

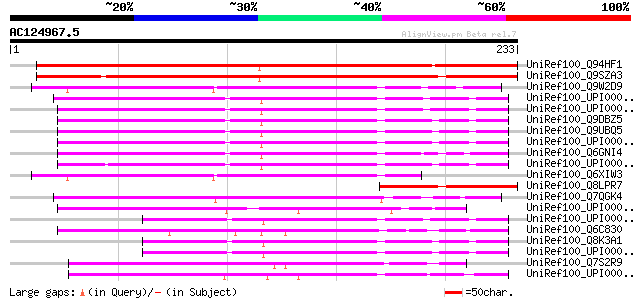
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.5 + phase: 0 /pseudo
(233 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q94HF1 Eukaryotic translation initiation factor 3 subu... 308 1e-82
UniRef100_Q9SZA3 Eukaryotic translation initiation factor 3 subu... 298 6e-80
UniRef100_Q9W2D9 Probable eukaryotic translation initiation fact... 111 2e-23
UniRef100_UPI0000361FC7 UPI0000361FC7 UniRef100 entry 107 2e-22
UniRef100_UPI00002F96BA UPI00002F96BA UniRef100 entry 107 2e-22
UniRef100_Q9DBZ5 Eukaryotic translation initiation factor 3 subu... 105 9e-22
UniRef100_Q9UBQ5 Eukaryotic translation initiation factor 3 subu... 105 9e-22
UniRef100_UPI00001816CA UPI00001816CA UniRef100 entry 105 1e-21
UniRef100_Q6GNI4 MGC82783 protein [Xenopus laevis] 99 9e-20
UniRef100_UPI000036BDEC UPI000036BDEC UniRef100 entry 98 2e-19
UniRef100_Q6XIW3 Similar to Drosophila melanogaster CG10306 [Dro... 98 2e-19
UniRef100_Q8LPR7 AT4g33250/F17M5_10 [Arabidopsis thaliana] 88 2e-16
UniRef100_Q7QGK4 ENSANGP00000015097 [Anopheles gambiae str. PEST] 87 3e-16
UniRef100_UPI00002350F5 UPI00002350F5 UniRef100 entry 79 9e-14
UniRef100_UPI0000361FC6 UPI0000361FC6 UniRef100 entry 72 1e-11
UniRef100_Q6C830 Yarrowia lipolytica chromosome D of strain CLIB... 72 1e-11
UniRef100_Q8K3A1 1200009C21Rik protein [Mus musculus] 70 3e-11
UniRef100_UPI00001816C0 UPI00001816C0 UniRef100 entry 70 4e-11
UniRef100_Q7S2R9 Hypothetical protein [Neurospora crassa] 68 2e-10
UniRef100_UPI000023E226 UPI000023E226 UniRef100 entry 66 8e-10
>UniRef100_Q94HF1 Eukaryotic translation initiation factor 3 subunit 11 [Oryza
sativa]
Length = 226
Score = 308 bits (788), Expect = 1e-82
Identities = 158/222 (71%), Positives = 183/222 (82%), Gaps = 2/222 (0%)
Query: 13 QGGTAYTVEQLVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQ 72
Q +YTVE+LVA N YNPDIL DLE +VNDQVS+QTY+L++NL LLRLYQFE E++S Q
Sbjct: 5 QAAESYTVEELVAVNPYNPDILNDLEGFVNDQVSNQTYNLDANLSLLRLYQFEPERLSVQ 64
Query: 73 IVARILVKALMAMPAPDFSLCLFLIPERVIANGRSSSRH*L-FCLTI*RFRQFWDEASKS 131
IV+RIL+KALMAMP PDFSLCLFLIPE V + + L L RFRQFWDEASK+
Sbjct: 65 IVSRILIKALMAMPGPDFSLCLFLIPEHVQMEEQFKTLIVLSHYLETARFRQFWDEASKN 124
Query: 132 RHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVI 191
R+I++ VPGFEQAIQ YAIHVLSLTYQK+PR VLAEAIN+EGL+LDKFLEH + NSGWVI
Sbjct: 125 RNILDVVPGFEQAIQSYAIHVLSLTYQKVPRPVLAEAINIEGLALDKFLEHHIANSGWVI 184
Query: 192 EKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRIFPIL 233
EK + QLIVLPRNEFN P LKKNTA++VP EHVTRIFP+L
Sbjct: 185 EKG-ARSQLIVLPRNEFNHPELKKNTAETVPFEHVTRIFPVL 225
>UniRef100_Q9SZA3 Eukaryotic translation initiation factor 3 subunit 11 [Arabidopsis
thaliana]
Length = 226
Score = 298 bits (764), Expect = 6e-80
Identities = 155/222 (69%), Positives = 182/222 (81%), Gaps = 6/222 (2%)
Query: 13 QGGTAYTVEQLVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQ 72
Q ++YTVEQLVA N +NP+ILPDLENYVN V+SQTYSLE NLCLLRLYQFE E+M++
Sbjct: 9 QEQSSYTVEQLVALNPFNPEILPDLENYVN--VTSQTYSLEVNLCLLRLYQFEPERMNTH 66
Query: 73 IVARILVKALMAMPAPDFSLCLFLIPERVIANGRSSSRH*L-FCLTI*RFRQFWDEASKS 131
IVARILVKALMAMP PDFSLCLFLIPERV + S L L RF+QFWDEA+K+
Sbjct: 67 IVARILVKALMAMPTPDFSLCLFLIPERVQMEEQFKSLIVLSHYLETGRFQQFWDEAAKN 126
Query: 132 RHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVI 191
RHI+EAVPGFEQAIQ YA H+LSL+YQK+PR+VLAEA+N++G SLDKF+E QV NSGW++
Sbjct: 127 RHILEAVPGFEQAIQAYASHLLSLSYQKVPRSVLAEAVNMDGASLDKFIEQQVTNSGWIV 186
Query: 192 EKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRIFPIL 233
EK G IVLP+NEFN P LKKNT ++VPLEH+ RIFPIL
Sbjct: 187 EKEGGS---IVLPQNEFNHPELKKNTGENVPLEHIARIFPIL 225
>UniRef100_Q9W2D9 Probable eukaryotic translation initiation factor 3 subunit 11
[Drosophila melanogaster]
Length = 222
Score = 111 bits (277), Expect = 2e-23
Identities = 71/219 (32%), Positives = 119/219 (53%), Gaps = 13/219 (5%)
Query: 11 KGQGGTAYTVEQLVA-FNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKM 69
K + G + T+++++ RYNPD L LE+YV DQ + TY LE+NL +L+LYQF +
Sbjct: 6 KMENGQSQTIQEMLGCIERYNPDHLKTLESYVQDQAKNNTYDLEANLAVLKLYQFNPHML 65
Query: 70 SSQIVARILVKALMAMPAPDFSL--CLFLIPERVIANGRSSSRH*LFCLTI*RFRQFWDE 127
+ I IL+K+L ++P DF + CL L+P+++ + L F FW
Sbjct: 66 NFDITYTILLKSLTSLPHTDFVMAKCL-LLPQQMKDENVQTIIDLADILERADFTLFWQR 124
Query: 128 ASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENS 187
A +R++ + GF +I+ + HV+ T+Q I + +L E + G D LE ++ +
Sbjct: 125 AEVNRNMFRHITGFHDSIRKFVSHVVGTTFQTIRKDLLKELL---GGIEDSTLESWIKRN 181
Query: 188 GWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHV 226
GW K+QG+G +IV + +D I KN + + ++V
Sbjct: 182 GW---KNQGQGLVIVAMQ---DDKIKTKNITEKIEFDNV 214
>UniRef100_UPI0000361FC7 UPI0000361FC7 UniRef100 entry
Length = 218
Score = 107 bits (268), Expect = 2e-22
Identities = 66/212 (31%), Positives = 107/212 (50%), Gaps = 14/212 (6%)
Query: 21 EQLVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVK 80
E L +RYNP+ L LE YV Q Y LE+NL +L+LYQF + + ++IL+K
Sbjct: 13 ELLRGIDRYNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAYFQTAVTSQILLK 72
Query: 81 ALMAMPAPDFSLCLFLIPERVIANGRSSSRH*LF---CLTI*RFRQFWDEASKSRHIVEA 137
AL +P DF+LC +I + R L+ L F+ FW ++R +++
Sbjct: 73 ALTNLPHTDFTLCKCMIDQ--THQEERPIRQILYLGNLLETCHFQSFWTNLEENRELIDG 130
Query: 138 VPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGK 197
+ GFE +++ + HV+ +TYQ I R +L E + G LD ++ + GW +
Sbjct: 131 ITGFEDSVRKFICHVVGITYQTIGRRLLGEML---GDPLDTQVKVWMNKHGWA---ENEE 184
Query: 198 GQLIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
GQ+ + ++E P KN + + E V+ I
Sbjct: 185 GQIFIFNQDESIKP---KNIVEKIDFESVSSI 213
>UniRef100_UPI00002F96BA UPI00002F96BA UniRef100 entry
Length = 218
Score = 107 bits (268), Expect = 2e-22
Identities = 66/210 (31%), Positives = 106/210 (50%), Gaps = 14/210 (6%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKAL 82
L +RYNP+ L LE YV Q Y LE+NL +L+LYQF + + ++IL+KAL
Sbjct: 15 LRGIDRYNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPPYFQTAVTSQILLKAL 74
Query: 83 MAMPAPDFSLCLFLIPERVIANGRSSSRH*LF---CLTI*RFRQFWDEASKSRHIVEAVP 139
+P DF+LC +I + R L+ L F+ FW ++R +++ +
Sbjct: 75 TNLPHTDFTLCKCMIDQ--THQEERPIRQILYLGNLLETCHFQSFWTNLEENRELIDGIA 132
Query: 140 GFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGKGQ 199
GFE +++ + HV+ +TYQ I R +LAE + G LD ++ + GW +GQ
Sbjct: 133 GFEDSVRKFICHVVGITYQTIGRCLLAEML---GDPLDTQVKLWMNKHGWT---ENEEGQ 186
Query: 200 LIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
+ + + E P KN + + E V+ I
Sbjct: 187 IFIFNQEESIKP---KNIVEKIDFESVSSI 213
>UniRef100_Q9DBZ5 Eukaryotic translation initiation factor 3 subunit 11 [Mus
musculus]
Length = 218
Score = 105 bits (262), Expect = 9e-22
Identities = 67/210 (31%), Positives = 106/210 (49%), Gaps = 14/210 (6%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKAL 82
L +RYNP+ L LE YV Q Y LE+NL +L+LYQF + + A+IL+KAL
Sbjct: 15 LKGIDRYNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAFFQTTVTAQILLKAL 74
Query: 83 MAMPAPDFSLCLFLIPERVIANGRSSSRH*LF---CLTI*RFRQFWDEASKSRHIVEAVP 139
+P DF+LC +I + R L+ L F+ FW ++ ++E +
Sbjct: 75 TNLPHTDFTLCKCMIDQ--AHQEERPIRQILYLGDLLETCHFQAFWQALDENMDLLEGIT 132
Query: 140 GFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGKGQ 199
GFE +++ + HV+ +TYQ I R +LAE + G D L+ + GW ++S GQ
Sbjct: 133 GFEDSVRKFICHVVGITYQHIDRWLLAEML---GDLTDNQLKVWMSKYGWSADES---GQ 186
Query: 200 LIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
+ + + E P KN + + + V+ I
Sbjct: 187 VFICSQEESIKP---KNIVEKIDFDSVSSI 213
>UniRef100_Q9UBQ5 Eukaryotic translation initiation factor 3 subunit 11 [Homo
sapiens]
Length = 218
Score = 105 bits (262), Expect = 9e-22
Identities = 67/210 (31%), Positives = 106/210 (49%), Gaps = 14/210 (6%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKAL 82
L +RYNP+ L LE YV Q Y LE+NL +L+LYQF + + A+IL+KAL
Sbjct: 15 LKGIDRYNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAFFQTTVTAQILLKAL 74
Query: 83 MAMPAPDFSLCLFLIPERVIANGRSSSRH*LF---CLTI*RFRQFWDEASKSRHIVEAVP 139
+P DF+LC +I + R L+ L F+ FW ++ ++E +
Sbjct: 75 TNLPHTDFTLCKCMIDQ--AHQEERPIRQILYLGDLLETCHFQAFWQALDENMDLLEGIT 132
Query: 140 GFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGKGQ 199
GFE +++ + HV+ +TYQ I R +LAE + G D L+ + GW ++S GQ
Sbjct: 133 GFEDSVRKFICHVVGITYQHIDRWLLAEML---GDLSDSQLKVWMSKYGWSADES---GQ 186
Query: 200 LIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
+ + + E P KN + + + V+ I
Sbjct: 187 IFICSQEESIKP---KNIVEKIDFDSVSSI 213
>UniRef100_UPI00001816CA UPI00001816CA UniRef100 entry
Length = 218
Score = 105 bits (261), Expect = 1e-21
Identities = 67/210 (31%), Positives = 105/210 (49%), Gaps = 14/210 (6%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKAL 82
L +RYNP+ L LE YV Q Y LE+NL +L+LYQF + + A+IL+KAL
Sbjct: 15 LKGIDRYNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAFFQTTVTAQILLKAL 74
Query: 83 MAMPAPDFSLCLFLIPERVIANGRSSSRH*LF---CLTI*RFRQFWDEASKSRHIVEAVP 139
+P DF+LC +I + R L+ L F+ FW ++ ++E +
Sbjct: 75 TNLPHTDFTLCKCMIDQ--AHQEERPIRQILYLGDLLETCHFQAFWQALDENMDLLEGIT 132
Query: 140 GFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGKGQ 199
GFE +++ + HV+ +TYQ I R +LAE + G D L + GW ++S GQ
Sbjct: 133 GFEDSVRKFICHVVGITYQHIDRWLLAEML---GDLTDNQLRVWMSKYGWSADES---GQ 186
Query: 200 LIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
+ + + E P KN + + + V+ I
Sbjct: 187 VFICSQEESIKP---KNIVEKIDFDSVSSI 213
>UniRef100_Q6GNI4 MGC82783 protein [Xenopus laevis]
Length = 218
Score = 99.0 bits (245), Expect = 9e-20
Identities = 68/210 (32%), Positives = 104/210 (49%), Gaps = 14/210 (6%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKAL 82
L +RYNP+ L LE YV Q Y LE+NL +L+LYQF + + A++L+KAL
Sbjct: 15 LRGIDRYNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAFFQTTVTAQVLLKAL 74
Query: 83 MAMPAPDFSLCLFLIPERVIANGRSSSRH*LF---CLTI*RFRQFWDEASKSRHIVEAVP 139
+P DF+LC +I + R L+ L F+ FW ++ +++ V
Sbjct: 75 TNLPHTDFTLCKCMIDQ--AHQEDRPIRQILYLGDLLETCHFQSFWQALDENLDLIDGVT 132
Query: 140 GFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGKGQ 199
GFE +++ + HV+ +TYQ I R +LAE + G + L + GW IE GK
Sbjct: 133 GFEDSVRKFICHVVGITYQHIDRWLLAEML---GDLSEPQLRVWMSKYGW-IESENGK-- 186
Query: 200 LIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
I + E N I KN + + + V+ I
Sbjct: 187 -IFVCNQEEN--IKPKNIVEKIDFDSVSGI 213
>UniRef100_UPI000036BDEC UPI000036BDEC UniRef100 entry
Length = 217
Score = 98.2 bits (243), Expect = 2e-19
Identities = 66/210 (31%), Positives = 107/210 (50%), Gaps = 15/210 (7%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKAL 82
L +RYNP+ L LE YV Q + + LE+NL +L+LYQF + + A+IL+KAL
Sbjct: 15 LKGIDRYNPENLATLERYVETQ-AKENDDLEANLAVLKLYQFNPAFFQTTVTAQILLKAL 73
Query: 83 MAMPAPDFSLCLFLIPERVIANGRSSSRH*LF---CLTI*RFRQFWDEASKSRHIVEAVP 139
+P DF+LC +I + R L+ L F+ FW ++ ++E +
Sbjct: 74 TNLPHTDFTLCKCMIDQ--AHQEERPIRQILYLGDLLETCHFQAFWQALDENMDLLEGIT 131
Query: 140 GFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGKGQ 199
GFE +++ + HV+ +TYQ I R +LAE + G D L+ + GW ++S GQ
Sbjct: 132 GFEDSVRKFICHVVGITYQHIDRWLLAEML---GDLSDSQLKVWMSKYGWSADES---GQ 185
Query: 200 LIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
+ + + E P KN + + + V+ I
Sbjct: 186 IFICSQEESIKP---KNIVEKIDFDSVSSI 212
>UniRef100_Q6XIW3 Similar to Drosophila melanogaster CG10306 [Drosophila yakuba]
Length = 185
Score = 98.2 bits (243), Expect = 2e-19
Identities = 60/182 (32%), Positives = 97/182 (52%), Gaps = 7/182 (3%)
Query: 11 KGQGGTAYTVEQLVA-FNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKM 69
K + G + T+++++ RYNPD L LE YV DQ + TY LE+NL +L+LYQF +
Sbjct: 6 KMENGQSQTIQEMLGCIERYNPDHLKTLEAYVQDQAKNNTYDLEANLAVLKLYQFNPHML 65
Query: 70 SSQIVARILVKALMAMPAPDFSL--CLFLIPERVIANGRSSSRH*LFCLTI*RFRQFWDE 127
+ I IL+K+L ++P DF + CL L+P+++ + L F FW
Sbjct: 66 NFDITYTILLKSLTSLPHTDFVMAKCL-LLPQQMKDENVQTIIDLADILERADFTLFWQR 124
Query: 128 ASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENS 187
A +R + + GF +I+ + HV+ T+Q I + +L E + G D LE ++ +
Sbjct: 125 AEVNRSMFRHIAGFHDSIRKFVSHVVGTTFQTIRKDLLKELL---GGIEDSTLESWIKRN 181
Query: 188 GW 189
GW
Sbjct: 182 GW 183
>UniRef100_Q8LPR7 AT4g33250/F17M5_10 [Arabidopsis thaliana]
Length = 61
Score = 87.8 bits (216), Expect = 2e-16
Identities = 41/63 (65%), Positives = 50/63 (79%), Gaps = 3/63 (4%)
Query: 171 VEGLSLDKFLEHQVENSGWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRIF 230
++G SLDKF+E QV NSGW++EK G IVLP+NEFN P LKKNT ++VPLEH+ RIF
Sbjct: 1 MDGASLDKFIEQQVTNSGWIVEKEGGS---IVLPQNEFNHPELKKNTGENVPLEHIARIF 57
Query: 231 PIL 233
PIL
Sbjct: 58 PIL 60
>UniRef100_Q7QGK4 ENSANGP00000015097 [Anopheles gambiae str. PEST]
Length = 221
Score = 87.0 bits (214), Expect = 3e-16
Identities = 62/208 (29%), Positives = 99/208 (46%), Gaps = 13/208 (6%)
Query: 21 EQLVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVK 80
E L + RYNP+ L +E YV +Q Y LE+NL L+LYQF ++ I IL+K
Sbjct: 17 EMLKSIERYNPEHLKVIEAYVEEQARENQYDLEANLACLKLYQFNPHLLNLDITYIILLK 76
Query: 81 ALMAMPAPDFSLC-LFLIPERVIANGRSSSRH*LFCLTI*RFRQFWDEASKSRHIVEAVP 139
AL P DF LC L+P ++ + L F FW +K + +
Sbjct: 77 ALTNFPHTDFVLCKCLLLPAQMNDETVKEIIYLADILEKCDFSLFWSRLAKHPENYQKIS 136
Query: 140 GFEQAIQGYAIHVLSLTYQKIPRTVLAEAI-NVEGLSLDKFLEHQVENSGWVIEKSQGKG 198
GF +I+ + HV+ +T+Q I R L + + NVE +K L ++ GW + +G
Sbjct: 137 GFHDSIRKFVCHVVGITFQTIERGYLMQLLGNVE----EKVLLSWLKRYGW-----KEEG 187
Query: 199 QLIVLPRNEFNDPILKKNTADSVPLEHV 226
L+ + E D I K+ + + +++
Sbjct: 188 GLVTIATQE--DNIKTKHITEKIEFDNL 213
>UniRef100_UPI00002350F5 UPI00002350F5 UniRef100 entry
Length = 250
Score = 79.0 bits (193), Expect = 9e-14
Identities = 61/214 (28%), Positives = 98/214 (45%), Gaps = 35/214 (16%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKAL 82
L +RYNP+ ++YV Q +T+ +NL LL+LYQF + + V ILVKAL
Sbjct: 19 LSGLDRYNPETTTVFQDYVVQQCEDRTFDCYANLALLKLYQFNPHLLQPETVTNILVKAL 78
Query: 83 MAMPAPDFSLCLFLIP----------------------ERVIANGRSSSRH*LFCLTI*R 120
P+P FSLCL L+P E V R SS L +
Sbjct: 79 TVFPSPAFSLCLALLPAYTQPFPSSEAEATAAQMSDFVESVQKLARLSS-----LLESAQ 133
Query: 121 FRQFWDEASKS---RHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGL-SL 176
+ QFW + +V V GFE+ ++ + ++++ VL + ++V +L
Sbjct: 134 YAQFWSTLNSDDLYADLVADVAGFEELVRIRIAVEVGKAFREVNAEVLEQWLDVRNSEAL 193
Query: 177 DKFLEHQVENSGWVIEKSQGKGQLIVLPRNEFND 210
+KF+ E W ++KS G ++ +P N+ N+
Sbjct: 194 EKFV---TEVCSWEVDKS-GSATVVKVPTNKENE 223
>UniRef100_UPI0000361FC6 UPI0000361FC6 UniRef100 entry
Length = 185
Score = 72.0 bits (175), Expect = 1e-11
Identities = 47/171 (27%), Positives = 83/171 (48%), Gaps = 14/171 (8%)
Query: 62 YQFESEKMSSQIVARILVKALMAMPAPDFSLCLFLIPERVIANGRSSSRH*LFC---LTI 118
YQF + + ++IL+KAL +P DF+LC +I + R L+ L
Sbjct: 21 YQFNPAYFQTAVTSQILLKALTNLPHTDFTLCKCMIDQT--HQEERPIRQILYLGNLLET 78
Query: 119 *RFRQFWDEASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDK 178
F+ FW ++R +++ + GFE +++ + HV+ +TYQ I R +L E + G LD
Sbjct: 79 CHFQSFWTNLEENRELIDGITGFEDSVRKFICHVVGITYQTIGRRLLGEML---GDPLDT 135
Query: 179 FLEHQVENSGWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
++ + GW +GQ+ + ++E P KN + + E V+ I
Sbjct: 136 QVKVWMNKHGWA---ENEEGQIFIFNQDESIKP---KNIVEKIDFESVSSI 180
>UniRef100_Q6C830 Yarrowia lipolytica chromosome D of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 234
Score = 72.0 bits (175), Expect = 1e-11
Identities = 61/220 (27%), Positives = 106/220 (47%), Gaps = 23/220 (10%)
Query: 23 LVAFNRYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQ--IVARILVK 80
L + +RYNP+ + L+ Y Q + Q +E+NL LL+L QF+ + ++ I+ IL
Sbjct: 19 LSSLDRYNPEKISILQEYATTQCADQHSDIEANLALLKLLQFQQQPNPNKEDIICNILSM 78
Query: 81 ALMAMPAPDFSLCLFLIPERVI----ANGRSSSRH*LF----CLTI*RFRQFW---DEAS 129
AL DF+ L ++P V+ A+ + S LF L RF +FW +
Sbjct: 79 ALANFLTSDFTTALHVLPSYVLDSPAADTLAESIQKLFHLYTLLDGCRFPEFWAVYERDD 138
Query: 130 KSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGW 189
I V FE ++ + ++ Q I + V +N LS +KF ++ V+ GW
Sbjct: 139 AHADITADVADFENLVRISITRAVDISSQAIHKDVFRSWLN---LSDNKFADY-VKELGW 194
Query: 190 VIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
+E G+ +V+P N+ N+ T +S+ +E ++R+
Sbjct: 195 KVE-----GETVVVPPNKENE-AKPATTTESIKIEQISRL 228
>UniRef100_Q8K3A1 1200009C21Rik protein [Mus musculus]
Length = 185
Score = 70.5 bits (171), Expect = 3e-11
Identities = 49/171 (28%), Positives = 83/171 (47%), Gaps = 14/171 (8%)
Query: 62 YQFESEKMSSQIVARILVKALMAMPAPDFSLCLFLIPERVIANGRSSSRH*LFC---LTI 118
YQF + + A+IL+KAL +P DF+LC +I + R L+ L
Sbjct: 21 YQFNPAFFQTTVTAQILLKALTNLPHTDFTLCKCMIDQA--HQEERPIRQILYLGDLLET 78
Query: 119 *RFRQFWDEASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDK 178
F+ FW ++ ++E + GFE +++ + HV+ +TYQ I R +LAE + G D
Sbjct: 79 CHFQAFWQALDENMDLLEGITGFEDSVRKFICHVVGITYQHIDRWLLAEML---GDLTDN 135
Query: 179 FLEHQVENSGWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
L+ + GW ++S GQ+ + + E P KN + + + V+ I
Sbjct: 136 QLKVWMSKYGWSADES---GQVFICSQEESIKP---KNIVEKIDFDSVSSI 180
>UniRef100_UPI00001816C0 UPI00001816C0 UniRef100 entry
Length = 185
Score = 70.1 bits (170), Expect = 4e-11
Identities = 49/171 (28%), Positives = 82/171 (47%), Gaps = 14/171 (8%)
Query: 62 YQFESEKMSSQIVARILVKALMAMPAPDFSLCLFLIPERVIANGRSSSRH*LFC---LTI 118
YQF + + A+IL+KAL +P DF+LC +I + R L+ L
Sbjct: 21 YQFNPAFFQTTVTAQILLKALTNLPHTDFTLCKCMIDQA--HQEERPIRQILYLGDLLET 78
Query: 119 *RFRQFWDEASKSRHIVEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDK 178
F+ FW ++ ++E + GFE +++ + HV+ +TYQ I R +LAE + G D
Sbjct: 79 CHFQAFWQALDENMDLLEGITGFEDSVRKFICHVVGITYQHIDRWLLAEML---GDLTDN 135
Query: 179 FLEHQVENSGWVIEKSQGKGQLIVLPRNEFNDPILKKNTADSVPLEHVTRI 229
L + GW ++S GQ+ + + E P KN + + + V+ I
Sbjct: 136 QLRVWMSKYGWSADES---GQVFICSQEESIKP---KNIVEKIDFDSVSSI 180
>UniRef100_Q7S2R9 Hypothetical protein [Neurospora crassa]
Length = 237
Score = 67.8 bits (164), Expect = 2e-10
Identities = 48/196 (24%), Positives = 86/196 (43%), Gaps = 20/196 (10%)
Query: 28 RYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKALMAMPA 87
RYNP+ LE Y+ Q + +N LL+LYQ +++ +++ ILVKA+ P+
Sbjct: 22 RYNPEAAGTLEAYLTQQCEEKFCDCNANRALLKLYQLNPDRIKDEVITNILVKAMTQFPS 81
Query: 88 PDFSLCLFLIPERVIANGRSSSRH*LFCLTI*R----------FRQFW---DEASKSRHI 134
P F L L L+ G +SS ++ R + +FW D +
Sbjct: 82 PQFDLALHLLSPSQSNPGPNSSSELTEAVSKLRALNAQLEGAEYARFWATLDSDDLYADL 141
Query: 135 VEAVPGFEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKS 194
+ GFE I+ ++ +Y++I VL + + + + + E GW +E
Sbjct: 142 TTDIAGFEDMIRVRIAQLVGQSYREIQFPVLESWLGLN--NSEATTQFITETCGWKVE-- 197
Query: 195 QGKGQLIVLPRNEFND 210
G ++ +P+N N+
Sbjct: 198 ---GDVVQIPKNADNE 210
>UniRef100_UPI000023E226 UPI000023E226 UniRef100 entry
Length = 231
Score = 65.9 bits (159), Expect = 8e-10
Identities = 51/209 (24%), Positives = 96/209 (45%), Gaps = 15/209 (7%)
Query: 28 RYNPDILPDLENYVNDQVSSQTYSLESNLCLLRLYQFESEKMSSQIVARILVKALMAMPA 87
RYNP+ + LE Y+ DQ + +N LL+LYQ +++ +++ +LVK + P+
Sbjct: 22 RYNPEAVGALETYLQDQCEQKFTDCNANRTLLKLYQLNPDRIKDEVITNVLVKTMTQFPS 81
Query: 88 PDFSLCLFLI-PERVIANGRSSSRH*LFCLT----I*RFRQFWDEASKS--RHIVEAVPG 140
FSL L LI P V+ + L L ++ +FW + ++ +P
Sbjct: 82 AQFSLALHLINPSSVVTGDLGEAITKLRSLNGQLEGAQYSRFWADLDDDMVADLIADIPD 141
Query: 141 FEQAIQGYAIHVLSLTYQKIPRTVLAEAINVEGLSLDKFLEHQVENSGWVIEKSQGKGQL 200
FE+ ++ ++S +++I L + GL+ D + E GW ++ ++G ++
Sbjct: 142 FEEVVRHRIALLVSQAFREIQIGHLESWL---GLNEDATKKFVTEVCGWTVD-AEGNVKV 197
Query: 201 IVLPRNEFNDPILKKNTADSVPLEHVTRI 229
P NE K + V +E +R+
Sbjct: 198 PSNPDNEAK----KAEIREDVNVEQFSRV 222
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.139 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 358,878,124
Number of Sequences: 2790947
Number of extensions: 13349279
Number of successful extensions: 42226
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 42168
Number of HSP's gapped (non-prelim): 40
length of query: 233
length of database: 848,049,833
effective HSP length: 123
effective length of query: 110
effective length of database: 504,763,352
effective search space: 55523968720
effective search space used: 55523968720
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC124967.5


