Q93ZL7 At1g48280/F11A17_25 [Arabidopsis thaliana]
Length = 200
Score = 192 bits (489), Expect = 2e-47
Identities = 95/164 (57%), Positives = 123/164 (74%)
Query: 565 IDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTLREAAFGYQDLKKLESEVSSYKD 624
++DV+ FV WLD EL L DERAVLKHF WPEKKADTL+EAA Y++LKKLE E+SSY D
Sbjct: 1 MEDVMKFVDWLDKELATLADERAVLKHFKWPEKKADTLQEAAVEYRELKKLEKELSSYSD 60
Query: 625 DPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKIKLG 684
DP + +ALKKM L +K E+ + L+R R S MR+ ++F+IPVEWMLD+G+I KIK
Sbjct: 61 DPNIHYGVALKKMANLLDKSEQRIRRLVRLRGSSMRSYQDFKIPVEWMLDSGMICKIKRA 120
Query: 685 SVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
S+KLAK YM RVA E+Q+ D++ + ++LQGVRFA+R HQ
Sbjct: 121 SIKLAKTYMNRVANELQSARNLDRESTKEALLLQGVRFAYRTHQ 164
>UniRef100_Q60EZ9 Hypothetical protein OJ1111_A10.14 [Oryza sativa]
Length = 617
Score = 83.2 bits (204), Expect = 3e-14
Identities = 78/309 (25%), Positives = 146/309 (47%), Gaps = 22/309 (7%)
Query: 126 LFTELDHMRSLLQESKEREAKLNAELVECR----KNQSEVDELVKKVALLEEEKSGLSEQ 181
L +E + +++ L ES + ++L A ++C+ K + + ++ +++A L+E S Q
Sbjct: 216 LQSENERLQAQLTESSKLASELEAARMKCKLLKKKLRQDAEQAKERIASLQEMAD--SWQ 273
Query: 182 LVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSME-SQLSCS 240
+ + E+K + LE E ELR +N L + +L RL +
Sbjct: 274 CKEIITEGKFSAEVEEKLSKLEELENEARELRVVNSRLQQENAHLARRLELTRLPPVPKP 333
Query: 241 DNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKN 300
N+ E K EA LR N+ L+K+VE L+T R ++VEEL YL+W+N+CLR EL+N
Sbjct: 334 INNME----VKALQEADHLRQENDKLAKEVEQLKTDRFSDVEELVYLKWINACLRYELRN 389
Query: 301 TCS------TLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQL 354
+ D K SPQS + I +++ +F+ ++ ++ + + L
Sbjct: 390 QDAPSGKNVARDLSKTLSPQS-EEKAKQLIMEYANAGPDEKNFDHIEFCSEYSSSRASSL 448
Query: 355 SQVE--CTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQ-SNC-FDSFE 410
+ + + S+++K+ + ++ R + G + + L +R S+C FD F
Sbjct: 449 GEPDDASIDVSLMNKHKNPKKKKFFSKLRKLVLGKEKENKTIPTLERRISISSCSFDEFN 508
Query: 411 CLKEIEKES 419
+ I+ S
Sbjct: 509 GRESIDSYS 517
>UniRef100_Q9S9T9 T32N4.10 protein [Arabidopsis thaliana]
Length = 681
Score = 80.9 bits (198), Expect = 1e-13
Identities = 62/256 (24%), Positives = 122/256 (47%), Gaps = 23/256 (8%)
Query: 437 RIPNPPPRPSCSISSKTKQEC--------SAQVQPPPPPPPPPPPMSFASRGNTAMVKRA 488
+ P PPP P S +S++ + C + P P PP PP + + T+ ++R+
Sbjct: 335 KAPAPPPPPPMSKASESGEFCQFSKTHSTNGDNAPSMPAPPAPPGSGRSLKKATSKLRRS 394
Query: 489 PQVVELYHSLMKR------DSRRDSSSGGLSDAPDVADV---RSSM---IGEIENRSSHL 536
Q+ LY +L + + + +S G + + + V RS M + E+ RSS+
Sbjct: 395 AQIANLYWALKGKLEGRGVEGKTKKASKGQNSVAEKSPVKVARSGMADALAEMTKRSSYF 454
Query: 537 LAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFD-WP 595
I+ D++ + + L ++ +++ +++ F ++ L L DE VL F+ +P
Sbjct: 455 QQIEEDVQKYAKSIEELKSSIHSFQTKDMKELLEFHSKVESILEKLTDETQVLARFEGFP 514
Query: 596 EKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTR 655
EKK + +R A Y+ L + E+ ++K +P P + L K+ K + + T+ RT+
Sbjct: 515 EKKLEVIRTAGALYKKLDGILVELKNWKIEP--PLNDLLDKIERYFNKFKGEIETVERTK 572
Query: 656 DSLMRNCKEFQIPVEW 671
D + K + I +++
Sbjct: 573 DEDAKMFKRYNINIDF 588
>UniRef100_Q6V8P9 Putative actin-binding protein [Malus domestica]
Length = 107
Score = 79.3 bits (194), Expect = 4e-13
Identities = 35/61 (57%), Positives = 50/61 (81%)
Query: 668 PVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIH 727
PV+W+LD+G++ KIKL SV+LA+KYMKRVA E+ S +K+P ++++LQGVR AFR+H
Sbjct: 16 PVDWLLDSGVVRKIKLSSVQLARKYMKRVASELDALSGPEKEPNREFILLQGVRSAFRVH 75
Query: 728 Q 728
Q
Sbjct: 76 Q 76
>UniRef100_Q9SAU9 F5F19.14 protein [Arabidopsis thaliana]
Length = 573
Score = 72.4 bits (176), Expect = 5e-11
Identities = 53/171 (30%), Positives = 91/171 (52%), Gaps = 18/171 (10%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
ELD +S +Q K+ KLN + +++ +++ L ++VA L+EE+ + L L
Sbjct: 219 ELDMAKSQVQVLKK---KLN---INTQQHVAQILSLKQRVARLQEEE--IKAVLPDLEAD 270
Query: 189 CGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDI 248
++R ++LE E+ EL N L + L+ +L S+ Q+ + E +
Sbjct: 271 KMMQR--------LRDLESEINELTDTNTRLQFENFELSEKLESV--QIIANSKLEEPEE 320
Query: 249 VAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
+ + + + LR NE+L K VE LQ R ++E+L YLRW+N+CLR EL+
Sbjct: 321 IETLREDCNRLRSENEELKKDVEQLQGDRCTDLEQLVYLRWINACLRYELR 371
>UniRef100_Q96258 AR791 [Arabidopsis thaliana]
Length = 573
Score = 72.4 bits (176), Expect = 5e-11
Identities = 53/171 (30%), Positives = 91/171 (52%), Gaps = 18/171 (10%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
ELD +S +Q K+ KLN + +++ +++ L ++VA L+EE+ + L L
Sbjct: 219 ELDMAKSQVQVLKK---KLN---INTQQHVAQILSLKQRVARLQEEE--IKPVLPDLEAD 270
Query: 189 CGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDI 248
++R ++LE E+ EL N L + L+ +L S+ Q+ + E +
Sbjct: 271 KMMQR--------LRDLESEINELTDTNTRLQFENFELSEKLESV--QIIANSKLEEPEE 320
Query: 249 VAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
+ + + + LR NE+L K VE LQ R ++E+L YLRW+N+CLR EL+
Sbjct: 321 IETLREDCNRLRSENEELKKDVEQLQGDRCTDLEQLVYLRWINACLRYELR 371
>UniRef100_Q9C946 Hypothetical protein T7P1.21 [Arabidopsis thaliana]
Length = 907
Score = 69.7 bits (169), Expect = 3e-10
Identities = 68/253 (26%), Positives = 108/253 (41%), Gaps = 41/253 (16%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPP-----------PPPPPPPMSFASRGNTAMVKR 487
P PPP P + +A PPPP PPPP S + +KR
Sbjct: 605 PPPPPPPMAMANG------AAGPPPPPPRMGMANGAAGPPPPPGAARSLRPKKAATKLKR 658
Query: 488 APQVVELYHSLMKRDSRRD------SSSG---GLSDAPDVADVRSSM---IGEIENRSSH 535
+ Q+ LY L + RD S SG G AP A + M + EI +S++
Sbjct: 659 STQLGNLYRILKGKVEGRDPNAKTGSGSGRKAGAGSAP--AGGKQGMADALAEITKKSAY 716
Query: 536 LLAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFD-W 594
L I+ADI +N L E+ +++ ++++F + ++ L L DE VL + +
Sbjct: 717 FLQIQADIAKYMTSINELKIEITKFQTKDMTELLSFHRRVESVLENLTDESQVLARCEGF 776
Query: 595 PEKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRT 654
P+KK + +R A Y L + +E+ + K +P L L +K+ER + T
Sbjct: 777 PQKKLEAMRMAVALYTKLHGMITELQNMKIEPPLN---------QLLDKVERYFTKIKET 827
Query: 655 RDSLMRNCKEFQI 667
+ NC E +
Sbjct: 828 MVDISSNCMELAL 840
Score = 45.8 bits (107), Expect = 0.005
Identities = 48/201 (23%), Positives = 80/201 (38%), Gaps = 25/201 (12%)
Query: 307 SDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIID 366
S ++ SP S S S S + CGS + V P + +V NS
Sbjct: 249 SSRVLSPSESFSDSKSSFGSRNSFCGSPTTPRSVL-----PESMMGSPGRVGDFANSASH 303
Query: 367 KNW------IESISEGSNRRR--HSIS-------GSNSSEEEVSVLSKRRQSNCFDSFEC 411
W +E +S +R H +S ++ E+ +SV+ + +Q D E
Sbjct: 304 LLWNMRVQALEKLSPIDVKRLAIHILSQKEAQEPNESNDEDVISVVEEIKQKK--DEIES 361
Query: 412 LKEIEKESVPMPLFVQQCAL---EKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPP 468
+ + + L + L + ++I + ++ K E S+Q+ PPPPPP
Sbjct: 362 IDVKMETEESVNLDEESVVLNGEQDTIMKISSLESTSESKLNHSEKYENSSQLFPPPPPP 421
Query: 469 PPPPPMSFASRGNTAMVKRAP 489
PPPPP+SF + + P
Sbjct: 422 PPPPPLSFIKTASLPLPSPPP 442
Score = 42.4 bits (98), Expect = 0.050
Identities = 41/147 (27%), Positives = 65/147 (43%), Gaps = 23/147 (15%)
Query: 339 LVKKPKKWPITSSDQ--LSQVECTNNSIIDKNWIESIS------EGSNRRRHSI--SGSN 388
L +K + P S+D+ +S VE K+ IESI E N S+ +G
Sbjct: 329 LSQKEAQEPNESNDEDVISVVEEIKQK---KDEIESIDVKMETEESVNLDEESVVLNGEQ 385
Query: 389 SSEEEVSVLSKRRQS--NCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPS 446
+ ++S L +S N + +E ++ P P ++ +L +P+PPP P
Sbjct: 386 DTIMKISSLESTSESKLNHSEKYENSSQLFPPPPPPPPPPPLSFIKTASLPLPSPPPTPP 445
Query: 447 CSISSKTKQECSAQVQPPPPPPPPPPP 473
+ + + PPPPPPPPPPP
Sbjct: 446 IADIAIS--------MPPPPPPPPPPP 464
Score = 35.4 bits (80), Expect = 6.1
Identities = 18/57 (31%), Positives = 24/57 (41%), Gaps = 8/57 (14%)
Query: 433 KRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPP--------PPPMSFASRGN 481
+ A+ P PPP P + + PPPPPPP PPPM + G+
Sbjct: 531 RAAVAPPPPPPPPGTAAAPPPPPPPPGTQAAPPPPPPPPMQNRAPSPPPMPMGNSGS 587
Score = 35.4 bits (80), Expect = 6.1
Identities = 16/39 (41%), Positives = 19/39 (48%)
Query: 434 RALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPP 472
RA P PPP P + ++ Q PPPPPPPP
Sbjct: 531 RAAVAPPPPPPPPGTAAAPPPPPPPPGTQAAPPPPPPPP 569
>UniRef100_UPI000043298F UPI000043298F UniRef100 entry
Length = 999
Score = 61.6 bits (148), Expect = 8e-08
Identities = 121/586 (20%), Positives = 231/586 (38%), Gaps = 84/586 (14%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSE-VDELVK-KVALLEEEKSGLSE 180
+ DL ++D++ L++ K+ +L E+ + + VDE+ K KV + +K G+
Sbjct: 313 NADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALDKCVDEMEKLKVENEKLKKEGMKV 372
Query: 181 QLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC- 239
+ L + L+ + + + + N E+ ++R N +L + L L S ++
Sbjct: 373 EATWLEENVNLKAKNTELEENLANTVNELDKMRSENADLLSELNRLKQELESGRKEIDQL 432
Query: 240 -SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTEL 298
S+ S D + K E L+ N+DL +V+GL++ R E+A L+ S L+ +L
Sbjct: 433 KSEIGSMKDALGKCVDEIEKLKTENKDLKSEVQGLESERDRLTNEVADLKPKISELQEKL 492
Query: 299 KNTCSTLDSDKLSSP---------QSVVSSSGDSISSFSDQCGS-ANSFN-LVKKPKKWP 347
+ LD K + + S+G I + S N N V++ +K
Sbjct: 493 TDASKKLDEAKTEDSDLRAEVDRLKKELESAGKEIDQLKAEMNSLKNGLNKCVEEMEKLT 552
Query: 348 ITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSS-EEEVSVLSKRRQSNCF 406
+S+ SQV + + + ++ NS+ ++E L+K+ N
Sbjct: 553 NENSELKSQV----------HGLRGEGDSLASELTNVKDENSALKDEKDQLNKQLAENKT 602
Query: 407 DSFECLK---EIEKESVPMPLFVQQCALEKRALRIPNPPPRPSC--------SISSKTKQ 455
++ K E+E E+ + ++ C E L+ N + S++ +T +
Sbjct: 603 ENERLKKQNDELESENTKIKKELESCKNENNNLKEENNKLKEELEKLGEQLKSLNDETNK 662
Query: 456 --------ECSAQVQPPPPPPPPPPPMSFASRGNTAMV-------KRAPQVVELYHSLMK 500
E Q+ P + A + + + K Q+ E+ SL K
Sbjct: 663 LRRELKEAEDKIQILEPQLRTTLDAEKTAAEKLKSDLQSCKDENDKLQTQINEMKRSLDK 722
Query: 501 RDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLIREVNDA 560
+ D + ++ + + + +EN+ S+L A K E V L R D
Sbjct: 723 MGTENDRLKREVDESRKKLEDMEAKVKSLENQLSNLSAEKE------ELVKELYRTRED- 775
Query: 561 VYENIDDVVAFVKWLDDELGFLVDERAVLKH--FDWPEKKADTLREAAFGYQDLKKLESE 618
+ L +EL DE A LK+ + E+ A+ +E A +L +E+
Sbjct: 776 -----------LNNLRNELEKQTDESANLKNDIENLNERNAELSKELAVAKDNLMGMETR 824
Query: 619 VSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKE 664
+S+ K + D K++ L + ++ D L R KE
Sbjct: 825 LSNLKKEN----DDMKNKIITLEDSIQEV--------DDLKRQLKE 858
Score = 45.8 bits (107), Expect = 0.005
Identities = 61/291 (20%), Positives = 115/291 (38%), Gaps = 36/291 (12%)
Query: 2 KEEIHNNPSENKTKVSKFSDQNQPPKLQTTK-------TTNPNNNNHSKPRLWGAHIVKG 54
K++++ +ENKT+ + QN + + TK N NNN + + K
Sbjct: 590 KDQLNKQLAENKTENERLKKQNDELESENTKIKKELESCKNENNNLKEENNKLKEELEK- 648
Query: 55 FSADKKTKQLPTKKQQNTTTITTSDVVTNQKNVNPFVPPHSRVKRSLMGDL-SCSQVHPH 113
K+ T K + + + + + L DL SC
Sbjct: 649 LGEQLKSLNDETNKLRRELKEAEDKIQILEPQLRTTLDAEKTAAEKLKSDLQSC------ 702
Query: 114 AFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEE 173
+ + L T+++ M+ L + +L E+ E RK +++++ KV LE
Sbjct: 703 ------KDENDKLQTQINEMKRSLDKMGTENDRLKREVDESRK---KLEDMEAKVKSLEN 753
Query: 174 EKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSM 233
+ S LS + L + L R ED + LE + E L ++ NL R + +
Sbjct: 754 QLSNLSAEKEELVKE--LYRTREDLNNLRNELEKQTDESANLKNDI----ENLNERNAEL 807
Query: 234 ESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEEL 284
+L+ + D + + S L+ N+D+ ++ L+ S + EV++L
Sbjct: 808 SKELAVA-----KDNLMGMETRLSNLKKENDDMKNKIITLEDS-IQEVDDL 852
Score = 42.4 bits (98), Expect = 0.050
Identities = 44/193 (22%), Positives = 82/193 (41%), Gaps = 23/193 (11%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSR 187
TE +++++ L ++ E+ KL E E ++N D + + L+E L + L R
Sbjct: 185 TENENLKTELDKADEQLDKLKTERNELQRN---FDTMKLENETLKENVKALKDDLEESKR 241
Query: 188 SC--------GLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
L+ +EE KD + L+ + L+ N EL + +L R S +E +L
Sbjct: 242 EVDEMKAVGDALKDKEELKDAEFRELQQNMQNLKTENGELKKENDDLRTRSSELEHKL-- 299
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
DN K E + N DL +++ L+ + +E+ L+ S L+ L
Sbjct: 300 -DN---------VKKELDKVESENADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALD 349
Query: 300 NTCSTLDSDKLSS 312
++ K+ +
Sbjct: 350 KCVDEMEKLKVEN 362
Score = 41.6 bits (96), Expect = 0.085
Identities = 46/182 (25%), Positives = 82/182 (44%), Gaps = 15/182 (8%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
E D +R LL+ESK+ + + + + E D+L+ + LE+ L+E ++ R+
Sbjct: 85 EKDKLRDLLEESKKLKEDNENLWAQLERLRGENDDLMGQKKALEDLNKQLNEDNESMKRT 144
Query: 189 CG-LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESD 247
G LE + + N+E E L N+ + KR L L+ E+ + D + E
Sbjct: 145 MGNLEARIDSLSNELSNVERERDALLDENESV---KRELERTLTENENLKTELDKADEQ- 200
Query: 248 IVAKFKAEAS-------LLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCL--RTEL 298
+ K K E + ++L NE L + V+ L+ E+ ++ V L + EL
Sbjct: 201 -LDKLKTERNELQRNFDTMKLENETLKENVKALKDDLEESKREVDEMKAVGDALKDKEEL 259
Query: 299 KN 300
K+
Sbjct: 260 KD 261
Score = 39.3 bits (90), Expect = 0.42
Identities = 67/323 (20%), Positives = 125/323 (37%), Gaps = 45/323 (13%)
Query: 58 DKKTKQLPTKKQQNTTTITTSDVVTNQKNVNPFVPPHSRVKRSLMGDLSCSQVHPHAFPT 117
D+ KQL K +N +D + ++ ++++K+ L SC +
Sbjct: 591 DQLNKQLAENKTENERLKKQNDELESE---------NTKIKKELE---SCKNEN-----N 633
Query: 118 HRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSG 177
+ ++ + L EL+ + L+ + KL EL E ++ ++ L+ EK+
Sbjct: 634 NLKEENNKLKEELEKLGEQLKSLNDETNKLRRELKEAEDKIQILEPQLRTT--LDAEKTA 691
Query: 178 LSEQLVALSRSCG-----LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSS 232
+E+L + +SC L+ Q + S + E L+R E + ++ ++ S
Sbjct: 692 -AEKLKSDLQSCKDENDKLQTQINEMKRSLDKMGTENDRLKREVDESRKKLEDMEAKVKS 750
Query: 233 MESQLSCSDNSSESDIVAKFKAEASLLRLTNE---------DLSKQVEGLQTSRLNEVEE 283
+E+QLS E + ++ L L NE +L +E L +E
Sbjct: 751 LENQLSNLSAEKEELVKELYRTREDLNNLRNELEKQTDESANLKNDIENLNERNAELSKE 810
Query: 284 LAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKP 343
LA + + T L N D K + + + DSI D +K+
Sbjct: 811 LAVAKDNLMGMETRLSNLKKENDDMK-----NKIITLEDSIQEVDD------LKRQLKEA 859
Query: 344 KKWPITSSDQLSQVECTNNSIID 366
KK S +L ++ TN + D
Sbjct: 860 KKELDKPSPELDTLKSTNKKLQD 882
>UniRef100_UPI000043298E UPI000043298E UniRef100 entry
Length = 1060
Score = 60.1 bits (144), Expect = 2e-07
Identities = 124/623 (19%), Positives = 232/623 (36%), Gaps = 116/623 (18%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSE-VDELVK-KVALLEEEKSGLSE 180
+ DL ++D++ L++ K+ +L E+ + + VDE+ K KV + +K G+
Sbjct: 383 NADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALDKCVDEMEKLKVENEKLKKEGMKV 442
Query: 181 QLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC- 239
+ L + L+ + + + + N E+ ++R N +L + L L S ++
Sbjct: 443 EATWLEENVNLKAKNTELEENLANTVNELDKMRSENADLLSELNRLKQELESGRKEIDQL 502
Query: 240 -SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTEL 298
S+ S D + K E L+ N+DL +V+GL++ R E+A L+ S L+ +L
Sbjct: 503 KSEIGSMKDALGKCVDEIEKLKTENKDLKSEVQGLESERDRLTNEVADLKPKISELQEKL 562
Query: 299 KNTCSTLDSDKLSSP---------QSVVSSSGDSISSFSDQCGS-ANSFN-LVKKPKKWP 347
+ LD K + + S+G I + S N N V++ +K
Sbjct: 563 TDASKKLDEAKTEDSDLRAEVDRLKKELESAGKEIDQLKAEMNSLKNGLNKCVEEMEKLT 622
Query: 348 ITSSDQLSQV------------ECTN----NSII--DKNWIESISEGSNRRRHSISGSNS 389
+S+ SQV E TN NS + +K+ + + + N
Sbjct: 623 NENSELKSQVHGLRGEGDSLASELTNVKDENSALKDEKDQLNKQLAENKTENERLKKQND 682
Query: 390 SEEEVSVLSKRRQSNCFDSFECLK--------EIEKESVPMPLFVQQCALEKRALRIPNP 441
E + K+ +C + LK E+EK + + +R LR
Sbjct: 683 ELESENTKIKKELESCKNENNNLKEENNKLKEELEKLGEQLKSLNDETNKLRRELRTTLD 742
Query: 442 PPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSLMKR 501
+ + Q C + K Q+ E+ SL K
Sbjct: 743 AEKTAAEKLKSDLQSCKDEND-----------------------KLQTQINEMKRSLDKM 779
Query: 502 DSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQ----GEFVNSL---- 553
+ D + ++ + + + +EN+ S+L A K ++ + E +N+L
Sbjct: 780 GTENDRLKREVDESRKKLEDMEAKVKSLENQLSNLSAEKEELVKELYRTREDLNNLRNEL 839
Query: 554 ------------------------------IREVNDAVYENIDDVVAFVKWLDDELGFLV 583
R+ ND + D + A + L+ +L L
Sbjct: 840 EKQTGVKDTMEKESTNLKEELKALKEELNKTRDENDRLKNENDKLNAEIARLNKQLDALK 899
Query: 584 DERAVLKH--FDWPEKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALS 641
DE A LK+ + E+ A+ +E A +L +E+ +S+ K + D K++ L
Sbjct: 900 DESANLKNDIENLNERNAELSKELAVAKDNLMGMETRLSNLKKEN----DDMKNKIITLE 955
Query: 642 EKMERTVYTLLRTRDSLMRNCKE 664
+ ++ D L R KE
Sbjct: 956 DSIQEV--------DDLKRQLKE 970
Score = 44.7 bits (104), Expect = 0.010
Identities = 67/310 (21%), Positives = 129/310 (41%), Gaps = 32/310 (10%)
Query: 2 KEEIHNNPSENKTKVSKFSDQNQPPKLQTTKTT-------NPNNNNHSKPRLWGAHIVK- 53
K++++ +ENKT+ + QN + + TK N NNN + + K
Sbjct: 660 KDQLNKQLAENKTENERLKKQNDELESENTKIKKELESCKNENNNLKEENNKLKEELEKL 719
Query: 54 -------GFSADKKTKQLPTKKQQNTTTITT--SDVVTNQKNVNPFVPPHSRVKRSLMGD 104
+K ++L T T SD+ + + + + +KRSL
Sbjct: 720 GEQLKSLNDETNKLRRELRTTLDAEKTAAEKLKSDLQSCKDENDKLQTQINEMKRSL-DK 778
Query: 105 LSCSQVHPHAFPTHRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEV-DE 163
+ R+ D+ ++ + + L + +L EL R++ + + +E
Sbjct: 779 MGTENDRLKREVDESRKKLEDMEAKVKSLENQLSNLSAEKEELVKELYRTREDLNNLRNE 838
Query: 164 LVKKVAL---LEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELH 220
L K+ + +E+E + L E+L AL R E D+ +N +L E+ RLNK+L
Sbjct: 839 LEKQTGVKDTMEKESTNLKEELKALKEELNKTRDENDR-LKNENDKLN-AEIARLNKQLD 896
Query: 221 MQK------RNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQ 274
K +N L+ ++LS + + D + + S L+ N+D+ ++ L+
Sbjct: 897 ALKDESANLKNDIENLNERNAELS-KELAVAKDNLMGMETRLSNLKKENDDMKNKIITLE 955
Query: 275 TSRLNEVEEL 284
S + EV++L
Sbjct: 956 DS-IQEVDDL 964
Score = 42.4 bits (98), Expect = 0.050
Identities = 44/193 (22%), Positives = 82/193 (41%), Gaps = 23/193 (11%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSR 187
TE +++++ L ++ E+ KL E E ++N D + + L+E L + L R
Sbjct: 255 TENENLKTELDKADEQLDKLKTERNELQRN---FDTMKLENETLKENVKALKDDLEESKR 311
Query: 188 SC--------GLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
L+ +EE KD + L+ + L+ N EL + +L R S +E +L
Sbjct: 312 EVDEMKAVGDALKDKEELKDAEFRELQQNMQNLKTENGELKKENDDLRTRSSELEHKL-- 369
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
DN K E + N DL +++ L+ + +E+ L+ S L+ L
Sbjct: 370 -DN---------VKKELDKVESENADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALD 419
Query: 300 NTCSTLDSDKLSS 312
++ K+ +
Sbjct: 420 KCVDEMEKLKVEN 432
Score = 41.6 bits (96), Expect = 0.085
Identities = 46/182 (25%), Positives = 82/182 (44%), Gaps = 15/182 (8%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
E D +R LL+ESK+ + + + + E D+L+ + LE+ L+E ++ R+
Sbjct: 155 EKDKLRDLLEESKKLKEDNENLWAQLERLRGENDDLMGQKKALEDLNKQLNEDNESMKRT 214
Query: 189 CG-LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESD 247
G LE + + N+E E L N+ + KR L L+ E+ + D + E
Sbjct: 215 MGNLEARIDSLSNELSNVERERDALLDENESV---KRELERTLTENENLKTELDKADEQ- 270
Query: 248 IVAKFKAEAS-------LLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCL--RTEL 298
+ K K E + ++L NE L + V+ L+ E+ ++ V L + EL
Sbjct: 271 -LDKLKTERNELQRNFDTMKLENETLKENVKALKDDLEESKREVDEMKAVGDALKDKEEL 329
Query: 299 KN 300
K+
Sbjct: 330 KD 331
Score = 37.7 bits (86), Expect = 1.2
Identities = 66/328 (20%), Positives = 133/328 (40%), Gaps = 56/328 (17%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSR 187
TE + ++ E + K+ EL C+ + + E K L+EE L EQL +L+
Sbjct: 672 TENERLKKQNDELESENTKIKKELESCKNENNNLKEENNK---LKEELEKLGEQLKSLND 728
Query: 188 SCGLERQE-------------------EDKDGSTQNLELEVVELR-----------RLNK 217
R+E + L+ ++ E++ RL +
Sbjct: 729 ETNKLRRELRTTLDAEKTAAEKLKSDLQSCKDENDKLQTQINEMKRSLDKMGTENDRLKR 788
Query: 218 ELHMQKRNL---TCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQV---E 271
E+ ++ L ++ S+E+QLS E + ++ L L NE L KQ +
Sbjct: 789 EVDESRKKLEDMEAKVKSLENQLSNLSAEKEELVKELYRTREDLNNLRNE-LEKQTGVKD 847
Query: 272 GLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQC 331
++ N EEL L+ +T +N ++DKL++ + ++ D++
Sbjct: 848 TMEKESTNLKEELKALK--EELNKTRDENDRLKNENDKLNAEIARLNKQLDALKD----- 900
Query: 332 GSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSE 391
SAN N ++ + S +L+ + +N + + + ++ + ++ ++ I S
Sbjct: 901 ESANLKNDIENLNERNAELSKELAVAK--DNLMGMETRLSNLKKENDDMKNKIITLEDSI 958
Query: 392 EEVSVLSKRRQSNCFDSFECLKEIEKES 419
+EV L ++ + E KE++K S
Sbjct: 959 QEVDDLKRQLK-------EAKKELDKPS 979
Score = 37.0 bits (84), Expect = 2.1
Identities = 48/193 (24%), Positives = 90/193 (45%), Gaps = 17/193 (8%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLS 179
+ +DL E+D ++ L+ + + +L AE+ + +++ V+++ L E S L
Sbjct: 573 KTEDSDLRAEVDRLKKELESAGKEIDQLKAEMNSLKNG---LNKCVEEMEKLTNENSELK 629
Query: 180 EQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
Q+ L R G E + +N L+ E +LNK+L + + RL +L
Sbjct: 630 SQVHGL-RGEGDSLASELTNVKDENSALK-DEKDQLNKQL-AENKTENERLKKQNDELE- 685
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
S+N+ + K E + L+ N L +++E L E+L L + LR EL+
Sbjct: 686 SENTKIKKELESCKNENNNLKEENNKLKEELEKLG-------EQLKSLNDETNKLRRELR 738
Query: 300 NTCSTLDSDKLSS 312
+TLD++K ++
Sbjct: 739 ---TTLDAEKTAA 748
>UniRef100_Q6CGN9 Similarity [Yarrowia lipolytica]
Length = 1411
Score = 58.9 bits (141), Expect = 5e-07
Identities = 82/423 (19%), Positives = 168/423 (39%), Gaps = 32/423 (7%)
Query: 57 ADKKTKQLPTKKQQNTTTITTSDVVT-NQKNVNPFVPPHSRVKRSLMGDLSCSQVHPHAF 115
A K+ +QL ++++ + +IT S T N + P PP R S + + + + A
Sbjct: 976 ARKQRQQLLEQEKRISDSITASFATTENSPAIAPTAPP-PRPHFSQLDERTLERNRERAE 1034
Query: 116 PTHRRQSSTDLFTELDH-MRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEE 174
H + + + ++ + +++ +A+ ++L + + EL+ E +
Sbjct: 1035 EEHLEREERERREMVQRDVQGRRKAAQKAQARAKSQLADEHAAREAAHELIHARVAAERK 1094
Query: 175 KSGLSE--QLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSS 232
++ + E + L +R+EE + + + LE VVE RR E +++ L
Sbjct: 1095 RATVEEAARKARLQAEAERQREEEAEAEAARQLEQRVVERRRKEVEQQRKEQEAEAELRG 1154
Query: 233 M-ESQLSCSDNSSE-SDIVAKFK----AEASLLRLTNEDLSKQVEGLQTSRLNEVEELAY 286
+ E+ S + SS+ S+ + K A+++ N + + G Q+ R E +A
Sbjct: 1155 VAETAKSVQEASSKLSNRIKKTNQARDVAAAIIHRLNGNKDDEESGSQSGREQETTPVAS 1214
Query: 287 LRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKW 346
+ S +T S+ + +S+G S+ +++ +P K
Sbjct: 1215 DAFSQSAYPQ------ATTAEYASSATDTASASTGSSMKERAEKMRQLRDKQKTMQPIK- 1267
Query: 347 PITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNCF 406
T + + E + S+ E +EG ++R S+ S+ E
Sbjct: 1268 --TEMNSATDNEANDKSVRSSKLSEKSTEGQRKKRKSVDESSGKRAETLGRHIEDAQKRP 1325
Query: 407 DSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPP 466
D + ++ K+ V + + KRA P S+ + K+ ++ PPPP
Sbjct: 1326 DGVKKRPDVTKK------IVSKKGISKRA------PSTAKMSMFERLKKGIPGEIPPPPP 1373
Query: 467 PPP 469
PPP
Sbjct: 1374 PPP 1376
>UniRef100_UPI0000432993 UPI0000432993 UniRef100 entry
Length = 795
Score = 55.5 bits (132), Expect = 6e-06
Identities = 73/337 (21%), Positives = 143/337 (41%), Gaps = 29/337 (8%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSE-VDELVK-KVALLEEEKSGLSE 180
+ DL ++D++ L++ K+ +L E+ + + VDE+ K KV + +K G+
Sbjct: 129 NADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALDKCVDEMEKLKVENEKLKKEGMKV 188
Query: 181 QLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC- 239
+ L + L+ + + + + N E+ ++R N +L + L L S ++
Sbjct: 189 EATWLEENVNLKAKNTELEENLANTVNELDKMRSENADLLSELNRLKQELESGRKEIDQL 248
Query: 240 -SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTEL 298
S+ S D + K E L+ N+DL +V+GL++ R E+A L+ S L+ +L
Sbjct: 249 KSEIGSMKDALGKCVDEIEKLKTENKDLKSEVQGLESERDRLTNEVADLKPKISELQEKL 308
Query: 299 KNTCSTLDSDKLSSP---------QSVVSSSGDSISSFSDQCGS-ANSFN-LVKKPKKWP 347
+ LD K + + S+G I + S N N V++ +K
Sbjct: 309 TDASKKLDEAKTEDSDLRAEVDRLKKELESAGKEIDQLKAEMNSLKNGLNKCVEEMEKLT 368
Query: 348 ITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSS-EEEVSVLSKRRQSNCF 406
+S+ SQV + + + ++ NS+ ++E L+K+ N
Sbjct: 369 NENSELKSQV----------HGLRGEGDSLASELTNVKDENSALKDEKDQLNKQLAENKT 418
Query: 407 DSFECLK---EIEKESVPMPLFVQQCALEKRALRIPN 440
++ K E+E E+ + ++ C E L+ N
Sbjct: 419 ENERLKKQNDELESENTKIKKELESCKNENNNLKEEN 455
Score = 49.3 bits (116), Expect = 4e-04
Identities = 51/213 (23%), Positives = 99/213 (45%), Gaps = 13/213 (6%)
Query: 120 RQSSTDLFTELD-HMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGL 178
++ + DL T+ D + SL E + + +L + K +SE+D+L K++A + E +
Sbjct: 564 KEENKDLKTKGDVQIASLNSELEALKKELEKLRADNSKYKSEIDDLGKQLASAKNELNDC 623
Query: 179 SEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLS 238
E++V L + R + DK G+ + L+R E + ++ ++ S+E+QLS
Sbjct: 624 REEIVVLKNANSALRSDLDKMGTEND------RLKREVDESRKKLEDMEAKVKSLENQLS 677
Query: 239 CSDNSSESDIVAKFKAEASLLRLTNEDLSKQV---EGLQTSRLNEVEELAYLRWVNSCLR 295
E + ++ L L NE L KQ + ++ N EEL L+ +
Sbjct: 678 NLSAEKEELVKELYRTREDLNNLRNE-LEKQTGVKDTMEKESTNLKEELKALK--EELNK 734
Query: 296 TELKNTCSTLDSDKLSSPQSVVSSSGDSISSFS 328
T +N ++DKL++ + ++ D++ S
Sbjct: 735 TRDENDRLKNENDKLNAEIARLNKQLDALKDES 767
Score = 42.4 bits (98), Expect = 0.050
Identities = 44/193 (22%), Positives = 82/193 (41%), Gaps = 23/193 (11%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSR 187
TE +++++ L ++ E+ KL E E ++N D + + L+E L + L R
Sbjct: 1 TENENLKTELDKADEQLDKLKTERNELQRN---FDTMKLENETLKENVKALKDDLEESKR 57
Query: 188 SC--------GLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
L+ +EE KD + L+ + L+ N EL + +L R S +E +L
Sbjct: 58 EVDEMKAVGDALKDKEELKDAEFRELQQNMQNLKTENGELKKENDDLRTRSSELEHKL-- 115
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
DN K E + N DL +++ L+ + +E+ L+ S L+ L
Sbjct: 116 -DN---------VKKELDKVESENADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALD 165
Query: 300 NTCSTLDSDKLSS 312
++ K+ +
Sbjct: 166 KCVDEMEKLKVEN 178
Score = 39.3 bits (90), Expect = 0.42
Identities = 95/529 (17%), Positives = 192/529 (35%), Gaps = 88/529 (16%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKN---------------------- 157
+ +DL E+D ++ L+ + + +L AE+ +
Sbjct: 319 KTEDSDLRAEVDRLKKELESAGKEIDQLKAEMNSLKNGLNKCVEEMEKLTNENSELKSQV 378
Query: 158 ---QSEVDELVKKVALLEEEKSGLSEQLVALSRSCG--------LERQEEDKDGSTQNLE 206
+ E D L ++ +++E S L ++ L++ L++Q ++ + ++
Sbjct: 379 HGLRGEGDSLASELTNVKDENSALKDEKDQLNKQLAENKTENERLKKQNDELESENTKIK 438
Query: 207 LEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDL 266
E+ + N L + L L + QL S +D K + E + L
Sbjct: 439 KELESCKNENNNLKEENNKLKEELEKLGEQL-----KSLNDETNKLRRELKEAEDKIQIL 493
Query: 267 SKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSSGDSISS 326
Q+ ++ ELA LR N ++K LD+ + + ++ ++
Sbjct: 494 EPQLSRARSENEKSQNELAVLR--NEANELKVKLDREMLDNTNMRNALKILEDQVLDLNK 551
Query: 327 FSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISG 386
D C N + +K+ K T D Q+ N+ + E + +
Sbjct: 552 KLDNCREEN--DALKEENKDLKTKGD--VQIASLNSEL----------EALKKELEKLRA 597
Query: 387 SNSS-EEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRP 445
NS + E+ L K+ S + +C +EI V + + + N +
Sbjct: 598 DNSKYKSEIDDLGKQLASAKNELNDCREEI----VVLKNANSALRSDLDKMGTENDRLKR 653
Query: 446 SCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSLMKRDSRR 505
S K ++ A+V+ S ++ + ++ V ELY + R
Sbjct: 654 EVDESRKKLEDMEAKVK------------SLENQLSNLSAEKEELVKELYRT-------R 694
Query: 506 DSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLIREVNDAVYENI 565
+ + ++ V+ +M E N L A+K ++ + R+ ND +
Sbjct: 695 EDLNNLRNELEKQTGVKDTMEKESTNLKEELKALKEEL--------NKTRDENDRLKNEN 746
Query: 566 DDVVAFVKWLDDELGFLVDERAVLKH--FDWPEKKADTLREAAFGYQDL 612
D + A + L+ +L L DE A LK+ + E+ A+ +E A +L
Sbjct: 747 DKLNAEIARLNKQLDALKDESANLKNDIENLNERNAELSKELAVAKDNL 795
>UniRef100_UPI0000432990 UPI0000432990 UniRef100 entry
Length = 1433
Score = 55.5 bits (132), Expect = 6e-06
Identities = 73/337 (21%), Positives = 143/337 (41%), Gaps = 29/337 (8%)
Query: 123 STDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSE-VDELVK-KVALLEEEKSGLSE 180
+ DL ++D++ L++ K+ +L E+ + + VDE+ K KV + +K G+
Sbjct: 309 NADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALDKCVDEMEKLKVENEKLKKEGMKV 368
Query: 181 QLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC- 239
+ L + L+ + + + + N E+ ++R N +L + L L S ++
Sbjct: 369 EATWLEENVNLKAKNTELEENLANTVNELDKMRSENADLLSELNRLKQELESGRKEIDQL 428
Query: 240 -SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTEL 298
S+ S D + K E L+ N+DL +V+GL++ R E+A L+ S L+ +L
Sbjct: 429 KSEIGSMKDALGKCVDEIEKLKTENKDLKSEVQGLESERDRLTNEVADLKPKISELQEKL 488
Query: 299 KNTCSTLDSDKLSSP---------QSVVSSSGDSISSFSDQCGS-ANSFN-LVKKPKKWP 347
+ LD K + + S+G I + S N N V++ +K
Sbjct: 489 TDASKKLDEAKTEDSDLRAEVDRLKKELESAGKEIDQLKAEMNSLKNGLNKCVEEMEKLT 548
Query: 348 ITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSS-EEEVSVLSKRRQSNCF 406
+S+ SQV + + + ++ NS+ ++E L+K+ N
Sbjct: 549 NENSELKSQV----------HGLRGEGDSLASELTNVKDENSALKDEKDQLNKQLAENKT 598
Query: 407 DSFECLK---EIEKESVPMPLFVQQCALEKRALRIPN 440
++ K E+E E+ + ++ C E L+ N
Sbjct: 599 ENERLKKQNDELESENTKIKKELESCKNENNNLKEEN 635
Score = 42.4 bits (98), Expect = 0.050
Identities = 44/193 (22%), Positives = 82/193 (41%), Gaps = 23/193 (11%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSR 187
TE +++++ L ++ E+ KL E E ++N D + + L+E L + L R
Sbjct: 181 TENENLKTELDKADEQLDKLKTERNELQRN---FDTMKLENETLKENVKALKDDLEESKR 237
Query: 188 SC--------GLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
L+ +EE KD + L+ + L+ N EL + +L R S +E +L
Sbjct: 238 EVDEMKAVGDALKDKEELKDAEFRELQQNMQNLKTENGELKKENDDLRTRSSELEHKL-- 295
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
DN K E + N DL +++ L+ + +E+ L+ S L+ L
Sbjct: 296 -DN---------VKKELDKVESENADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALD 345
Query: 300 NTCSTLDSDKLSS 312
++ K+ +
Sbjct: 346 KCVDEMEKLKVEN 358
Score = 41.6 bits (96), Expect = 0.085
Identities = 46/182 (25%), Positives = 82/182 (44%), Gaps = 15/182 (8%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
E D +R LL+ESK+ + + + + E D+L+ + LE+ L+E ++ R+
Sbjct: 81 EKDKLRDLLEESKKLKEDNENLWAQLERLRGENDDLMGQKKALEDLNKQLNEDNESMKRT 140
Query: 189 CG-LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESD 247
G LE + + N+E E L N+ + KR L L+ E+ + D + E
Sbjct: 141 MGNLEARIDSLSNELSNVERERDALLDENESV---KRELERTLTENENLKTELDKADEQ- 196
Query: 248 IVAKFKAEAS-------LLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCL--RTEL 298
+ K K E + ++L NE L + V+ L+ E+ ++ V L + EL
Sbjct: 197 -LDKLKTERNELQRNFDTMKLENETLKENVKALKDDLEESKREVDEMKAVGDALKDKEEL 255
Query: 299 KN 300
K+
Sbjct: 256 KD 257
Score = 41.2 bits (95), Expect = 0.11
Identities = 43/175 (24%), Positives = 84/175 (47%), Gaps = 14/175 (8%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEV-DELVKKVAL---LEEEK 175
R+ D+ ++ + + L + +L EL R++ + + +EL K+ + +E+E
Sbjct: 1045 RKKLEDMEAKVKSLENQLSNLSAEKEELVKELYRTREDLNNLRNELEKQTGVKDTMEKES 1104
Query: 176 SGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQK------RNLTCR 229
+ L E+L AL R E D+ +N +L E+ RLNK+L K +N
Sbjct: 1105 TNLKEELKALKEELNKTRDENDR-LKNENDKLN-AEIARLNKQLDALKDESANLKNDIEN 1162
Query: 230 LSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEEL 284
L+ ++LS + + D + + S L+ N+D+ ++ L+ S + EV++L
Sbjct: 1163 LNERNAELS-KELAVAKDNLMGMETRLSNLKKENDDMKNKIITLEDS-IQEVDDL 1215
Score = 41.2 bits (95), Expect = 0.11
Identities = 56/259 (21%), Positives = 110/259 (41%), Gaps = 30/259 (11%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKN-----------QSEVDELVKKVALLEEEKS 176
T + ++S LQ K+ KL ++ E +++ + EVDE KK+ +E +
Sbjct: 997 TAAEKLKSDLQSCKDENDKLQTQINEMKRSLDKMGTENDRLKREVDESRKKLEDMEAKVK 1056
Query: 177 GLSEQLVALSRSCGLERQEEDKD-----GSTQNLELEVVELRRLNKELHMQKRNLTCRLS 231
L QL LS E++E K+ NL E+ + + + + NL L
Sbjct: 1057 SLENQLSNLS----AEKEELVKELYRTREDLNNLRNELEKQTGVKDTMEKESTNLKEELK 1112
Query: 232 SMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVN 291
+++ +L+ + + ++ + K E L L+KQ++ L+ N ++ L N
Sbjct: 1113 ALKEELNKTRDEND-----RLKNENDKLNAEIARLNKQLDALKDESANLKNDIENLNERN 1167
Query: 292 SCLRTEL---KNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSF-NLVKKPKKWP 347
+ L EL K+ +++ +LS+ + + I + D + +K+ KK
Sbjct: 1168 AELSKELAVAKDNLMGMET-RLSNLKKENDDMKNKIITLEDSIQEVDDLKRQLKEAKKEL 1226
Query: 348 ITSSDQLSQVECTNNSIID 366
S +L ++ TN + D
Sbjct: 1227 DKPSPELDTLKSTNKKLQD 1245
Score = 40.0 bits (92), Expect = 0.25
Identities = 48/204 (23%), Positives = 83/204 (40%), Gaps = 33/204 (16%)
Query: 121 QSSTDLFTELDHMRSLLQESKERE-------AKLNAELVECRKN-----------QSEVD 162
++ TDL ++LD + L+ K + A LN+EL +K +SE+D
Sbjct: 840 KNMTDLTSQLDRQKRSLEAEKSAKDRGDVQIASLNSELEALKKELEKLRADNSKYKSEID 899
Query: 163 ELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQ 222
+L K++A + E + E++V L + R E D S ++ + RL EL
Sbjct: 900 DLGKQLASAKNELNDCREEIVVLKNANSALRSELDPLRSLKD------DYSRLTTELDGL 953
Query: 223 KRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVE 282
K T L D S D K + E R+ + L ++ +T+
Sbjct: 954 KSENTKLL---------QDKRSLEDEFGKLRGEGDGQRVEIDRLRTTLDAEKTAAEKLKS 1004
Query: 283 ELAYLRWVNSCLRTELKNTCSTLD 306
+L + N L+T++ +LD
Sbjct: 1005 DLQSCKDENDKLQTQINEMKRSLD 1028
Score = 40.0 bits (92), Expect = 0.25
Identities = 69/306 (22%), Positives = 134/306 (43%), Gaps = 33/306 (10%)
Query: 126 LFTELDHMRS----LLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQ 181
L TELD ++S LLQ+ + E + E + E+D L L+ EK+ +E+
Sbjct: 946 LTTELDGLKSENTKLLQDKRSLEDEFGKLRGEGDGQRVEIDRLR---TTLDAEKTA-AEK 1001
Query: 182 LVALSRSCG-----LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQ 236
L + +SC L+ Q + S + E L+R E + ++ ++ S+E+Q
Sbjct: 1002 LKSDLQSCKDENDKLQTQINEMKRSLDKMGTENDRLKREVDESRKKLEDMEAKVKSLENQ 1061
Query: 237 LSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQV---EGLQTSRLNEVEELAYLRWVNSC 293
LS E + ++ L L NE L KQ + ++ N EEL L+
Sbjct: 1062 LSNLSAEKEELVKELYRTREDLNNLRNE-LEKQTGVKDTMEKESTNLKEELKALK--EEL 1118
Query: 294 LRTELKNTCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQ 353
+T +N ++DKL++ + ++ D++ SAN N ++ + S +
Sbjct: 1119 NKTRDENDRLKNENDKLNAEIARLNKQLDALKD-----ESANLKNDIENLNERNAELSKE 1173
Query: 354 LSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNCFDSFECLK 413
L+ + +N + + + ++ + ++ ++ I S +EV L ++ + E K
Sbjct: 1174 LAVAK--DNLMGMETRLSNLKKENDDMKNKIITLEDSIQEVDDLKRQLK-------EAKK 1224
Query: 414 EIEKES 419
E++K S
Sbjct: 1225 ELDKPS 1230
Score = 37.0 bits (84), Expect = 2.1
Identities = 63/301 (20%), Positives = 120/301 (38%), Gaps = 52/301 (17%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLS 179
+ +DL E+D ++ L+ + + +L AE+ + +++ V+++ L E S L
Sbjct: 499 KTEDSDLRAEVDRLKKELESAGKEIDQLKAEMNSLKNG---LNKCVEEMEKLTNENSELK 555
Query: 180 EQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
Q+ L G +L E+ ++ N L +K L +L
Sbjct: 556 SQVHGLR-------------GEGDSLASELTNVKDENSALKDEKDQLNKQL--------- 593
Query: 240 SDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
++N +E++ + K + L N + K++E + N EE N+ L+ EL+
Sbjct: 594 AENKTENE---RLKKQNDELESENTKIKKELESCKNENNNLKEE-------NNKLKEELE 643
Query: 300 NTCSTLDS--DKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQV 357
L S D+ + + + + D I Q A S N S ++L+ +
Sbjct: 644 KLGEQLKSLNDETNKLRRELKEAEDKIQILEPQLSRARSEN---------EKSQNELAVL 694
Query: 358 ECTNNSIIDKNWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNCFDSFECLKEIEK 417
N + K E + + R I E++V L+K + NC + + LKE K
Sbjct: 695 RNEANELKVKLDREMLDNTNMRNALKI-----LEDQVLDLNK-KLDNCREENDALKEENK 748
Query: 418 E 418
+
Sbjct: 749 D 749
Score = 36.6 bits (83), Expect = 2.7
Identities = 64/343 (18%), Positives = 128/343 (36%), Gaps = 72/343 (20%)
Query: 117 THRRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKN-----------QSEVDELV 165
T+ ++ L EL+ R K KLNAE+ K +++++ L
Sbjct: 1105 TNLKEELKALKEELNKTRDENDRLKNENDKLNAEIARLNKQLDALKDESANLKNDIENLN 1164
Query: 166 KKVALLEEEKSGLSEQLVAL-SRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKR 224
++ A L +E + + L+ + +R L+++ +D LE + E+ L ++L K+
Sbjct: 1165 ERNAELSKELAVAKDNLMGMETRLSNLKKENDDMKNKIITLEDSIQEVDDLKRQLKEAKK 1224
Query: 225 NLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEEL 284
L K E L+ TN+ L ++ + LN +L
Sbjct: 1225 ELD-----------------------KPSPELDTLKSTNKKLQDDLDNARNESLNLKNDL 1261
Query: 285 AYLRWVNSCLRTEL----------KNTCSTLDSD--KLSSPQSVVSSSGDSISSFSDQCG 332
L+ + L+TEL + + L+ D ++ + +++ + D C
Sbjct: 1262 DNLQNDYNNLQTELADVKEERDTFRERAAALEKDLVRVKRENEELVEQNETLRTELDDCR 1321
Query: 333 SANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWIESISEGSNRRRHSISGSNSS-- 390
N+ L+K+ +K + V+ +N I ++ E + E N+ + S +
Sbjct: 1322 GENN-RLLKELEKL------KSENVKLQDNLINARSEGERLKEDLNKLKKDYSDLRTDLT 1374
Query: 391 ----------------EEEVSVLSKRRQSNCFDSFECLKEIEK 417
++E+ L D ++C KE EK
Sbjct: 1375 KAREDRDVRKEKDMELDKEIDELKAVNAKLKSDLYDCQKENEK 1417
Score = 35.0 bits (79), Expect = 8.0
Identities = 44/180 (24%), Positives = 75/180 (41%), Gaps = 26/180 (14%)
Query: 128 TELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSR 187
TE + ++ E + K+ EL C+ + + E K L+EE L EQL +L+
Sbjct: 598 TENERLKKQNDELESENTKIKKELESCKNENNNLKEENNK---LKEELEKLGEQLKSLND 654
Query: 188 SCG-LERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSES 246
L R+ ++ + Q LE ++ R N++ +++L+ N +
Sbjct: 655 ETNKLRRELKEAEDKIQILEPQLSRARSENEK--------------SQNELAVLRNEANE 700
Query: 247 DIVAKFKAEASLLRLTN-----EDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNT 301
K K + +L TN + L QV L N EE L+ N L+T+L +T
Sbjct: 701 ---LKVKLDREMLDNTNMRNALKILEDQVLDLNKKLDNCREENDALKEENKDLKTKLSDT 757
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.128 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,191,046,237
Number of Sequences: 2790947
Number of extensions: 51889806
Number of successful extensions: 476609
Number of sequences better than 10.0: 5073
Number of HSP's better than 10.0 without gapping: 1231
Number of HSP's successfully gapped in prelim test: 4069
Number of HSP's that attempted gapping in prelim test: 409456
Number of HSP's gapped (non-prelim): 27020
length of query: 732
length of database: 848,049,833
effective HSP length: 135
effective length of query: 597
effective length of database: 471,271,988
effective search space: 281349376836
effective search space used: 281349376836
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC124968.5


