

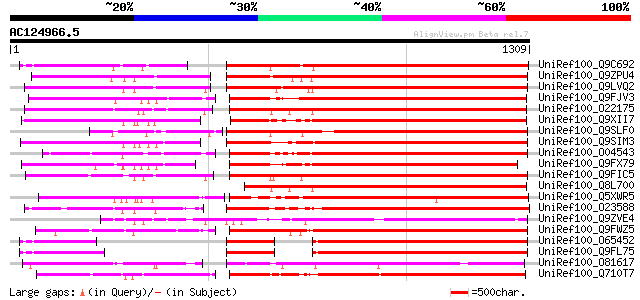
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.5 - phase: 0
(1309 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C692 Polyprotein, putative [Arabidopsis thaliana] 731 0.0
UniRef100_Q9ZPU4 Putative retroelement pol polyprotein [Arabidop... 726 0.0
UniRef100_Q9LVQ2 Retroelement pol polyprotein-like [Arabidopsis ... 719 0.0
UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis ... 706 0.0
UniRef100_O22175 Putative retroelement pol polyprotein [Arabidop... 691 0.0
UniRef100_Q9XII7 Putative retroelement pol polyprotein [Arabidop... 672 0.0
UniRef100_Q9SLF0 Putative retroelement pol polyprotein [Arabidop... 671 0.0
UniRef100_Q9SIM3 Putative retroelement pol polyprotein [Arabidop... 668 0.0
UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana] 655 0.0
UniRef100_Q9FX79 Putative retroelement polyprotein [Arabidopsis ... 654 0.0
UniRef100_Q9FIC5 Retroelement pol polyprotein-like [Arabidopsis ... 654 0.0
UniRef100_Q8L700 Hypothetical protein [Arabidopsis thaliana] 650 0.0
UniRef100_Q5XWR5 Putative retroelement pol polyprotein-like [Sol... 649 0.0
UniRef100_O23588 Retrotransposon like protein [Arabidopsis thali... 634 e-180
UniRef100_Q9ZVE4 Putative retroelement pol polyprotein [Arabidop... 629 e-178
UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis ... 611 e-173
UniRef100_O65452 LTR retrotransposon like protein [Arabidopsis t... 605 e-171
UniRef100_Q9FL75 Retroelement pol polyprotein-like [Arabidopsis ... 605 e-171
UniRef100_O81617 F8M12.17 protein [Arabidopsis thaliana] 603 e-170
UniRef100_Q710T7 Gag-pol polyprotein [Populus deltoides] 581 e-164
>UniRef100_Q9C692 Polyprotein, putative [Arabidopsis thaliana]
Length = 1468
Score = 731 bits (1888), Expect = 0.0
Identities = 381/783 (48%), Positives = 525/783 (66%), Gaps = 27/783 (3%)
Query: 548 QTYL-FYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMN 606
Q+YL F GEC+L+AAYLIN+ P+ +L+ KSP+++L + P YS LRVFG LC+A N N
Sbjct: 690 QSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYAHNQN 749
Query: 607 IQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQ--------- 656
+ KF RS+ +FVGYP QKG+R++D++ ++ +VSRDV F ET FPY
Sbjct: 750 HKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSCNEEDE 809
Query: 657 ----DIQSPPF-NNAISINTQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTP 711
D PPF AI T I N + + I PE + +S V++S+
Sbjct: 810 RVLVDCVGPPFIEEAIGPRTIIGRNIGEATVGPNVATGPIIPEINQESSSPSEFVSLSSL 869
Query: 712 EDDASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNV------- 764
+ +S+ + LS+ P+ P R S R P+ LK+++ +V
Sbjct: 870 DPFLASSTVQTADLPLSSTTPA----PIQLRRSSRQTQKPMKLKNFVTNTVSVESISPEA 925
Query: 765 TSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESN 824
+S +P+E Y+ ++SH+AFL + EP +Y++AM WR+AM+ EI++L N
Sbjct: 926 SSSSLYPIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVN 985
Query: 825 NTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAK 884
T+ + LP GK A+G KW+YKIKY SDG+I+RYKARLV G Q +G+DY +TFAPVAK
Sbjct: 986 QTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAK 1045
Query: 885 LVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSIY 944
+ TVRL L +AA ++W ++Q DV+NAFL GDL EEVYMKLP GF VC+L+KS+Y
Sbjct: 1046 MSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLY 1105
Query: 945 GLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAI 1004
GLKQA R WFSK S+ L Q GF QS+SDYSLF+YN D + VLVYVDD+II+G+ +A+
Sbjct: 1106 GLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGIFVHVLVYVDDLIISGSCPDAV 1165
Query: 1005 SKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFP 1064
++ K +L F +KDLG L Y LGIEVSR+ +G +L QRKY LDI+S+ G+ G RPS FP
Sbjct: 1166 AQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFP 1225
Query: 1065 MEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAA 1124
+EQ+ +L + LSD + YRR VGRL+YL VTRP++ Y+V+TL+QFMQ+P H++AA
Sbjct: 1226 LEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPELSYSVHTLAQFMQNPRQDHWNAA 1285
Query: 1125 TQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTK 1184
+V+RYLK + G+G+ LS++S++ + G+ DSD+A CP TRRS TGYF LG PISWKTK
Sbjct: 1286 IRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKTK 1345
Query: 1185 KQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVF 1244
KQPT+SRSSAEAEYR++A L+ EL WLK +L DLG+ H Q + I+ DS++AI ++ NPV
Sbjct: 1346 KQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPVQ 1405
Query: 1245 HERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEIS 1304
HERTKH+E+DCHF+R+ I G+IA S++ S QLADI TK LG + L KLG++++
Sbjct: 1406 HERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGILDVH 1465
Query: 1305 IPP 1307
PP
Sbjct: 1466 APP 1468
Score = 180 bits (457), Expect = 2e-43
Identities = 135/495 (27%), Positives = 223/495 (44%), Gaps = 93/495 (18%)
Query: 26 MSDIPPSENSPLSSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVT 85
M+D PP S I V T +P +DN V+ P+L +NY W+
Sbjct: 1 MADAPPPP-------PSVIEVRRTISPYDLTA--ADNSGAVISHPILKTNNYEEWACGFK 51
Query: 86 MALRAKNKFGFVDGSLTIPKK-KEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQ 144
ALR++ KFGF+DG++ P D+ W N L+ SW+ ++ +E+ +I + + A
Sbjct: 52 TALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLTNISHRDVARD 111
Query: 145 IWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICG 204
+W ++ RFS SN PK ++K ++ KQEGM V Y+ +L +WD +NS + C CG
Sbjct: 112 LWEQIRKRFSVSNGPKNQKMKADLATCKQEGMTVEGYYGKLNKIWDNINSYRPLRICKCG 171
Query: 205 NAKSIL--DQQ---NQDRAMEFLQGVHD-RFSAVRSQILLMDPFPSIQRIYNIVRQEEKQ 258
L DQ+ D ++L G+++ +F +RS + P P ++ +YNIVRQEE
Sbjct: 172 RCICNLGTDQEKYREDDMVHQYLYGLNETKFHTIRSSLTSRVPLPGLEEVYNIVRQEEDM 231
Query: 259 QE---------------INFRPVPAVESAALQTSKAPYRTPGKRQRPFCEHCNRHGHTIT 303
+ RP V S S+ + + C HCNR GH+
Sbjct: 232 VNNRSSNEERTDVTAFAVQMRPRSEVISEKFANSEK------LQNKKLCTHCNRGGHSPE 285
Query: 304 TCYQIHGFPS-NPSKPQKKTETSPSTSANQ------------------------------ 332
C+ + G+P +P+ K+ ++ STS +
Sbjct: 286 NCFVLIGYPEWWGDRPRGKSNSNGSTSRGRGRFGPGFNGGQPRPTYVNVVMTGPFPSSEH 345
Query: 333 ---------------LSSAQYHKLLTLLAKEDNVGSSVNLAGT----AFTCIPF-SWIID 372
L+ Q+ ++ LL N G S N + + TC F SWI+D
Sbjct: 346 VNRVITDSDRDAVSGLTDEQWRGVVKLL----NAGRSDNKSNAHETQSGTCSLFTSWILD 401
Query: 373 SGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFK 432
+GAS+H+ +L L S + + + + DG++ + GT+ LIL +V++V +
Sbjct: 402 TGASHHMTGNLELLSDMRSM-SPVLIILADGNKRVAVSEGTVRLGSHLILKSVFYVKELE 460
Query: 433 FNLMSVTQLTESLNC 447
+L+SV Q+ + +C
Sbjct: 461 SDLISVGQMMDENHC 475
>UniRef100_Q9ZPU4 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1501
Score = 726 bits (1875), Expect = 0.0
Identities = 376/804 (46%), Positives = 513/804 (63%), Gaps = 48/804 (5%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ + F GE +LTAAYLIN+ P+ IL ++P++VL GS P YS LRVFG C+ +
Sbjct: 702 FQASLPIKFWGESILTAAYLINRTPSSILSGRTPYEVLHGSKPVYSQLRVFGSACYVHRV 761
Query: 606 NI-QHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
+ KF +RS+ IFVGYPF +KG+++YD++ + VSRDV F E VFPY + N
Sbjct: 762 TRDKDKFGQRSRSCIFVGYPFGKKGWKVYDIERNEFLVSRDVIFREEVFPYAGV-----N 816
Query: 665 NAISINTQILDNEFDDLFTGSPTHPNIPPENSHNDN---SNDTIVTIST----------- 710
++ +T + DD + P ++ + + D +V ++
Sbjct: 817 SSTLASTSLPTVSEDDDWAIPPLEVRGSIDSVETERVVCTTDEVVLDTSVSDSEIPNQEF 876
Query: 711 -PEDDASSNPPSFSAESLSNNPPST---------EMNPPNHRHSQRIRNPPIHLKDYIC- 759
P+D S+P S S N P + ++PP R S+R +PP L DY+
Sbjct: 877 VPDDTPPSSPLSVSPSGSPNTPTTPIVVPVASPIPVSPPKQRKSKRATHPPPKLNDYVLY 936
Query: 760 -----------------KINNVTSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYS 802
+ + V K FPL +Y+S + S+SHRA+L I +N EPK +
Sbjct: 937 NAMYTPSSIHALPADPSQSSTVPGKSLFPLTDYVSDAAFSSSHRAYLAAITDNVEPKHFK 996
Query: 803 QAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARL 862
+A++ W DAM E+ ALE N TW + LP GK AIG +W++K KY+SDG+++RYKARL
Sbjct: 997 EAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFKTKYNSDGTVERYKARL 1056
Query: 863 VAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYM 922
V +G QV+G DY +TFAPV ++ TVR LL A W +YQ DV+NAFL GDL EEVYM
Sbjct: 1057 VVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMDVHNAFLHGDLEEEVYM 1116
Query: 923 KLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQ 982
KLPPGF H VC+L KS+YGLKQA R WF K S +L++ GF QS DYSLF+Y +
Sbjct: 1117 KLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGFVQSYEDYSLFSYTRNN 1176
Query: 983 TTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQ 1042
+ VL+YVDD++I GN+ + K K +L++ FS+KDLG L Y LGIEVSR +GIFL Q
Sbjct: 1177 IELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIEVSRGPEGIFLSQ 1236
Query: 1043 RKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDI 1102
RKY LD+++DSG G RP+ P+EQ+ L +DG LSDP YRR VGRLLYL TRP++
Sbjct: 1237 RKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVGRLLYLLHTRPEL 1296
Query: 1103 QYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPT 1162
Y+V+ L+QFMQ+P +HFDAA +V+RYLKGS G+G+ L+A + L Y DSDW CP
Sbjct: 1297 SYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILLNADPDLTLEVYCDSDWQSCPL 1356
Query: 1163 TRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDH 1222
TRRS + Y +LG +PISWKTKKQ T+S SSAEAEYR+++ E++WL+ LL +LGI+
Sbjct: 1357 TRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSYALKEIKWLRKLLKELGIEQ 1416
Query: 1223 PQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIF 1282
P +YCDS+AAIHIA NPVFHERTKHIE DCH VR+ ++ G+I ++R+++QLAD+F
Sbjct: 1417 STPARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDAVRDGIITTQHVRTTEQLADVF 1476
Query: 1283 TKPLGGDAYKRILGKLGVIEISIP 1306
TK LG + + ++ KLGV + P
Sbjct: 1477 TKALGRNQFLYLMSKLGVQNLHTP 1500
Score = 213 bits (542), Expect = 3e-53
Identities = 152/519 (29%), Positives = 252/519 (48%), Gaps = 72/519 (13%)
Query: 54 PCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKE-DILT 112
P + SDNP V+ + L GDNY W+ + AL+AK K GF++G++ P + +
Sbjct: 31 PYTLASSDNPGAVISSVELNGDNYNQWATEMLNALQAKRKTGFINGTIPRPPPNDPNYEN 90
Query: 113 WQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALK 172
W N ++ WI S+ +++ ++ + A +W DL+ RFS N +I+Q++ +S+ +
Sbjct: 91 WTAVNSMIVGWIRTSIEPKVKATVTFISDAHLLWKDLKQRFSVGNKVRIHQIRAQLSSCR 150
Query: 173 QEGMYVSLYFTQLKSLWDELN-----SIALVNPCICGNAKSILDQQNQDRAMEFLQGVHD 227
Q+G V Y+ +L +LW+E N ++ C CG ++ +++ +F+ G+ +
Sbjct: 151 QDGQAVIEYYGRLSNLWEEYNIYKPVTVCTCGLCRCGATSEPTKEREEEKIHQFVLGLDE 210
Query: 228 -RFSAVRSQILLMDPFPSIQRIYN-IVRQ----------EEKQQEINF---RPVPAVESA 272
RF + + ++ MDP PS+ IY+ ++R+ E+K++ + F R S
Sbjct: 211 SRFGGLCATLINMDPLPSLGEIYSRVIREEQRLASVHVREQKEEAVGFLARREQLDHHSR 270
Query: 273 ALQTSKAPYRTPGKRQ------RPFCEHCNRHGHTITTCYQIHGFP-------------- 312
+S T G R R C +C R GH C+QI GFP
Sbjct: 271 VDASSSRSEHTGGSRSNSIIKGRVTCSNCGRTGHEKKECWQIVGFPDWWSERNGGRGSNG 330
Query: 313 -------SNPSKPQKKTETSPSTSANQL----SSAQYHKLLTLLAKE-DNVGSSVN---- 356
SN + Q + + +TS+N + ++ ++L+ L KE N GS+ N
Sbjct: 331 RGRGGRGSNGGRGQGQVMAAHATSSNSSVFPEFTEEHMRVLSQLVKEKSNSGSTSNNNSD 390
Query: 357 -LAG-TAFTCIPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTI 414
L+G T I I+DSGAS+H+ +LS ++ PV V DGS+A +G +
Sbjct: 391 RLSGKTKLGDI----ILDSGASHHMTGTLSSLTNVVPV-PPCPVGFADGSKAFALSVGVL 445
Query: 415 NCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARN 474
S ++ LTNV VP+ L+SV++L + C A F+ + C QD+++K +IG G R
Sbjct: 446 TLSNTVSLTNVLFVPSLNCTLISVSKLLKQTQCLATFTDTLCFLQDRSSKTLIGSGEERG 505
Query: 475 ELYYLNQDLVSNKH----DKD----HYCLSHPLSSLFSS 505
+YYL + H D D H L HP S+ SS
Sbjct: 506 GVYYLTDVTPAKIHTANVDSDQALWHQRLGHPSFSVLSS 544
>UniRef100_Q9LVQ2 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1491
Score = 719 bits (1856), Expect = 0.0
Identities = 381/808 (47%), Positives = 502/808 (61%), Gaps = 49/808 (6%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ + F GE V+TAAYLIN+ P+ I SP+++L G P Y LRVFG C+A +
Sbjct: 685 FQASLPIKFWGEAVMTAAYLINRTPSSIHNGLSPYELLHGCKPDYDQLRVFGSACYAHRV 744
Query: 606 NI-QHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPY-----QDIQ 659
+ KF ERS+ IFVGYPF QKG+++YD+ T + VSRDV F E VFPY I
Sbjct: 745 TRDKDKFGERSRLCIFVGYPFGQKGWKVYDLSTNEFIVSRDVVFRENVFPYATNEGDTIY 804
Query: 660 SPPFNNAISIN------TQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPED 713
+PP I+ + T + D D+ P P + S +D +DT ++ TP D
Sbjct: 805 TPPVTCPITYDEDWLPFTTLEDRGSDENSLSDP--PVCVTDVSESDTEHDTPQSLPTPVD 862
Query: 714 D-----ASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPP-----------IHLKDY 757
D S P S S+ PST ++PP + I N P LKDY
Sbjct: 863 DPLSPSTSVTPTQTPTNSSSSTSPSTNVSPPQQDTTPIIENTPPRQGKRQVQQLARLKDY 922
Query: 758 I-----CKIN--------------NVTSKINFPLENYLSLSNLSNSHRAFLINIIENKEP 798
I C N ++ +PL +Y+ S H+ FL I N EP
Sbjct: 923 ILYNASCTPNTPHVLSPSTSQSSSSIQGNSQYPLTDYIFDECFSAGHKVFLAAITANDEP 982
Query: 799 KSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRY 858
K + +A+K W DAM KE+ ALE N TW + LP GK AIG +W+YK K+++DG+++RY
Sbjct: 983 KHFKEAVKVKVWNDAMYKEVDALEVNKTWDIVDLPTGKVAIGSQWVYKTKFNADGTVERY 1042
Query: 859 KARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSE 918
KARLV +G +Q++G DY +TFAPV K+ TVR LL + A W +YQ DV+NAFL GDL E
Sbjct: 1043 KARLVVQGNNQIEGEDYTETFAPVVKMTTVRTLLRLVAANQWEVYQMDVHNAFLHGDLEE 1102
Query: 919 EVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTY 978
EVYMKLPPGF H VC+L KS+YGLKQA R WF K S L + GF Q DYS F+Y
Sbjct: 1103 EVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDALKRFGFIQGYEDYSFFSY 1162
Query: 979 NCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGI 1038
+C + VLVYVDD+II GN+E + K K++L + FS+KDLG L Y LGIEVSR GI
Sbjct: 1163 SCKGIELRVLVYVDDLIICGNDEYMVQKFKEYLGRCFSMKDLGKLKYFLGIEVSRGPDGI 1222
Query: 1039 FLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVT 1098
FL QRKY LDI+SDSG G RP+ P+EQ+ L +DG L DP +RR VGRLLYL T
Sbjct: 1223 FLSQRKYALDIISDSGTLGARPAYTPLEQNHHLASDDGPLLQDPKPFRRLVGRLLYLLHT 1282
Query: 1099 RPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWA 1158
RP++ Y+V+ LSQFMQ+P +H +AA +++RYLKGS G+G+ LS++ + L Y DSD+
Sbjct: 1283 RPELSYSVHVLSQFMQAPREAHLEAAMRIVRYLKGSPGQGILLSSNKDLTLEVYCDSDFQ 1342
Query: 1159 GCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDL 1218
CP TRRS + Y +LG +PISWKTKKQ T+S SSAEAEYR+++ E++WL LL +L
Sbjct: 1343 SCPLTRRSLSAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSVALKEIKWLNKLLKEL 1402
Query: 1219 GIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQL 1278
GI P ++CDS+AAI IA NPVFHERTKHIE DCH VR+ ++ G+I ++R+S+QL
Sbjct: 1403 GITLAAPTRLFCDSKAAISIAANPVFHERTKHIERDCHSVRDAVRDGIITTHHVRTSEQL 1462
Query: 1279 ADIFTKPLGGDAYKRILGKLGVIEISIP 1306
ADIFTK LG + + ++ KLG+ + P
Sbjct: 1463 ADIFTKALGRNQFIYLMSKLGIQNLHTP 1490
Score = 228 bits (582), Expect = 7e-58
Identities = 148/526 (28%), Positives = 258/526 (48%), Gaps = 62/526 (11%)
Query: 37 LSSISSPITVHS--TTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKF 94
+SS + P T T P + SDNP ++ + +LTGDNY WS + AL+AK K
Sbjct: 7 VSSATHPRTNQQPDVTKVSPYTLASSDNPGAMISSVMLTGDNYNEWSTEMLNALQAKRKT 66
Query: 95 GFVDGSLTIPK-KKEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRF 153
GF++GS++ P D WQ N ++ WI S+ +++ ++ + A Q+WS+L+ RF
Sbjct: 67 GFINGSISKPPLDNPDYENWQAVNSMIVGWIRASIEPKVKSTVTFISDAHQLWSELKQRF 126
Query: 154 SQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELN-----SIALVNPCICGNAKS 208
S N +++Q+K ++A +Q+G V Y+ +L LW+E ++ C CG
Sbjct: 127 SVGNKVRVHQIKAQLAACRQDGQPVIDYYGRLCKLWEEFQIYKPITVCKCGLCTCGATLE 186
Query: 209 ILDQQNQDRAMEFLQGVHD-RFSAVRSQILLMDPFPSIQRIYN-IVRQEEKQQEINFRPV 266
++ +++ +F+ G+ D RF + + ++ MDPFPS+ IY+ +VR+E++ + R
Sbjct: 187 PSKEREEEKIHQFVLGLDDSRFGGLSATLIAMDPFPSLGEIYSRVVREEQRLASVQIRE- 245
Query: 267 PAVESAALQTSKAPYRTPGK--------RQRP-FCEHCNRHGHTITTCYQIHGFP----- 312
+ T ++ G+ R R C HC R GH C+QI GFP
Sbjct: 246 QQQSAIGFLTRQSEVTADGRTDSSIIKSRDRSVLCSHCGRSGHEKKDCWQIVGFPDWWTE 305
Query: 313 ---------------------SNPSKPQKKTETSPSTSANQLSSAQYH----KLLTLLAK 347
+N + + + + +T++N S ++ +++T + +
Sbjct: 306 RTNGGGRGSSSRGRGGRSSGSNNSGRGRGQVTAAHATTSNLSSFPEFTPDQLRVITQMIQ 365
Query: 348 EDNVGSSVNLAGTAFTCIPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQAL 407
N G+S L+G I+D+GAS+H+ LSL ++ + + SV D +
Sbjct: 366 NKNNGTSDKLSGKMKL---GDVILDTGASHHMTGQLSLLTNIVTIPS-CSVGFADDRKTF 421
Query: 408 VKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMI 467
+GT S ++ L+NV +VP +L+SV++L + + C A+F+ + CV QD+ ++ +I
Sbjct: 422 AISMGTFKLSETVSLSNVLYVPALNCSLISVSKLVKQIKCLALFTDTICVLQDRFSRTLI 481
Query: 468 GRGSARNELYYLNQDLVSNKHDKD--------HYCLSHPLSSLFSS 505
G G R+ +YYL + H D H L HP S+ SS
Sbjct: 482 GTGEERDGVYYLTDAATTTVHKVDVTTDHALWHQRLGHPSFSVLSS 527
>UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1475
Score = 706 bits (1822), Expect = 0.0
Identities = 356/758 (46%), Positives = 501/758 (65%), Gaps = 61/758 (8%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQ-HKFD 612
F G+C+L+A +LIN+LPTP+L KSP ++L P Y+SL+VFGCLC+ Q HKF
Sbjct: 767 FWGDCILSAVFLINRLPTPLLSNKSPFELLHLKVPDYTSLKVFGCLCYESTSPQQRHKFA 826
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQ 672
R++ +F+GYP KGY++ D++T I++SR V F+ETVFP+ D
Sbjct: 827 PRARACVFLGYPSGYKGYKLLDLETNTIHISRHVVFYETVFPFTD-------------KT 873
Query: 673 ILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPP 732
I+ + DL P H NI NPP
Sbjct: 874 IIPRDVFDLV--DPVHENI-------------------------------------ENPP 894
Query: 733 STEMNPPNHRHSQRIRNPPIHLKDYICK-INNVTSKINFPLENYLSLSNLSNSHRAFLIN 791
ST + P S+R PP +L+DY C + +VT + +PL Y++ + LS A++
Sbjct: 895 STSESAPK-VSSKRESRPPGYLQDYFCNAVPDVTKDVRYPLNAYINYTQLSEEFTAYICA 953
Query: 792 IIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHS 851
+ + EP +Y+QA K EW DAM EI ALES NTW +C LP+GK IGCKW++K+K ++
Sbjct: 954 VNKYPEPCTYAQAKKIKEWLDAMEIEIDALESTNTWSVCSLPQGKKPIGCKWVFKVKLNA 1013
Query: 852 DGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAF 911
DGS++R+KARLVAKGY+Q +G+DY+DTF+PVAK+ TV+ LLS+AAIK W L+Q D++NAF
Sbjct: 1014 DGSLERFKARLVAKGYTQREGLDYYDTFSPVAKMTTVKTLLSVAAIKEWSLHQLDISNAF 1073
Query: 912 LQGDLSEEVYMKLPPGFSHKK-----KPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGF 966
L GDL EE+YM LPPG+S K+ + V KL KS+YGLKQASRQW+ KFS+TL + GF
Sbjct: 1074 LNGDLKEEIYMTLPPGYSMKQGGVLPQNPVLKLQKSLYGLKQASRQWYLKFSSTLKKLGF 1133
Query: 967 RQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYI 1026
++S +D++LFT + I +LVYVDDI+I GNN+ I ++KK LA++F ++DLG + Y
Sbjct: 1134 KKSHADHTLFTRISGKAYIALLVYVDDIVIAGNNDENIEELKKDLAKAFKLRDLGPMKYF 1193
Query: 1027 LGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRL-RPNDGTPLSDPTVY 1085
LG+E++R+K+GI +CQRKYT+++L D+G+ GCRPS PME L+L + ND + +P VY
Sbjct: 1194 LGLEIARTKEGISVCQRKYTMELLEDTGLLGCRPSTIPMEPSLKLSQHNDEHVIDNPEVY 1253
Query: 1086 RRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASS 1145
RR VG+L+YLT+TRPDI YA+N L QF SP +SH AA +V+ YLKG++G GLF S+ S
Sbjct: 1254 RRLVGKLMYLTITRPDITYAINRLCQFSSSPKNSHLKAAQKVVHYLKGTIGLGLFYSSKS 1313
Query: 1146 SINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLS 1205
+ L Y D+DW C +RRST+G LG + ISWK+KKQ S SSAE+EYR++A S
Sbjct: 1314 DLCLKAYTDADWGSCVDSRRSTSGICMFLGDSLISWKSKKQNMASSSSAESEYRAMAMGS 1373
Query: 1206 SELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSG 1265
E+ WL LL++ + +P+ ++CDS AAIHIA N VFHERTKHIE DCH R++I+ G
Sbjct: 1374 REIAWLVKLLAEFQVKQTKPVPLFCDSTAAIHIANNAVFHERTKHIENDCHITRDRIEQG 1433
Query: 1266 LIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEI 1303
++ ++ ++ QLAD+ TKPL + ++GK+ ++ I
Sbjct: 1434 MLKTMHVDTTSQLADVLTKPLFPTLFNSLIGKMSLLSI 1471
Score = 256 bits (653), Expect = 4e-66
Identities = 172/544 (31%), Positives = 259/544 (46%), Gaps = 83/544 (15%)
Query: 48 STTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKK 107
S + P + + D+P LV+ +L G N+ SW A+ ++L AKNK FVDG+L P +
Sbjct: 57 SDAHYSPFALSNGDSPGNTLVSEVLDGTNFSSWKIAMFVSLYAKNKIAFVDGTLPRPPES 116
Query: 108 E-DILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQ 166
+ W RCN +V SWILNSV+ +I SIL A +IW DL RF +N P+ YQL Q
Sbjct: 117 DPSFRVWSRCNSMVKSWILNSVTKQIYKSILRFNDAAEIWKDLDTRFHITNLPRSYQLTQ 176
Query: 167 SISALKQEGMYVSLYFTQLKSLWDELNSIALVNPC-ICGNAKSILDQQNQDRAMEFLQGV 225
I +L+Q M +S Y+T LK+LWD+L+ + V+ C C + + ++FL G+
Sbjct: 177 QIWSLQQGTMSLSDYYTALKTLWDDLDGASCVSTCKNCTCCIATASMIEHSKIVKFLSGL 236
Query: 226 HDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRP-------VPAVESAALQTSK 278
++ +S +RSQI++ P + IYN++ Q+ Q+ I P V A +S +
Sbjct: 237 NESYSTIRSQIIMKKTIPDLAEIYNLLDQDHSQRNIVTMPTNASTFNVSAPQSDQFAVNL 296
Query: 279 APYRTPGKRQRPFCEHCNRHGHTITTCYQIHGFPSNPSKPQKKT----ETSPSTSAN--- 331
A + + C HC GH TCY+IHG+P KKT E S AN
Sbjct: 297 AKSFGTQPKPKVQCSHCGYTGHNADTCYKIHGYPVGFKHKDKKTVTPSEKPKSVVANLAL 356
Query: 332 ---------------------QLSSAQYHKLLTLLAKE----------DNVGSSVNLAGT 360
+S +Q ++ + + + S+ N G+
Sbjct: 357 TDGKVSVTQGIGPDGIVELVGSMSKSQIQDVIAYFSTQLHNPAKPITVASFASTNNDNGS 416
Query: 361 AFTCIPFS---------------------WIIDSGASNHICTSLSLFSSYYP-VNNQISV 398
FT I FS WIIDSGA++H+ +LF S ++N++++
Sbjct: 417 TFTGISFSPSTLRLLCSLTSSKKVLSLNTWIIDSGATHHVSYDRNLFESLSDGLSNEVTL 476
Query: 399 QQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVF 458
P GS + IG I + +L L NV ++P F+ NL+SV+Q T+ + C F CV
Sbjct: 477 --PTGSNVKIAGIGVIKLNSNLTLKNVLYIPEFRLNLLSVSQQTKDMKCKIYFDEDCCVI 534
Query: 459 QDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYCLSHPL-SSLFSSKHCNKF---DLW 514
QD ++ IGRG+ LY L+ V C S + SS+ ++CN LW
Sbjct: 535 QDPIKEQKIGRGNQIGGLYVLDTSSVE--------CTSVDINSSVTEKQYCNAVVDSALW 586
Query: 515 HLRL 518
H RL
Sbjct: 587 HSRL 590
>UniRef100_O22175 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1496
Score = 691 bits (1783), Expect = 0.0
Identities = 367/809 (45%), Positives = 500/809 (61%), Gaps = 60/809 (7%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAK-NMNIQHKFD 612
F GE +LTA +LIN+ P+ +LK K+P+++L G PSY LR FGCLC+A + KF
Sbjct: 683 FWGESILTATHLINRTPSAVLKGKTPYELLFGERPSYDMLRSFGCLCYAHIRPRNKDKFT 742
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQ---------SPPF 663
RS+ +F+GYP +K +R+YD++T +I+ SRDV+FHE ++PY +PP
Sbjct: 743 SRSRKCVFIGYPHGKKAWRVYDLETGKIFASRDVRFHEDIYPYATATQSNVPLPPPTPPM 802
Query: 664 NNA---ISINTQILDNEFDDLFTGSPTHPNI---PPENSHNDNSNDT---------IVTI 708
N + I+TQ+ D + SP PP + S T IV
Sbjct: 803 VNDDWFLPISTQVDSTNVDSSSSSSPAQSGSIDQPPRSIDQSPSTSTNPVPEEIGSIVPS 862
Query: 709 STPEDDASSNPPSFSA----ESLSNNPPSTEMNPPNHR---HSQRIRNPPIHLKDYICKI 761
S+P + SA E LS ST +P R + + LKD++
Sbjct: 863 SSPSRSIDRSTSDLSASDTTELLSTGESSTPSSPGLPELLGKGCREKKKSVLLKDFVTNT 922
Query: 762 ----------------------------NNVTSKINFPLENYLSLSNLSNSHRAFLINII 793
++V+ K +PL ++L+ S S +H AF+ I+
Sbjct: 923 TSKKKTASHNIHSPSQVLPSGLPTSLSADSVSGKTLYPLSDFLTNSGYSANHIAFMAAIL 982
Query: 794 ENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDG 853
++ EPK + A+ EW +AM+KEI ALE+N+TW + LP GK AI KW+YK+KY+SDG
Sbjct: 983 DSNEPKHFKDAILIKEWCEAMSKEIDALEANHTWDITDLPHGKKAISSKWVYKLKYNSDG 1042
Query: 854 SIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQ 913
+++R+KARLV G Q +G+D+ +TFAPVAKL TVR +L++AA K+W ++Q DV+NAFL
Sbjct: 1043 TLERHKARLVVMGNHQKEGVDFKETFAPVAKLTTVRTILAVAAAKDWEVHQMDVHNAFLH 1102
Query: 914 GDLSEEVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDY 973
GDL EEVYM+LPPGF VC+L KS+YGLKQA R WFSK ST L GF QS DY
Sbjct: 1103 GDLEEEVYMRLPPGFKCSDPSKVCRLRKSLYGLKQAPRCWFSKLSTALRNIGFTQSYEDY 1162
Query: 974 SLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSR 1033
SLF+ T I VLVYVDD+I+ GNN +AI + K L + F +KDLG L Y LG+EVSR
Sbjct: 1163 SLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDRFKSQLHKCFHMKDLGKLKYFLGLEVSR 1222
Query: 1034 SKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLL 1093
G L QRKY LDI+ ++G+ GC+PS P+ + +L G ++P YRR VGR +
Sbjct: 1223 GPDGFCLSQRKYALDIVKETGLLGCKPSAVPIALNHKLASITGPVFTNPEQYRRLVGRFI 1282
Query: 1094 YLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYA 1153
YLT+TRPD+ YAV+ LSQFMQ+P +H++AA +++RYLKGS +G+FL + SS+ + Y
Sbjct: 1283 YLTITRPDLSYAVHILSQFMQAPLVAHWEAALRLVRYLKGSPAQGIFLRSDSSLIINAYC 1342
Query: 1154 DSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKY 1213
DSD+ CP TRRS + Y LG +PISWKTKKQ T+S SSAEAEYR++A EL+WLK
Sbjct: 1343 DSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTVSYSSAEAEYRAMAYTLKELKWLKA 1402
Query: 1214 LLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIR 1273
LL DLG+ H P+ ++CDS+AAIHIA NPVFHERTKHIE DCH VR+ + LI +I
Sbjct: 1403 LLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTKHIESDCHKVRDAVLDKLITTEHIY 1462
Query: 1274 SSDQLADIFTKPLGGDAYKRILGKLGVIE 1302
+ DQ+AD+ TK L ++R+L LGV +
Sbjct: 1463 TEDQVADLLTKSLPRPTFERLLSTLGVTD 1491
Score = 239 bits (609), Expect = 5e-61
Identities = 162/527 (30%), Positives = 253/527 (47%), Gaps = 58/527 (11%)
Query: 37 LSSISSPITVHSTTNPDPC---VIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNK 93
+SS S+ +T S++N +I+ SDNP ++ + +L +NY WS + LRAK K
Sbjct: 1 MSSASALVTSTSSSNTSSTTAYLINASDNPGALISSVVLKENNYAEWSEELQNFLRAKQK 60
Query: 94 FGFVDGSLTIPKKKEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRF 153
GF+DGS+ P ++ W N ++ WI S+ IR ++ + A Q+W +L+ RF
Sbjct: 61 LGFIDGSIPKPAADPELSLWIAINSMIVGWIRTSIDPTIRSTVGFVSEASQLWENLRRRF 120
Query: 154 SQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICGNAKSILDQQ 213
S N + LK I+A Q+G V Y+ +L LW+EL + C C A I ++
Sbjct: 121 SVGNGVRKTLLKDEIAACTQDGQPVLAYYGRLIKLWEELQNYKSGRECKCEAASDIEKER 180
Query: 214 NQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAA 273
DR +FL G+ RFS++RS I ++P P + ++Y+ V +EE+ + R V++ A
Sbjct: 181 EDDRVHKFLLGLDSRFSSIRSSITDIEPLPDLYQVYSRVVREEQNLNAS-RTKDVVKTEA 239
Query: 274 LQTSKAPYRTPGKRQRP--FCEHCNRHGHTITTCYQIHGFP-----SNPSKPQKKT---- 322
+ S TP R + FC HCNR GH +T C+ +HG+P NP + Q T
Sbjct: 240 IGFSVQSSTTPRFRDKSTLFCTHCNRKGHEVTQCFLVHGYPDWWLEQNPQENQPSTRGRG 299
Query: 323 --------------ETSPST----SANQLSSA----------QYHKLLTLLAKEDNVGSS 354
++P+T AN +A Q +L++LL + SS
Sbjct: 300 SNGRGSSSGRGGNRSSAPTTRGRGRANNAQAAAPTVSGDGNDQIAQLISLLQAQRPSSSS 359
Query: 355 VNLAGTAFTCIPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTI 414
L+G TC+ +ID+GAS+H+ S+ + + V +PDG + GT+
Sbjct: 360 ERLSGN--TCLT-DGVIDTGASHHMTGDCSILVDVFDITPS-PVTKPDGKASQATKCGTL 415
Query: 415 NCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARN 474
S L +V VP F L+SV++L + + AIF+ + C QD+ + +IG G R
Sbjct: 416 LLHDSYKLHDVLFVPDFDCTLISVSKLLKQTSSIAIFTDTFCFLQDRFLRTLIGAGEERE 475
Query: 475 ELYYLNQDLVSNKH----------DKDHYCLSHPLSS-LFSSKHCNK 510
+YY L H D H L HP +S L S CN+
Sbjct: 476 GVYYFTGVLAPRVHKASSDFAISGDLWHRRLGHPSTSVLLSLPECNR 522
>UniRef100_Q9XII7 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1454
Score = 672 bits (1735), Expect = 0.0
Identities = 338/754 (44%), Positives = 495/754 (64%), Gaps = 51/754 (6%)
Query: 556 GECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQ-HKFDER 614
G+CVLTA +LIN+ P+ +L K+P+++L G+ P Y LR FGCLC++ Q HKF R
Sbjct: 741 GDCVLTAVFLINRTPSQLLMNKTPYEILTGTAPVYEQLRTFGCLCYSSTSPKQRHKFQPR 800
Query: 615 SKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQIL 674
S+ +F+GYP KGY++ D+++ +++SR+VQFHE VFP ++P +++ + T ++
Sbjct: 801 SRACLFLGYPSGYKGYKLMDLESNTVFISRNVQFHEEVFPL--AKNPGSESSLKLFTPMV 858
Query: 675 DNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPPST 734
P I + +H+ PS +S+ PP
Sbjct: 859 -----------PVSSGIISDTTHS---------------------PSSLPSQISDLPPQI 886
Query: 735 EMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHRAFLINIIE 794
SQR+R PP HL DY C N + S +P+ + +S S +S SH ++ NI +
Sbjct: 887 S--------SQRVRKPPAHLNDYHC--NTMQSDHKYPISSTISYSKISPSHMCYINNITK 936
Query: 795 NKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGS 854
P +Y++A + EW +A+ EI A+E NTW + LP+GK A+GCKW++ +K+ +DG+
Sbjct: 937 IPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGN 996
Query: 855 IDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQG 914
++RYKARLVAKGY+Q +G+DY DTF+PVAK+ T++LLL ++A K W L Q DV+NAFL G
Sbjct: 997 LERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNG 1056
Query: 915 DLSEEVYMKLPPGFSHKK-----KPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQS 969
+L EE++MK+P G++ +K V +L +SIYGLKQASRQWF KFS++L+ GF+++
Sbjct: 1057 ELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKT 1116
Query: 970 ISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGI 1029
D++LF D + VLVYVDDI+I +E A +++ + L Q F ++DLG+L Y LG+
Sbjct: 1117 HGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGL 1176
Query: 1030 EVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHV 1089
EV+R+ GI +CQRKY L++L +GM C+P PM +L++R +DG + D YRR V
Sbjct: 1177 EVARTTAGISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIV 1236
Query: 1090 GRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINL 1149
G+L+YLT+TRPDI +AVN L QF +P ++H AA +VL+Y+KG+VG+GLF SASS + L
Sbjct: 1237 GKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTL 1296
Query: 1150 VGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQ 1209
G+ADSDWA C +RRSTT + +G + ISW++KKQ T+SRSSAEAEYR+LA + E+
Sbjct: 1297 KGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMV 1356
Query: 1210 WLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAP 1269
WL LL L P PI +Y DS AAI+IA NPVFHERTKHI++DCH VRE++ +G +
Sbjct: 1357 WLFTLLVSLQASPPVPI-LYSDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKL 1415
Query: 1270 SYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEI 1303
++R+ DQ+ADI TKPL ++ + K+ ++ I
Sbjct: 1416 LHVRTEDQVADILTKPLFPYQFEHLKSKMSILNI 1449
Score = 255 bits (652), Expect = 5e-66
Identities = 159/511 (31%), Positives = 251/511 (49%), Gaps = 63/511 (12%)
Query: 30 PPSENSP---LSSISSPITVHSTTNP--DPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAV 84
PP NSP S S +T + +P P +H +D+P +++ L NYG WS A+
Sbjct: 34 PP--NSPPVNRSGASRALTSSESGDPTQSPFFLHSADHPGLNIISHRLDETNYGDWSVAM 91
Query: 85 TMALRAKNKFGFVDGSLTIPKKKE-DILTWQRCNDLVASWILNSVSTEIRPSILYAETAE 143
++L AKNK GF+DG+L+ P + + + W RCN +V SW+LNSVS +I SIL A
Sbjct: 92 LISLDAKNKTGFIDGTLSRPLESDLNFRLWSRCNSMVKSWLLNSVSPQIYRSILRMNDAS 151
Query: 144 QIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSI-ALVNPCI 202
IW DL RF+ +N P+ Y L Q I +Q + +S Y+T+LK+LWD+L+S AL PC
Sbjct: 152 DIWRDLNSRFNVTNLPRTYNLTQEIQDFRQGTLSLSEYYTRLKTLWDQLDSTEALDEPCT 211
Query: 203 CGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEIN 262
CG A + + Q + ++FL G+++ ++ VR QI+ PS+ +Y+I+ Q+ QQ +
Sbjct: 212 CGKAMRLQQKAEQAKIVKFLAGLNESYAIVRRQIIAKKALPSLGEVYHILDQDNSQQSFS 271
Query: 263 FRPVPAVESAALQTSKAPYRTPG--------KRQRPFCEHCNRHGHTITTCYQIHGFP-- 312
P + +++P P + RP C NR GH CY+ HGFP
Sbjct: 272 NVVAPPAAFQVSEITQSPSMDPTVCYVQNGPNKGRPICSFYNRVGHIAERCYKKHGFPPG 331
Query: 313 --------SNPSKPQ---------KKTETSPSTSANQLSSAQYHKLLTLLAKE------- 348
KP+ + TS + LS Q + + + + +
Sbjct: 332 FTPKGKAGEKLQKPKPLAANVAESSEVNTSLESMVGNLSKEQLQQFIAMFSSQLQNTPPS 391
Query: 349 ----------DNVGSSVNLAGTAF---------TCIPFSWIIDSGASNHICTSLSLFSSY 389
DN+G + + +F T +W+IDSGA++H+ SLFSS
Sbjct: 392 TYATASTSQSDNLGICFSPSTYSFIGILTVARHTLSSATWVIDSGATHHVSHDRSLFSSL 451
Query: 390 YPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDA 449
+ +V P G + +GT+ + ++L NV +P F+ NL+S++ LT+ +
Sbjct: 452 -DTSVLSAVNLPTGPTVKISGVGTLKLNDDILLKNVLFIPEFRLNLISISSLTDDIGSRV 510
Query: 450 IFSSSGCVFQDQATKKMIGRGSARNELYYLN 480
IF + C QD +M+G+G LY L+
Sbjct: 511 IFDKNSCEIQDLIKGRMLGQGRRVANLYLLD 541
>UniRef100_Q9SLF0 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 671 bits (1730), Expect = 0.0
Identities = 361/795 (45%), Positives = 489/795 (61%), Gaps = 61/795 (7%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ F GECVLTAAYLIN+ P+ +L +P++ L P + LRVFG LC+A N
Sbjct: 565 FQANLPIQFWGECVLTAAYLINRTPSSVLNDSTPYERLHKKQPRFDHLRVFGSLCYAHNR 624
Query: 606 NI-QHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPY--------- 655
N KF ERS+ +FVGYP QKG+R++D++ + +VSRDV F E FP+
Sbjct: 625 NRGGDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVFSELEFPFRISHEQNVI 684
Query: 656 ----QDIQSPPFNNAISINTQILDN--EFDDLFTGSPTHPNIPPENSHNDNSN--DTIV- 706
+ + +P + I + N + SP P+ S + S+ DT V
Sbjct: 685 EEEEEALWAPIVDGLIEEEVHLGQNAGPTPPICVSSPISPSATSSRSEHSTSSPLDTEVV 744
Query: 707 ---TISTPEDDASSNPPSFSAESLSNNPPSTEMN------PPNHRHSQRIRNPPIHLKDY 757
ST + S+P + LS P+T PP R S R + PP+ LKD+
Sbjct: 745 PTPATSTTSASSPSSPTNLQFLPLSRAKPTTAQAVAPPAVPPPRRQSTRNKAPPVTLKDF 804
Query: 758 ICK---INNVTSKIN---FPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWR 811
+ SK+N + L+ S SH ++
Sbjct: 805 VVNTTVCQESPSKLNSILYQLQKRDDTRRFSASHTTYV---------------------- 842
Query: 812 DAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQ 871
I A E N+TW + LP GK AIG +W+YK+K++SDGS++RYKARLVA G Q +
Sbjct: 843 -----AIDAQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKE 897
Query: 872 GIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHK 931
G DY +TFAPVAK+ TVRL L +A +NW ++Q DV+NAFL GDL EEVYMKLPPGF
Sbjct: 898 GEDYGETFAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGFEAS 957
Query: 932 KKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYV 991
VC+L K++YGLKQA R WF K +T L + GF+QS++DYSLFT I +L+YV
Sbjct: 958 HPNKVCRLRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYV 1017
Query: 992 DDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILS 1051
DD+IITGN++ A + K++LA F +KDLG L Y LGIEV+RS GI++CQRKY LDI+S
Sbjct: 1018 DDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIYICQRKYALDIIS 1077
Query: 1052 DSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQ 1111
++G+ G +P++FP+EQ+ +L + L+DP YRR VGRL+YL VTR D+ ++V+ L++
Sbjct: 1078 ETGLLGVKPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSVHILAR 1137
Query: 1112 FMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYF 1171
FMQ P H+ AA +V+RYLK G+G+FL S + G+ DSDWAG P +RRS TGYF
Sbjct: 1138 FMQEPREDHWAAALRVVRYLKADPGQGVFLRRSGDFQITGWCDSDWAGDPMSRRSVTGYF 1197
Query: 1172 TMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCD 1231
G +PISWKTKKQ T+S+SSAEAEYR+++ L+SEL WLK LL LG+ H QP+ + CD
Sbjct: 1198 VQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCD 1257
Query: 1232 SQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAY 1291
S++AI+IA NPVFHERTKHIEID HFVR++ G+I P ++ ++ QLADIFTKPLG D +
Sbjct: 1258 SKSAIYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFTKPLGRDCF 1317
Query: 1292 KRILGKLGVIEISIP 1306
KLG+ + P
Sbjct: 1318 SAFRIKLGIRNLYAP 1332
Score = 84.0 bits (206), Expect = 3e-14
Identities = 98/392 (25%), Positives = 163/392 (41%), Gaps = 79/392 (20%)
Query: 201 CICGNAKSILDQQNQDRAMEFLQGVHDR-FSAVRSQILLMDPFPSIQRIYNIVRQEEK-- 257
C+C + +D+ EFL G+ D F VRS ++ P ++ +YNIVRQEE
Sbjct: 80 CVCDLGALQEKDREEDKVHEFLSGLDDALFRTVRSSLVSRIPVQPLEEVYNIVRQEEDLL 139
Query: 258 ---------QQEINFRPVPAVESAALQTSKAPYRTPG--KRQRPFCEHCNRHGHTITTCY 306
Q+E+N + A Q Y+ G K + C+HCNR GH +CY
Sbjct: 140 RNGANVLDDQREVN--------AFAAQMRPKLYQGRGDEKDKSMVCKHCNRSGHASESCY 191
Query: 307 QIHGFP----SNPSKPQKKTETSPSTSAN---QLSSAQYHKLLTL------------LAK 347
+ G+P P +T T+++ +A Y +T+ L
Sbjct: 192 AVIGYPEWWGDRPRSRSLQTRGRGGTNSSGGRGRGAAAYANRVTVPNHDTYEQANYALTD 251
Query: 348 EDNVGSSVNLAGTAFTCIPFSWIIDSG--ASNHICTSLSLFSSYYPVNNQISVQQPDGSQ 405
ED G + L + + I I++SG A+ T + + I + DG +
Sbjct: 252 EDRDGVN-GLTDSQWRTI--KSILNSGKDAATEKLTDM----------DPILIVLADGRE 298
Query: 406 ALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKK 465
+ GT+ +L++ +V++V F+ +L+S+ QL + C S V QD+ ++
Sbjct: 299 RISVKEGTVRLGSNLVMISVFYVEEFQSDLISIGQLMDENRCVLQMSDRFLVVQDRTSRM 358
Query: 466 MIGRGSARNELYYLNQD----LVSNKHDKD----HYCLSHPLSSL----------FSSKH 507
++G G ++ V+ K +K+ H + HP + + SS H
Sbjct: 359 VMGAGRRVGGTFHFRSTEIAASVTVKEEKNYELWHSRMGHPAARVVSLIPESSVSVSSTH 418
Query: 508 CNK-FDLWHLRLVALQLRNKMEL-WNENIDIF 537
NK D+ H A Q RN L N+ + IF
Sbjct: 419 LNKACDVCHR---AKQTRNSFPLSINKTLRIF 447
>UniRef100_Q9SIM3 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1461
Score = 668 bits (1723), Expect = 0.0
Identities = 343/766 (44%), Positives = 483/766 (62%), Gaps = 55/766 (7%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ + G+CVLTA +LIN+ P+ +L K+P +VL G P YS L+ FGCLC++
Sbjct: 742 FQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLPDYSQLKTFGCLCYSSTS 801
Query: 606 NIQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
+ Q HKF RS+ +F+GYPF KGY++ D+++ +++SR+V+FHE +FP Q
Sbjct: 802 SKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVEFHEELFPLASSQQ---- 857
Query: 665 NAISINTQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSA 724
S T + TP D P S
Sbjct: 858 ------------------------------------SATTASDVFTPMD------PLSSG 875
Query: 725 ESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNS 784
S++++ PS +++P +RI P HL+DY C N + P+ + LS S +S S
Sbjct: 876 NSITSHLPSPQISPSTQISKRRITKFPAHLQDYHCYFVNKDD--SHPISSSLSYSQISPS 933
Query: 785 HRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWI 844
H ++ NI + P+SY +A S EW A+ +EI A+E +TW + LP GK A+GCKW+
Sbjct: 934 HMLYINNISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWV 993
Query: 845 YKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQ 904
+ +K+H+DGS++R+KAR+VAKGY+Q +G+DY +TF+PVAK+ TV+LLL ++A K W L Q
Sbjct: 994 FTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQ 1053
Query: 905 FDVNNAFLQGDLSEEVYMKLPPGFSHKKKPC-----VCKLNKSIYGLKQASRQWFSKFST 959
D++NAFL GDL E +YMKLP G++ K VC+L KSIYGLKQASRQWF KFS
Sbjct: 1054 LDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSN 1113
Query: 960 TLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKD 1019
+L+ GF + D++LF I +LVYVDDI+I E A + + L SF +++
Sbjct: 1114 SLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRE 1173
Query: 1020 LGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPL 1079
LG L Y LG+EV+R+ +GI L QRKY L++L+ + M C+PS PM ++RL NDG L
Sbjct: 1174 LGPLKYFLGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLL 1233
Query: 1080 SDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGL 1139
D +YRR VG+L+YLT+TRPDI +AVN L QF +P ++H A +VL+Y+KG+VG+GL
Sbjct: 1234 EDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGL 1293
Query: 1140 FLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYR 1199
F SA + L GY D+DW CP +RRSTTG+ +GS+ ISW++KKQPT+SRSSAEAEYR
Sbjct: 1294 FYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYR 1353
Query: 1200 SLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVR 1259
+LA S E+ WL LL L + PI +Y DS AA++IA NPVFHERTKHIEIDCH VR
Sbjct: 1354 ALALASCEMAWLSTLLLALRVHSGVPI-LYSDSTAAVYIATNPVFHERTKHIEIDCHTVR 1412
Query: 1260 EKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISI 1305
EK+ +G + ++++ DQ+ADI TKPL + +L K+ + I +
Sbjct: 1413 EKLDNGQLKLLHVKTKDQVADILTKPLFPYQFAHLLSKMSIQNIFV 1458
Score = 254 bits (648), Expect = 2e-65
Identities = 166/518 (32%), Positives = 242/518 (46%), Gaps = 65/518 (12%)
Query: 27 SDIPPSENSPLSSISSPITVHSTTNP--DPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAV 84
SD PP S + S + +P P +H +D+P +++ L YG WS A+
Sbjct: 36 SDSPPMARSQTAGASRGVFSSGFDDPTQSPFFLHSADHPGLSIISHRLDETTYGDWSVAM 95
Query: 85 TMALRAKNKFGFVDGSLTIPKKKE-DILTWQRCNDLVASWILNSVSTEIRPSILYAETAE 143
++L AKNK GFVDGSL P + + + W RCN +V SW+LNSVS +I SIL A
Sbjct: 96 RISLDAKNKLGFVDGSLPRPLESDPNFRLWSRCNSMVKSWLLNSVSPQIYRSILRLNDAT 155
Query: 144 QIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSI-ALVNPCI 202
IW DL DRF+ +N P+ Y L Q I L+Q M +S Y+T LK+LWD+L+S AL +PC
Sbjct: 156 DIWRDLFDRFNLTNLPRTYNLTQEIQDLRQGTMSLSEYYTLLKTLWDQLDSTEALDDPCT 215
Query: 203 CGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEIN 262
CG A + + + + M+FL G+++ ++ VR QI+ PS+ +Y+I+ Q+ Q+
Sbjct: 216 CGKAVRLYQKAEKAKIMKFLAGLNESYAIVRRQIIAKKALPSLAEVYHILDQDNSQKGFF 275
Query: 263 FRPVPAVESAALQTSKAPYRTP--------GKRQRPFCEHCNRHGHTITTCYQIHGFP-- 312
P + S +P +P + RP C CNR GH CY+ HGFP
Sbjct: 276 NVVAPPAAFQVSEVSHSPITSPEIMYVQSGPNKGRPTCSFCNRVGHIAERCYKKHGFPPG 335
Query: 313 --------SNPSKPQ---KKTETSP-------STSANQLSSAQYHKLLTLLAK------- 347
P KPQ + SP T A S Q L+ L +
Sbjct: 336 FTPKGKSSDKPPKPQAVAAQVTLSPDKMTGQLETLAGNFSPDQIQNLIALFSSQLQPQIV 395
Query: 348 ---------EDNVGSSVNLAGTAFTCIPF----------------SWIIDSGASNHICTS 382
E + SV +G F+ + +W+IDSGA++H+
Sbjct: 396 SPQTASSQHEASSSQSVAPSGILFSPSTYCFIGILAVSHNSLSSDTWVIDSGATHHVSHD 455
Query: 383 LSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLT 442
LF + + V P G + +GT+ + +IL NV +P F+ NL+S++ LT
Sbjct: 456 RKLFQT-LDTSIVSFVNLPTGPNVRISGVGTVLINKDIILQNVLFIPEFRLNLISISSLT 514
Query: 443 ESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLN 480
L IF S C QD +G G LY L+
Sbjct: 515 TDLGTRVIFDPSCCQIQDLTKGLTLGEGKRIGNLYVLD 552
>UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana]
Length = 1315
Score = 655 bits (1690), Expect = 0.0
Identities = 338/758 (44%), Positives = 481/758 (62%), Gaps = 38/758 (5%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM-NIQHKFD 612
+ G+C+LTA YLIN+LP PIL+ K P +VL + P+Y ++VFGCLC+A +HKF
Sbjct: 588 YWGDCILTAVYLINRLPAPILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFS 647
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQ 672
R+K F+GYP KGY++ D++T I VSR V FHE +FP+ S +Q
Sbjct: 648 PRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVVFHEELFPFLG----------SDLSQ 697
Query: 673 ILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPP 732
N F DL +P P + +D+ N P D +SS E L + P
Sbjct: 698 EEQNFFPDL------NPTPPMQRQSSDHVN--------PSDSSSS------VEILPSANP 737
Query: 733 STEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHRAFLINI 792
+ + P+ + S R P +L+DY C ++V S + +LS +++ + FL +
Sbjct: 738 TNNVPEPSVQTSHRKAKKPAYLQDYYC--HSVVSSTPHEIRKFLSYDRINDPYLTFLACL 795
Query: 793 IENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSD 852
+ KEP +Y++A K WRDAM E LE +TW +C LP K IGC+WI+KIKY+SD
Sbjct: 796 DKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYNSD 855
Query: 853 GSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFL 912
GS++RYKARLVA+GY+Q +GIDY++TF+PVAKL +V+LLL +AA L Q D++NAFL
Sbjct: 856 GSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISNAFL 915
Query: 913 QGDLSEEVYMKLPPGFSHKKKP-----CVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFR 967
GDL EE+YM+LP G++ ++ VC+L KS+YGLKQASRQW+ KFS+TL+ GF
Sbjct: 916 NGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGLGFI 975
Query: 968 QSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYIL 1027
QS D++ F D + VLVY+DDIII NN+ A+ +K + F ++DLG L Y L
Sbjct: 976 QSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELKYFL 1035
Query: 1028 GIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRR 1087
G+E+ RS KGI + QRKY LD+L ++G GC+PS PM+ + + G + YRR
Sbjct: 1036 GLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGPYRR 1095
Query: 1088 HVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSI 1147
+GRL+YL +TRPDI +AVN L+QF +P +H A ++L+Y+KG++G+GLF SA+S +
Sbjct: 1096 LIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFYSATSEL 1155
Query: 1148 NLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSE 1207
L YA++D+ C +RRST+GY LG + I WK++KQ +S+SSAEAEYRSL+ + E
Sbjct: 1156 QLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVATDE 1215
Query: 1208 LQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLI 1267
L WL L +L + +P ++CD++AAIHIA N VFHERTKHIE DCH VRE++ GL
Sbjct: 1216 LVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRERLLKGLF 1275
Query: 1268 APSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISI 1305
+I + Q+AD FTKPL + R++ K+G++ I +
Sbjct: 1276 ELYHINTELQIADPFTKPLYPSHFHRLISKMGLLNIFV 1313
Score = 239 bits (610), Expect = 4e-61
Identities = 150/446 (33%), Positives = 231/446 (51%), Gaps = 46/446 (10%)
Query: 84 VTMALRAKNKFGFVDGSLTIPKKKED---ILTWQRCNDLVASWILNSVSTEIRPSILYAE 140
+T ++ AKNK GFVDGS IPK +D W+RCN +V SW+LNSVS EI SILY
Sbjct: 1 MTTSIEAKNKLGFVDGS--IPKPDDDDPYCKIWRRCNSMVKSWLLNSVSKEIYTSILYFP 58
Query: 141 TAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNP 200
TA IW DL RF +S+ P++Y+L+Q I +L+Q + +S Y T+ ++LW+EL S+ V
Sbjct: 59 TAAAIWKDLYTRFHKSSLPRLYKLRQQIHSLRQGNLDLSSYHTRTQTLWEELTSLQAVPR 118
Query: 201 CICGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQE 260
+ + +L ++ +R ++FL G++D + VRSQIL+ PS+ ++N++ Q+E Q+
Sbjct: 119 TV----EDLLIERETNRVIDFLMGLNDCYDTVRSQILMKKTLPSLSEVFNMIDQDETQRS 174
Query: 261 INFRPVPAVESAALQTSKAPYR------TPGKRQRPFCEHCNRHGHTITTCYQIHGFPSN 314
P + S+ S + T K++RP C +C+R GH TCY+ HG+P++
Sbjct: 175 ARISTTPGMTSSVFPVSNQSSQSALNGDTYQKKERPVCSYCSRPGHVEDTCYKKHGYPTS 234
Query: 315 PSKPQKKTETSPSTSANQLSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCIPFSWIIDSG 374
QK + PS SAN + + V ++ +++ T ++
Sbjct: 235 FKSKQKFVK--PSISANAAIGS------------EEVVNNTSVSTGDLTTSQIQQLVSFL 280
Query: 375 ASNHICTSLSLFSSYYPVNNQISV-QQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKF 433
+S S + P + ISV P S + G+++ LIL +V +P FKF
Sbjct: 281 SSKLQPPS----TPVQPEVHSISVSSDPSSSSTVCPISGSVHLGRHLILNDVLFIPQFKF 336
Query: 434 NLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHY 493
NL+SV+ LT+S+ C F + CV QD + M+G G LY ++ D
Sbjct: 337 NLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMGKQVANLYIVDLD----------- 385
Query: 494 CLSHP-LSSLFSSKHCNKFDLWHLRL 518
LSHP S + DLWH RL
Sbjct: 386 SLSHPGTDSSITVASVTSHDLWHKRL 411
>UniRef100_Q9FX79 Putative retroelement polyprotein [Arabidopsis thaliana]
Length = 1413
Score = 654 bits (1687), Expect = 0.0
Identities = 330/734 (44%), Positives = 476/734 (63%), Gaps = 51/734 (6%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQ-HKFD 612
+ G+C+LTA ++IN+ P+P++ K+ ++L P Y+ L+ FGCLC+A Q HKF+
Sbjct: 715 YWGDCILTAVFIINRTPSPVISNKTLFEMLTKKVPDYTHLKSFGCLCYASTSPKQRHKFE 774
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQ 672
+R++ F+GYP KGY++ D+++ I++SR+V F+E +FP++
Sbjct: 775 DRARTCAFLGYPSGYKGYKLLDLESHTIFISRNVVFYEDLFPFK---------------- 818
Query: 673 ILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPP 732
T P + N+ S+ I +D+ + P E+ ++N P
Sbjct: 819 --------------TKP------AENEESSVFFPHIYVDRNDSHPSQPLPVQETSASNVP 858
Query: 733 STEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHRAFLINI 792
+ + N R+ PP +LKDY C N+VTS + P+ LS S+LS+ + F+ +
Sbjct: 859 AEKQN-------SRVSRPPAYLKDYHC--NSVTSSTDHPISEVLSYSSLSDPYMIFINAV 909
Query: 793 IENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSD 852
+ EP +Y+QA + EW DAM EI ALE N TW++C LP GK A+GCKW+YKIK ++D
Sbjct: 910 NKIPEPHTYAQARQIKEWCDAMGMEITALEDNGTWVVCSLPVGKKAVGCKWVYKIKLNAD 969
Query: 853 GSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFL 912
GS++RYKARLVAKGY+Q +G+DY DTF+PVAKL TV+LL+++AA K W L Q D++NAFL
Sbjct: 970 GSLERYKARLVAKGYTQTEGLDYVDTFSPVAKLTTVKLLIAVAAAKGWSLSQLDISNAFL 1029
Query: 913 QGDLSEEVYMKLPPGFSHKKKP-----CVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFR 967
G L EE+YM LPPG+S ++ VC+L KS+YGLKQASRQW+ KFS +L GF
Sbjct: 1030 NGSLDEEIYMTLPPGYSPRQGDSFPPNAVCRLKKSLYGLKQASRQWYLKFSESLKALGFT 1089
Query: 968 QSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYIL 1027
QS D++LFT + + VLVYVDDIII + + ++ L +S ++DLG L Y L
Sbjct: 1090 QSSGDHTLFTRKSKNSYMAVLVYVDDIIIASSCDRETELLRDALQRSSKLRDLGTLRYFL 1149
Query: 1028 GIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRR 1087
G+E++R+ GI +CQRKYTL++L+++G+ GC+ S PME + +L DG + D YR+
Sbjct: 1150 GLEIARNTDGISICQRKYTLELLAETGLLGCKSSSVPMEPNQKLSQEDGELIDDAEHYRK 1209
Query: 1088 HVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSI 1147
VG+L+YLT TRPDI YAV+ L QF +P H A +++ YLKG+VG+GLF SA+ +
Sbjct: 1210 LVGKLMYLTFTRPDITYAVHRLCQFTSAPRVPHLKAVYKIIYYLKGTVGQGLFYSANVDL 1269
Query: 1148 NLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSE 1207
L G+ADSD++ C +R+ TTGY LG++ ++WK+KKQ IS SSAEAEY++++ E
Sbjct: 1270 KLSGFADSDFSSCSDSRKLTTGYCMFLGTSLVAWKSKKQEVISMSSAEAEYKAMSMAVRE 1329
Query: 1208 LQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLI 1267
+ WL++LL DL ID + +YCD+ AAIHIA NPVFHERTKHIE D H +REKI GLI
Sbjct: 1330 MMWLRFLLEDLWIDVSEASVLYCDNTAAIHIANNPVFHERTKHIERDYHHIREKIILGLI 1389
Query: 1268 APSYIRSSDQLADI 1281
++R+ +QLADI
Sbjct: 1390 RTLHVRTENQLADI 1403
Score = 242 bits (618), Expect = 5e-62
Identities = 160/524 (30%), Positives = 250/524 (47%), Gaps = 90/524 (17%)
Query: 29 IPPSENSPLSSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMAL 88
+PP + + + S + ++ + P +H++D+P +V+ L G NY WS A+ +AL
Sbjct: 28 LPPIQEALFAPFGSSVLMNPDSVHSPLFLHNADHPGISIVSVQLDGANYNQWSSAMKIAL 87
Query: 89 RAKNKFGFVDGSLTIPKKKEDILT-WQRCNDLVASWILNSVSTEIRPSILYAETAEQIWS 147
AKNK F+DGS P++ +L W RCN +V SWILNSV+ EI SIL + A QIW+
Sbjct: 88 DAKNKIAFIDGSCPRPEEGNHLLRIWSRCNSMVKSWILNSVNREIYGSILSFDDAAQIWN 147
Query: 148 DLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICGNAK 207
DL +RF +N P+ +QL Q I L+Q M +S Y+T LK+L D L+ PC C K
Sbjct: 148 DLHNRFHMTNLPRTFQLVQQIQDLRQGSMNLSTYYTTLKTLRDNLDGAEASVPCHCCK-K 206
Query: 208 SILDQQ-------NQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQE 260
S + Q N+ R ++FL G+++++S +R QI++ P P + +YNI+ Q++ Q++
Sbjct: 207 STCESQIFAKSNVNRGRIIKFLAGLNEKYSIIRGQIIMKKPLPDLAEVYNILDQDDSQRQ 266
Query: 261 INFRPVPAVESAALQTSKAPYR-----------TPG-------KRQRPFCEHCNRHGHTI 302
+ V SAA Q +K + PG K ++ C H GHT
Sbjct: 267 FSNN----VASAAFQVTKDDVQPGALASSSNMPQPGMLGAVQKKDKKSICSHYGYTGHTS 322
Query: 303 TTCYQIHGFP-------------------SNPSKPQ-----KKTETSPSTSA------NQ 332
CY++HG+P S KP + T S +T A
Sbjct: 323 ERCYKLHGYPVGWKKGKSFYEKIAQASQSSQAPKPNSAVTAQVTGNSQNTPAGLESLIGN 382
Query: 333 LSSAQYHKLLTLLAKE-----------------DNVGSSVNLAGTAFTCIPF-------- 367
+S Q L+ L + + +N S + + + F+ I
Sbjct: 383 MSKDQIQNLIALFSSQLQPASPVLNTAPMSTSHNNDPSGITFSSSTFSFIGILTVSETEM 442
Query: 368 ---SWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTN 424
+WI+DSGA++H+C +F + + Q V P G+ V +G I + LIL N
Sbjct: 443 THGTWIVDSGATHHVCHVKDMFLN-LDTSVQHHVNLPTGTTIRVGGVGNIAVNADLILKN 501
Query: 425 VYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIG 468
V ++P F+ NL+SV+ LT + +F + CV D IG
Sbjct: 502 VLYIPEFRLNLLSVSALTTDIGARVVFDPTCCVVHDLTKGSTIG 545
>UniRef100_Q9FIC5 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1462
Score = 654 bits (1686), Expect = 0.0
Identities = 352/797 (44%), Positives = 490/797 (61%), Gaps = 44/797 (5%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQ-HKFD 612
F C LTAAYLIN+ PTP+L+ K+P ++L PP + +RVFGC+C+ N KF+
Sbjct: 651 FWSYCALTAAYLINRTPTPLLQGKTPFELLYNRPPPVNHIRVFGCICYVHNQKHGGDKFE 710
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQ--------DIQSPPFN 664
RS IF+GYPF +KG+R+Y+ +T I VSRDV F ET FP+ D Q P N
Sbjct: 711 SRSNKSIFLGYPFAKKGWRVYNFETGVISVSRDVVFRETEFPFPASVFDSTPDSQLSPSN 770
Query: 665 --------------NAISINTQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTIST 710
+SI T + + + + IP + S + N +++
Sbjct: 771 ADQSFFLPSELQAPTPVSITTTLELTQSSSSTNLNDDNFRIPSDESSSVNEMSDNEDLNS 830
Query: 711 PEDDASSNPPSFSAESLSNNPPST-------------------EMNPPNHRHSQRIRNPP 751
P + SS S ++ SL +P S E P +R + P
Sbjct: 831 PTTNESSPFLSPASPSLPLSPASLSLPLSPAAPSPSLPKIAEPEPEPELLGKGKRKKTQP 890
Query: 752 IHLKDYICKINNVT--SKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVE 809
+ L DY + + S +PL+NY+S S S +++A++ I EPKSY +A+
Sbjct: 891 VRLADYATTLLHQPHPSVTPYPLDNYVSSSQFSAAYQAYVFAISLGIEPKSYKEAILDEN 950
Query: 810 WRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQ 869
WR A++ EI +LE+ TW + LP GK A+GCKW++++KY SDG+++R+KARLV G Q
Sbjct: 951 WRCAVSDEIVSLENLGTWTVEDLPPGKKALGCKWVFRLKYKSDGTLERHKARLVVLGNKQ 1010
Query: 870 VQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFS 929
+GIDY +TFAPVAK+VTVR L A +W ++Q DV+NAFL GDL EEVY+K PPGF
Sbjct: 1011 TEGIDYSETFAPVAKMVTVRAFLQQVASLDWEVHQMDVHNAFLHGDLDEEVYIKFPPGFG 1070
Query: 930 HKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLV 989
VC+L K++YGLKQA R WF+K +T L GF Q ISDYSLFT + + +LV
Sbjct: 1071 SDDNRKVCRLRKALYGLKQAPRCWFAKLTTALNDYGFIQDISDYSLFTMERNGIRLHILV 1130
Query: 990 YVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDI 1049
YVDD+IITG++ + I+K K +L+ F +KDLG L Y LGIEV+RS GI+LCQRKY +DI
Sbjct: 1131 YVDDLIITGSSLDVITKFKGYLSSCFYMKDLGILRYFLGIEVARSPAGIYLCQRKYAIDI 1190
Query: 1050 LSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTL 1109
++++G+ G P+ P+EQ+ +L G +SDP+ YRR VGRL+YL TRP++ YA++ L
Sbjct: 1191 ITETGLLGVWPASHPLEQNHKLALAFGDTISDPSRYRRLVGRLIYLGTTRPELSYAIHML 1250
Query: 1110 SQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTG 1169
SQFM P + H +AA +V+RYLK S G+G+ L +++ + L G+ DS + CP T+RS TG
Sbjct: 1251 SQFMSDPKADHMEAALRVVRYLKSSPGQGILLRSNTPLVLTGWCDSGFDSCPITQRSLTG 1310
Query: 1170 YFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIY 1229
+F LG +PISWKTKK +SRSSAEAEYR++A SEL WL+ LL LGI +PI +Y
Sbjct: 1311 WFIQLGGSPISWKTKKHDVVSRSSAEAEYRAMADTVSELLWLRALLPALGISCNEPIMLY 1370
Query: 1230 CDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGD 1289
DS +AI +A NPV+H RTKH+ D HFVR++I G IA ++ ++ QLADI TK LG
Sbjct: 1371 SDSLSAISLAANPVYHARTKHVGRDVHFVRDEIIRGTIATKHVSTTSQLADIMTKALGRR 1430
Query: 1290 AYKRILGKLGVIEISIP 1306
+ L KLG+ + P
Sbjct: 1431 EFDAFLLKLGICNLHTP 1447
Score = 167 bits (423), Expect = 2e-39
Identities = 132/509 (25%), Positives = 223/509 (42%), Gaps = 58/509 (11%)
Query: 41 SSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGS 100
SS T P + DNP T++ PLL G NY W+ + +AL+A+ KFGF DG+
Sbjct: 10 SSSTTTQPRRTISPYDLTSGDNPGTLISKPLLRGPNYDEWATNLRLALKARKKFGFADGT 69
Query: 101 LTIP-KKKEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAP 159
+ P + D W N LV SW+ ++ + S+ + + + +W+ +Q RF N
Sbjct: 70 IPQPDETNPDFDDWIANNALVVSWMKLTIHESLATSMSHLDDSHDMWTHIQKRFGVKNGQ 129
Query: 160 KIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICGNAKSILDQQNQDRAM 219
+I +LK ++ +Q+G + Y+ +L LW L + + ++ +D+
Sbjct: 130 RIQRLKTELATCRQKGTPIETYYGKLSQLWRSLADYQQAK-----TMEEVRKEREEDKLH 184
Query: 220 EFLQGVHD-RFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEI----NFRPVPAVESAAL 274
+FL G+ + + AV+S +L P PS++ YN + Q+E+ + + + R + A
Sbjct: 185 QFLMGLDESMYGAVKSALLSRVPLPSLEEAYNTLTQDEESKSLSRLHDERNDEKMRRNAT 244
Query: 275 QTSKAPYRTPGKRQRPFCEHCNRHGHTITTCYQIHGFPS---------NPSKPQKKTETS 325
+S+ + G+ G ++ G S + + T +
Sbjct: 245 SSSRNQSASMGR------------GSSVVPATSFKGKQSFGRGASANHVANIGESTTAAT 292
Query: 326 PSTSANQLSSA----------QYHKLLTLLAKEDNVGSSVNLAGTAFTCIPFSWIIDSGA 375
S S +QL+ A + K L + KE N S+ + F SWIIDSGA
Sbjct: 293 SSMSGSQLTEADRVGISGLNDEQWKQLRQMLKERNFNSTNTKSSKFFL---ESWIIDSGA 349
Query: 376 SNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNL 435
+NH+ +L + I ++ PDG G + SL L V+ V +L
Sbjct: 350 TNHMTGTLEFLRDVCDM-PPIMIKLPDGRLTTSTKHGRVYLGSSLDLQEVFFVDGLHCHL 408
Query: 436 MSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYL-NQDLVSNKHDKD--- 491
+SV+QLT + +C + C+ QD+ T +IG G +N LY+ + V++ D
Sbjct: 409 ISVSQLTRAKSCVFQITDKVCIIQDRITLTLIGAGKQQNGLYFFRGTETVASMTRMDSSS 468
Query: 492 ---HYCLSHPLSSL-----FSSKHCNKFD 512
H L HP S + FS + FD
Sbjct: 469 QLWHCRLGHPSSKVLKLLSFSDSTGHAFD 497
>UniRef100_Q8L700 Hypothetical protein [Arabidopsis thaliana]
Length = 776
Score = 650 bits (1677), Expect = 0.0
Identities = 348/770 (45%), Positives = 472/770 (61%), Gaps = 60/770 (7%)
Query: 593 LRVFGCLCFAK-NMNIQHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHET 651
LR FGCLC+A + KF RS+ +F+GYP +K +R+YD++T +I+ SRDV+FHE
Sbjct: 2 LRSFGCLCYAHIRPRNKDKFTSRSRKCVFIGYPHGKKAWRVYDLETGKIFASRDVRFHED 61
Query: 652 VFPYQDIQ---------SPPFNNA---ISINTQILDNEFDDLFTGSPTHPNI---PPENS 696
++PY +PP N + I+TQ+ D + SP PP +
Sbjct: 62 IYPYATATQSNVPLPPPTPPMVNDDWFLPISTQVDSTNVDSSSSSSPAQSGSIDQPPRSI 121
Query: 697 HNDNSNDT---------IVTISTPEDDASSNPPSFSA----ESLSNNPPSTEMNPPNHR- 742
S T IV S+P + SA E LS ST +P
Sbjct: 122 DQSPSTSTNPVPEEIGSIVPSSSPSRSIDRSTSDLSASDTTELLSTGESSTPSSPGLPEL 181
Query: 743 --HSQRIRNPPIHLKDYICKI----------------------------NNVTSKINFPL 772
R + + LKD++ ++V+ K +PL
Sbjct: 182 LGKGCREKKKSVLLKDFVTNTTSKKKTASHNIHSPSQVLPSGLPTSLSADSVSGKTLYPL 241
Query: 773 ENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPL 832
++L+ S S +H AF+ I+++ EPK + A+ EW +AM+KEI ALE+N+TW + L
Sbjct: 242 SDFLTNSGYSANHIAFMAAILDSNEPKHFKDAILIKEWCEAMSKEIDALEANHTWDITDL 301
Query: 833 PEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLL 892
P GK AI KW+YK+KY+SDG+++R+KARLV G Q +G+D+ +TFAPVAKL TVR +L
Sbjct: 302 PHGKKAISSKWVYKLKYNSDGTLERHKARLVVMGNHQKEGVDFKETFAPVAKLTTVRTIL 361
Query: 893 SIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQ 952
++AA K+W ++Q DV+NAFL GDL EEVYM+LPPGF VC+L KS+YGLKQA R
Sbjct: 362 AVAAAKDWEVHQMDVHNAFLHGDLEEEVYMRLPPGFKCSDPSKVCRLRKSLYGLKQAPRC 421
Query: 953 WFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLA 1012
WFSK ST L GF QS DYSLF+ T I VLVYVDD+I+ GNN +AI + K L
Sbjct: 422 WFSKLSTALRNIGFTQSYEDYSLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDRFKSQLH 481
Query: 1013 QSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLR 1072
+ F +KDLG L Y LG+EVSR G L QRKY LDI+ ++G+ GC+PS P+ + +L
Sbjct: 482 KCFHMKDLGKLKYFLGLEVSRGPDGFCLSQRKYALDIVKETGLLGCKPSAVPIALNHKLA 541
Query: 1073 PNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLK 1132
G ++P YRR VGR +YLT+TRPD+ YAV+ LSQFMQ+P +H++AA +++RYLK
Sbjct: 542 SITGPVFTNPEQYRRLVGRFIYLTITRPDLSYAVHILSQFMQAPLVAHWEAALRLVRYLK 601
Query: 1133 GSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRS 1192
GS +G+FL + SS+ + Y DSD+ CP TRRS + Y LG +PISWKTKKQ T+S S
Sbjct: 602 GSPAQGIFLRSDSSLIINAYCDSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTVSYS 661
Query: 1193 SAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIE 1252
SAEAEYR++A EL+WLK LL DLG+ H P+ ++CDS+AAIHIA NPVFHERTKHIE
Sbjct: 662 SAEAEYRAMAYTLKELKWLKALLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTKHIE 721
Query: 1253 IDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIE 1302
DCH VR+ + LI +I + DQ+AD+ TK L ++R+L LGV +
Sbjct: 722 SDCHKVRDAVLDKLITTEHIYTEDQVADLLTKSLPRPTFERLLSTLGVTD 771
>UniRef100_Q5XWR5 Putative retroelement pol polyprotein-like [Solanum tuberosum]
Length = 1476
Score = 649 bits (1673), Expect = 0.0
Identities = 343/768 (44%), Positives = 480/768 (61%), Gaps = 57/768 (7%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQHKFDE 613
F GECVL+A ++IN++P+ +L KSP +++ P S +RV GCLC A N+
Sbjct: 734 FWGECVLSAVHIINRIPSSVLHNKSPFELMYKRSPDLSYMRVIGCLCHATNLVNTS---- 789
Query: 614 RSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQI 673
QKGY++YD++ + +VSRD+ F+E VFP+Q + +
Sbjct: 790 ------------TQKGYKLYDLEHQHFFVSRDMVFNEAVFPFQ-------------SPAL 824
Query: 674 LDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPED--DASSNPPSFSAESLSNNP 731
D +F SP P +SH ++++ I T E+ +S P + S + L
Sbjct: 825 ADPHDTPVFLASP------PCSSHTEDADAVQPAIITSEEIIPVASPPSAVSDDHLHP-- 876
Query: 732 PSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHRAFLIN 791
PP R S R PPI KD+I + ++ +P+ + + S LS++++ ++ +
Sbjct: 877 ------PPERRRSYRTGKPPIWQKDFITTSTSRSNHCLYPISDNIDYSCLSSTYQCYIAS 930
Query: 792 IIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHS 851
EP+ Y QA W AM +EIQALE N TW + LP+GK AIGCKW+YKIKY +
Sbjct: 931 SSVETEPQFYYQAANDCRWVHAMKEEIQALEDNKTWEVVSLPKGKKAIGCKWVYKIKYKA 990
Query: 852 DGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAF 911
G I+R+KARLVAKGY+Q +G+DY +TF+PV K+VT+R +L++A K W + Q DV NAF
Sbjct: 991 SGEIERFKARLVAKGYNQKEGLDYQETFSPVVKMVTLRTVLTLAVSKGWDIQQMDVYNAF 1050
Query: 912 LQGDLSEEVYMKLPPGFSHKKK--PCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQS 969
LQGDL EEVYM+LP GF + K P VC+L KS+YGLKQASRQW K +T L+ GF+QS
Sbjct: 1051 LQGDLIEEVYMQLPQGFQYDKTGDPKVCRLLKSLYGLKQASRQWNVKLTTALLAAGFQQS 1110
Query: 970 ISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGI 1029
DYSL + VL+YVDD++ITG++ I K+ L +F IKDLG L Y LG+
Sbjct: 1111 HLDYSLMLKRTADGIVIVLIYVDDLLITGSSLQLIDDAKQVLKANFKIKDLGTLRYFLGM 1170
Query: 1030 EVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPND----------GTPL 1079
E +R+ G+ + QRKY L+++SD G+ G +PS P+E HL+L + + L
Sbjct: 1171 EFARNASGMLMHQRKYALELISDLGLGGSKPSVTPVELHLKLTTREFDLHVGSSGADSLL 1230
Query: 1080 SDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGL 1139
+DPT Y+R VGRLLYLT+TRPDI +AV LSQFM +P SH +AA +V++Y+K + G GL
Sbjct: 1231 ADPTEYQRLVGRLLYLTITRPDISFAVQHLSQFMHAPKVSHMEAAIRVVKYVKQAPGLGL 1290
Query: 1140 FLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYR 1199
+++ ++ L Y D+DW C TR+S TGY GS +SWK+KKQPTISRSSAEAEYR
Sbjct: 1291 YMAVQTADTLQAYCDADWGSCINTRKSITGYMIQFGSALLSWKSKKQPTISRSSAEAEYR 1350
Query: 1200 SLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVR 1259
SLA+ +EL WL L +L + P+++YCDS+AAI IA NPVFHERTKHI+IDCHF+R
Sbjct: 1351 SLASTVAELVWLTGLFKELDMPLSLPVSLYCDSKAAIQIAANPVFHERTKHIDIDCHFIR 1410
Query: 1260 EKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISIPP 1307
EK+++GL+ Y+ + +Q ADI TK L + ++ KLG+ I IPP
Sbjct: 1411 EKVQAGLVMIHYLPTQEQPADILTKGLSSAQHSYLVSKLGLKNIFIPP 1458
Score = 238 bits (607), Expect = 9e-61
Identities = 169/571 (29%), Positives = 269/571 (46%), Gaps = 112/571 (19%)
Query: 72 LTG-DNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKEDI--LTWQRCNDLVASWILNSV 128
LTG +NY WSRA+ + L KNK GF+DGSL KE++ W RCN +V SW++N+V
Sbjct: 21 LTGMENYSLWSRAMQLTLLTKNKMGFIDGSLRRDDFKEELEKKQWDRCNAMVLSWLMNNV 80
Query: 129 STEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSL 188
ST++ IL+ A +W+DL++RF + N +I+ L ++I Q VS+Y+++LK L
Sbjct: 81 STDLVSGILFRSNATLVWNDLKERFDKVNMSRIFHLHKAIVTHVQGVSPVSVYYSKLKDL 140
Query: 189 WDELNSIALVNPCICGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRI 248
WDE +SI C C + D + + ++FL G++D + RSQIL+M+P PS+ +
Sbjct: 141 WDEYDSILPPPSCDCEKSVDYTDSMLRQKLLQFLMGLNDNYGQARSQILMMNPSPSVNQC 200
Query: 249 YNIVRQEEKQQEIN-----------FRPVPAVESAALQTSKAP-----------YRTPGK 286
Y ++ Q+E Q+ ++ F P Q S+ + G
Sbjct: 201 YAMIVQDESQRSLSGSGQTIDPTALFTHRPGGSGFGSQGSQGSGNGSSNGNSHRFHKGGN 260
Query: 287 RQRPFCE----------------HCNRHGHTITTCYQIHGFPSN------------PSKP 318
FC HCN GHT TCYQ+ G+P++ PS P
Sbjct: 261 IYCDFCNMKGHIRANCNKLKHCTHCNMQGHTKDTCYQLIGYPADYKGKKKANIVTAPSLP 320
Query: 319 QKK---------------------------------TETSPSTSAN-------QLSSAQY 338
Q + +S S + N Q + +QY
Sbjct: 321 QMQHNNFNNNLNYPMQYTGDGIGHFVSPMQFTGNTNGHSSGSIAGNFGPGSVPQFTPSQY 380
Query: 339 HKLLTLLAKEDNVGSSVNLAG-------TAFTCIPFSWIIDSGASNHICTSLSLFSSYYP 391
+ +L +L K SS N+AG +WI+DSGA++H+ ++ +L +
Sbjct: 381 NNILQMLNKPMLSESSANVAGIFAGSSHCNSNTHSSAWIVDSGATDHMVSNTTLLNHGLS 440
Query: 392 VNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIF 451
V++ VQ P G A+V H G+ + ++ NV VPTF+FNL+SV++LT+ LNC IF
Sbjct: 441 VSHPGKVQLPTGDSAVVTHSGSSQLTGGDVVKNVLCVPTFQFNLLSVSKLTKELNCCVIF 500
Query: 452 SSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYCLSHPLSSLFSSKHCNKF 511
+ QD T K+ G N LY ++ H H+ S +L + K C +
Sbjct: 501 FPDFFIIQDLFTGKVKEIGEEINGLY------ITRPH--QHHDTSK--KTLAAIKGCEEA 550
Query: 512 DLWHLRL--VALQLRNKMELWNENIDIFLMS 540
++WH RL + + + K+++++ + L S
Sbjct: 551 EMWHKRLGHIPMSVLRKIKMFDSPQKLVLPS 581
>UniRef100_O23588 Retrotransposon like protein [Arabidopsis thaliana]
Length = 1433
Score = 634 bits (1636), Expect = e-180
Identities = 328/772 (42%), Positives = 473/772 (60%), Gaps = 63/772 (8%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ F G+CVLTA +LIN+LPTP+L KSP++ L PP+Y SL+ FGCLC++
Sbjct: 717 FQSNIPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAYESLKTFGCLCYSSTS 776
Query: 606 NIQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
Q HKF+ R++ +F+GYP KGY++ D++T + +SR V FHE +FP+
Sbjct: 777 PKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFHEDIFPF--------- 827
Query: 665 NAISINTQILDN--EFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSF 722
I++ I D+ +F L ++P E T + + P D SS+
Sbjct: 828 ----ISSTIKDDIKDFFPLLQFPARTDDLPLEQ--------TSIIDTHPHQDVSSSKALV 875
Query: 723 SAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLS 782
+ LS +R + PP HL+D+ C NN T +
Sbjct: 876 PFDPLS----------------KRQKKPPKHLQDFHC-YNNTTEPFH------------- 905
Query: 783 NSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCK 842
AF+ NI P+ YS+A W DAM +EI A+ NTW + LP K AIGCK
Sbjct: 906 ----AFINNITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCK 961
Query: 843 WIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPL 902
W++ IK+++DGSI+RYKARLVAKGY+Q +G+DY +TF+PVAKL +VR++L +AA W +
Sbjct: 962 WVFTIKHNADGSIERYKARLVAKGYTQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSV 1021
Query: 903 YQFDVNNAFLQGDLSEEVYMKLPPGFSHK-----KKPCVCKLNKSIYGLKQASRQWFSKF 957
+Q D++NAFL GDL EE+YMK+PPG++ +C+L+KSIYGLKQASRQW+ K
Sbjct: 1022 HQLDISNAFLNGDLDEEIYMKIPPGYADLVGEALPPHAICRLHKSIYGLKQASRQWYLKL 1081
Query: 958 STTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSI 1017
S TL GF++S +D++LF + + VLVYVDDI+I N+++A+++ L F +
Sbjct: 1082 SNTLKGMGFQKSNADHTLFIKYANGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKL 1141
Query: 1018 KDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGT 1077
+DLG Y LGIE++RS+KGI +CQRKY L++LS +G G +PS P++ ++L DG
Sbjct: 1142 RDLGAAKYFLGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGV 1201
Query: 1078 PLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGK 1137
PL+D T YR+ VG+L+YL +TRPDI YAVNTL QF +P S H A +VLRYLKG+VG+
Sbjct: 1202 PLTDSTSYRKLVGKLMYLQITRPDIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQ 1261
Query: 1138 GLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAE 1197
GLF SA +L GY DSD+ C +RR Y +G +SWK+KKQ T+S S+AEAE
Sbjct: 1262 GLFYSADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAE 1321
Query: 1198 YRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHF 1257
+R+++ + E+ WL L D + P +YCD+ AA+HI N VFHERTK +E+DC+
Sbjct: 1322 FRAMSQGTKEMIWLSRLFDDFKVPFIPPAYLYCDNTAALHIVNNSVFHERTKFVELDCYK 1381
Query: 1258 VREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISIPPPT 1309
RE ++SG + ++ + +Q+AD TK + + +++GK+GV I P P+
Sbjct: 1382 TREAVESGFLKTMFVETGEQVADPLTKAIHPAQFHKLIGKMGVCNIFAPLPS 1433
Score = 232 bits (592), Expect = 5e-59
Identities = 156/516 (30%), Positives = 245/516 (47%), Gaps = 110/516 (21%)
Query: 38 SSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFV 97
+SI S HS P +H SD+P +V+ +L G NY +WS A+ M+L AKNK FV
Sbjct: 56 ASIESYDNAHS-----PYFLHSSDHPGLNIVSHILDGTNYNNWSIAMRMSLDAKNKLSFV 110
Query: 98 DGSLTIPKKKEDILT-WQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQS 156
DGSL P + + W RCN +V +W+LN V+ ++W+DL RF S
Sbjct: 111 DGSLPRPDVSDRMFKIWSRCNSMVKTWLLNVVT--------------EMWNDLFSRFRVS 156
Query: 157 NAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNS--IALVNPCICGNAKSILDQQN 214
N P+ YQL+QSI LKQ + +S Y+T+ K+LW++L + + V C C + K +L++
Sbjct: 157 NLPRKYQLEQSIHTLKQGNLDLSTYYTKKKTLWEQLANTRVLTVRKCNCEHVKELLEEAE 216
Query: 215 QDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAAL 274
R ++FL G++D F+ +R QIL M P P + IYN++ Q+E Q+ + P + +AA
Sbjct: 217 TSRIIQFLMGLNDNFAHIRGQILNMKPRPGLTEIYNMLDQDESQRLVG-NPTLSNPTAAF 275
Query: 275 QTSKAPY------RTPGKRQRPFCEHCNRHGHTITTCYQIHGFP---------------- 312
Q +P G ++P C +CN+ GH + CY+ HG+P
Sbjct: 276 QVQASPIIDSQVNMAQGSYKKPKCSYCNKLGHLVDKCYKKHGYPPGSKWTKGQTIGSTNL 335
Query: 313 -SNPSKPQKKTETSPSTSANQLSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCI------ 365
S +P +T + S + S+ Q +++ L+ + ++ S+ + T+ I
Sbjct: 336 ASTQLQPVNETPNEKTDSYEEFSTDQIQTMISYLSTKLHIASASPMPTTSSASISASPSV 395
Query: 366 ---------------------------------PFSWIIDSGASNHICTSLSLFSSYYPV 392
P W+IDSGA++H+ + L+ ++ +
Sbjct: 396 PMISQISGTFLSLFSNAYYDMLISSVSQEPAVSPRGWVIDSGATHHVTHNRDLYLNFRSL 455
Query: 393 NNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFS 452
N V+ P+ + IG I S ++ L NV ++P FKFNL+S +LT+ L
Sbjct: 456 ENTF-VRLPNDCTVKIAGIGFIQLSDAISLHNVLYIPEFKFNLIS--ELTKEL------- 505
Query: 453 SSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKH 488
MIGRGS LY L D N H
Sbjct: 506 -------------MIGRGSQVGNLYVL--DFNENNH 526
>UniRef100_Q9ZVE4 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1102
Score = 629 bits (1622), Expect = e-178
Identities = 414/1190 (34%), Positives = 595/1190 (49%), Gaps = 213/1190 (17%)
Query: 228 RFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAAL----QTSKAPY-- 281
RF+ +RS+I DP PS R+Y+ V + ++ ++ R S A+ +T AP
Sbjct: 14 RFAPIRSKITDEDPLPSHNRVYSRVIRGQQNLDVA-RSKETTNSEAINFSVKTPSAPQVA 72
Query: 282 --RTPGKRQRPFCEHCNRHGHTITTCYQIHGFP------------------------SNP 315
P R R C HC+R GH +T C+ +HGFP + P
Sbjct: 73 AVYAPKPRDRS-CTHCHRQGHDVTDCFLVHGFPEWYYEQKGGSRVSSDNREVVSRLENKP 131
Query: 316 SKPQKKTETSPSTSANQLSSA-----------QYHKLLTLLAKEDNVGSSVNLAGTAFTC 364
+K + ++ +++SA Q +L++LL + +S L+G TC
Sbjct: 132 AKREGRSSKGNGRGRGRVNSARAPLSSSNGSDQITQLISLLQAQRPKSTSERLSGN--TC 189
Query: 365 IPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTN 424
+ IIDSGAS+H+ S+ + + +V +PDG + T+ S S L +
Sbjct: 190 LT-DVIIDSGASHHMTGDCSILVDVFDIIPS-AVTKPDGKASCATKCVTLLLSSSYKLQD 247
Query: 425 VYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLV 484
V VP F L+SV++L + ++ +IG G R +YY LV
Sbjct: 248 VLFVPDFDCTLISVSKLLKQTG--------------PFSRTLIGAGEVRERVYYFTGVLV 293
Query: 485 SNKHDKD----------HYCLSHPLSSLFSSK---HCNKFDLWHLRLVALQLRNKMELWN 531
++ H H L HP +S S H + DL + R K L +
Sbjct: 294 ASAHKTSSDSTSSGALWHRRLGHPSTSFLLSLPECHQSSKDLGKIDSCDTCSRAKQTLPS 353
Query: 532 ENIDIFLMSQELY-------------------AFKQTYLFYFGGECVLTA---------- 562
+ M+ + ++ Q + CV T
Sbjct: 354 LIRNFCAMADRQFRKPVRSIRSDNGTEFMCHTSYFQEHGILHETSCVDTPQQNARVERKH 413
Query: 563 AYLINKLPTPILK--FKSPHQV---------LLGSPPSYSSLRVFGCLCFAK-NMNIQHK 610
+++N T + + F +P V L G PSY LR FGCLC+A + K
Sbjct: 414 RHILNVARTCLFQGNFPTPSPVLKGKTPYEVLFGKQPSYDMLRTFGCLCYAHIRPRDKDK 473
Query: 611 FDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISIN 670
F RS+ IF+GYP HET P
Sbjct: 474 FASRSRKCIFIGYP------------------------HETATP---------------- 493
Query: 671 TQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNN 730
TH +I P ++ +D +N + TP+ + +P S S+ + +N
Sbjct: 494 ---------------NTHDSIDPTSTSSDENNTPPEPV-TPQAEQPHSPSSISSPHIVHN 537
Query: 731 PPSTEMNPPNHRH-------------SQRIRNPPIHLKDYIC-KINNVTSKINFPLENYL 776
S N H R ++PP++LKDY+ K+++ + L +
Sbjct: 538 KGSVHSRHLNEDHDSSSPGLPELLGKGHRPKHPPVYLKDYVAHKVHSSPHTSSPGLSDSN 597
Query: 777 SLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGK 836
+S +H AF+ I+++ E + + EW DAM KEI+ALE+N+TW + LP GK
Sbjct: 598 VSPTVSANHIAFMAAILDSNEQNHFKDDVLIKEWCDAMQKEIEALEANHTWDVTDLPHGK 657
Query: 837 SAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAA 896
AI KW+YK+K++SDG+++R+KARLV G Q +GID+ +TFAPVAK+ TVRLLL++AA
Sbjct: 658 KAISSKWVYKLKFNSDGTLERHKARLVVMGNHQKEGIDFKETFAPVAKMTTVRLLLAVAA 717
Query: 897 IKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSK 956
K+W ++Q DV+NAFL GDL +E S+YGLKQA R WF+K
Sbjct: 718 AKDWDVFQMDVHNAFLHGDLEQE----------------------SLYGLKQAPRCWFAK 755
Query: 957 FSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFS 1016
ST L + GF QS DYSLF+ N D T I LVYVDD II GNN AI K+ L + F
Sbjct: 756 LSTALRKLGFTQSYEDYSLFSLNRDGTVIHFLVYVDDFIIVGNNLKAIDHFKEHLHKCFH 815
Query: 1017 IKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDG 1076
+KDLG L Y LG+EVSR G L Q+KY LDI++++G+ G +PS PME H +L
Sbjct: 816 MKDLGKLKYFLGLEVSRGADGFCLSQQKYALDIINEAGLLGYKPSAVPMELHHKLGSISS 875
Query: 1077 TPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVG 1136
+P YRR V R +YLT+TRPD+ YAV+ LSQFMQ+P +H+ A +++RYLKGS
Sbjct: 876 PVFDNPAQYRRLVDRFIYLTITRPDLSYAVHILSQFMQTPLEAHWHATLRLVRYLKGSPD 935
Query: 1137 KGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEA 1196
+G+ L + +++L Y DSD+ CP TRRS + Y LG PISWKTKKQ T+S SSAEA
Sbjct: 936 QGILLRSDRALSLTAYCDSDYNPCPRTRRSLSAYVLYLGDTPISWKTKKQDTVSSSSAEA 995
Query: 1197 EYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCH 1256
EYR++A E++WLK L++ LG+DH QPI ++CDSQAAIHIA NPVFHERTKHIE DCH
Sbjct: 996 EYRAMAYTLKEIKWLKALMTTLGVDHTQPILLFCDSQAAIHIAANPVFHERTKHIEKDCH 1055
Query: 1257 FVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISIP 1306
VR+ + +I+ +I ++ D+ TK L ++R+L LG +P
Sbjct: 1056 QVRDAVTDKVISTPHISTT----DLLTKALPRPTFERLLSTLGTCNYDLP 1101
>UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis thaliana]
Length = 1404
Score = 611 bits (1576), Expect = e-173
Identities = 336/776 (43%), Positives = 477/776 (61%), Gaps = 35/776 (4%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQH-KFD 612
F G+ VLTA YLIN+ PT +L SP +VL + P LRVFGC+CF Q K D
Sbjct: 637 FWGDAVLTACYLINRTPTKVLSDLSPFEVLNNTKPFIDHLRVFGCVCFVLIPGEQRSKLD 696
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVF-----PYQDIQSPPFNNAI 667
+S +F+GY QKGY+ +D + ++SRDV+F E +++++ + +
Sbjct: 697 AKSTKCMFLGYSTTQKGYKCFDPTKNRTFISRDVKFLENQDYNNKKDWENLKDLTHSTSD 756
Query: 668 SINTQ--ILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVT------ISTPEDDASSNP 719
+ T +LD+ +D + + P + + + N+ + ED ++
Sbjct: 757 RVETLKFLLDHLGNDSTSTTQHQPEMTQDQEDLNQENEEVSLQHQENLTHVQEDPPNTQE 816
Query: 720 PSFSAESLSNN------PPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLE 773
S + + ++ P PP R S RIR K++ ++ + P +
Sbjct: 817 HSEHVQEIQDDSSEDEEPTQVLPPPPPLRRSTRIRRK----KEFFN-----SNAVAHPFQ 867
Query: 774 NYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLP 833
SL+ + H+AFL I E+ P++Y +AM+ EWRDA+A EI A++ N+TW LP
Sbjct: 868 ATCSLALVPLDHQAFLSKISEHWIPQTYEEAMEVKEWRDAIADEINAMKRNHTWDEDDLP 927
Query: 834 EGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLS 893
+GK + +W++ IKY S+G I+RYK RLVA+G++Q G DY +TFAPVAKL TVR++L+
Sbjct: 928 KGKKTVSSRWVFTIKYKSNGDIERYKTRLVARGFTQTYGSDYMETFAPVAKLHTVRVVLA 987
Query: 894 IAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPC--VCKLNKSIYGLKQASR 951
+A +W L+Q DV NAFLQG+L ++VYM PPG PC V +L K+IYGLKQ+ R
Sbjct: 988 LATNLSWGLWQMDVKNAFLQGELEDDVYMTPPPGLEDTI-PCDKVLRLRKAIYGLKQSPR 1046
Query: 952 QWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFL 1011
W+ K S TL GF++S SD++LFT Q + VL+YVDD+IITG+N++ I K FL
Sbjct: 1047 AWYHKLSRTLKDHGFKKSESDHTLFTLQSPQGIVVVLIYVDDLIITGDNKDGIDSTKTFL 1106
Query: 1012 AQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRL 1071
F IKDLG L Y LGIEV RS G+FL QRKYTLD+L+++G +P+ P+E ++
Sbjct: 1107 KSCFDIKDLGELKYFLGIEVCRSNAGLFLSQRKYTLDLLNETGFMDAKPARTPLEDGYKV 1166
Query: 1072 R---PNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVL 1128
+ D +YR+ VG+L+YLT TRPDI +AVN +SQ M+ P H++ ++L
Sbjct: 1167 NRKGEKEDEKFGDAPLYRKLVGKLIYLTNTRPDICFAVNQVSQHMKVPMVYHWNMVERIL 1226
Query: 1129 RYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPT 1188
RYLKGS G+G+++ +SS +VGY D+D+AG RRS TGY T +G N +WKTKKQ
Sbjct: 1227 RYLKGSSGQGIWMGKNSSTEIVGYCDADYAGDRGDRRSKTGYCTFIGGNLATWKTKKQKV 1286
Query: 1189 ISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERT 1248
+S SSAE+EYR++ L++EL WLK LL DLGI+ PIT++CD++AAI+IA N VFHERT
Sbjct: 1287 VSCSSAESEYRAMRKLTNELTWLKALLKDLGIEQHMPITMHCDNKAAIYIASNSVFHERT 1346
Query: 1249 KHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEIS 1304
KHIE+DCH VREKI G+ P Y RS DQLADIFTK I GKLG++++S
Sbjct: 1347 KHIEVDCHKVREKIIEGVTLPCYTRSEDQLADIFTKAASLKVCNFIHGKLGLVDLS 1402
Score = 149 bits (377), Expect = 4e-34
Identities = 123/482 (25%), Positives = 226/482 (46%), Gaps = 53/482 (10%)
Query: 66 VLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKEDILT---------WQRC 116
V+ T +L G NY +WSR L + + V S + KE+ T W +
Sbjct: 7 VITTVILQGGNYLTWSRTTKTVLCGRGLWSHVISSQAPKEDKEEEETETISPEEEKWFQE 66
Query: 117 NDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRF-SQSNAPKIYQLKQSISALKQEG 175
+ V + + NS+ T I Y ETA+++W L++ + ++SN +++++K++I+ L QE
Sbjct: 67 DQAVLALLQNSLETSILEGYSYCETAKELWDTLKNVYGNESNLTRVFEVKKAINELSQED 126
Query: 176 MYVSLYFTQLKSLWDELNSI--ALVNPCICGNAKSILDQQNQDRAMEFLQGVHDRFSAVR 233
+ + +F + +SLW EL S+ ++P K + +++ QD+ L ++ ++ +
Sbjct: 127 LEFTKHFGKFRSLWSELKSLRPGTLDP------KILHERREQDKVFGLLLTLNPGYNDLI 180
Query: 234 SQILLMDPFPSIQRIYNIVRQEEKQQEI--NFRPVPAVESAALQTSKAPYRTPGKRQRPF 291
+L + PS+ + + +++E+ + + + +K Y+ ++
Sbjct: 181 KHLLRSEKLPSLDEVCSKIQKEQGSTGLFGGKSELITANKGEVVANKGVYKNEDRKLLT- 239
Query: 292 CEHCNRHGHTITTCYQIHG------FPSNPSKPQKKT--ETSPSTSANQLSSAQYHKLLT 343
C+HC + GHT C+ +H F + + ++T E S + S+ +S + +
Sbjct: 240 CDHCKKKGHTKDKCWLLHPHLKPAKFKDSRAHFSQETHEEQSQAGSSKGETSTSFGDYVR 299
Query: 344 LLAKEDNVGSSVNLAGTAFT----CIPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQ 399
E + S V+L + T S +IDSGAS+H+ ++ +L + P + +
Sbjct: 300 KSDLEALIKSIVSLKESGITFSSQTSSGSIVIDSGASHHMISNSNLLDNIEPALGHVIIA 359
Query: 400 QPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQ 459
+G + ++ IG + + + +P F NL+SV + T LNC AIF + FQ
Sbjct: 360 --NGDKVPIEGIGNLKLFNKD--SKAFFMPKFTSNLLSVKRTTRDLNCYAIFGPNDVYFQ 415
Query: 460 DQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYCLSHPLSSLFSSKH---CNKFDLWHL 516
D T K+IG G ++ ELY L +DL N SS FSSK + LWH
Sbjct: 416 DIETGKVIGEGGSKGELYVL-EDLSPNS------------SSCFSSKSHLGISFNTLWHA 462
Query: 517 RL 518
RL
Sbjct: 463 RL 464
>UniRef100_O65452 LTR retrotransposon like protein [Arabidopsis thaliana]
Length = 1109
Score = 605 bits (1561), Expect = e-171
Identities = 295/543 (54%), Positives = 397/543 (72%), Gaps = 1/543 (0%)
Query: 764 VTSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALES 823
+ + FP +S + ++SH+AFL + EP +Y++AM WR+AM+ EI++L
Sbjct: 567 IFQETEFPYSK-MSCNRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRV 625
Query: 824 NNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVA 883
N T+ + LP GK A+G KW+YKIKY SDG+I+RYKARLV G Q +G+DY +TFAPVA
Sbjct: 626 NQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVA 685
Query: 884 KLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSI 943
K+ TVRL L +AA ++W ++Q DV+NAFL GDL EEVYMKLP GF VC+L+KS+
Sbjct: 686 KMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSL 745
Query: 944 YGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENA 1003
YGLKQA R WFSK S+ L Q GF QS+SDYSLF+YN D + VLVYVDD+II+G+ +A
Sbjct: 746 YGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDA 805
Query: 1004 ISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDF 1063
+++ K +L F +KDLG L Y LGIEVSR+ +G +L QRKY LDI+S+ G+ G RPS F
Sbjct: 806 VAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAF 865
Query: 1064 PMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDA 1123
P+EQ+ +L + LSD + YRR VGRL+YL VTRP++ Y+V+TL+QFMQ+P H++A
Sbjct: 866 PLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLAVTRPELSYSVHTLAQFMQNPRQDHWNA 925
Query: 1124 ATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKT 1183
A +V+RYLK + G+G+ LS++S++ + G+ DSD+A CP TRRS TGYF LG PISWKT
Sbjct: 926 AIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKT 985
Query: 1184 KKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPV 1243
KKQPTISRSSAEAEYR++A L+ EL WLK +L DLG+ H Q + I+ DS++AI ++ NPV
Sbjct: 986 KKQPTISRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPV 1045
Query: 1244 FHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEI 1303
HERTKH+E+DCHF+R+ I G+IA S++ S QLADI TK LG + L KLG++++
Sbjct: 1046 QHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGILDV 1105
Query: 1304 SIP 1306
P
Sbjct: 1106 HAP 1108
Score = 119 bits (299), Expect = 5e-25
Identities = 59/121 (48%), Positives = 85/121 (69%), Gaps = 2/121 (1%)
Query: 548 QTYL-FYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMN 606
Q+YL F GEC+L+AAYLIN+ P+ +L+ KSP+++L + P YS LRVFG LC+A N N
Sbjct: 466 QSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYAHNQN 525
Query: 607 IQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNN 665
+ KF RS+ +FVGYP QKG+R++D++ ++ +VSRDV F ET FPY + F +
Sbjct: 526 HKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSCNRFTS 585
Query: 666 A 666
+
Sbjct: 586 S 586
Score = 105 bits (262), Expect = 9e-21
Identities = 61/195 (31%), Positives = 98/195 (49%), Gaps = 10/195 (5%)
Query: 26 MSDIPPSENSPLSSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVT 85
M+D PP S I V T P +DN V+ P+L +NY W+
Sbjct: 1 MADAPPPP-------PSVIEVRRTILPYDLTA--ADNSGAVISHPILKTNNYEEWACGFK 51
Query: 86 MALRAKNKFGFVDGSLTIPKK-KEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQ 144
ALR++ KFGF+DG++ P D+ W N L+ SW+ ++ +E+ +I + + A
Sbjct: 52 TALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLTNISHRDVARD 111
Query: 145 IWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICG 204
+W ++ RFS SN PK ++K ++ KQEGM V Y+ +L +WD +NS + C
Sbjct: 112 LWEQIRKRFSVSNGPKNQKMKADLATCKQEGMTVEGYYGKLNKIWDNINSYRPLRICASH 171
Query: 205 NAKSILDQQNQDRAM 219
+ L+ + R+M
Sbjct: 172 HMTGNLELLSGMRSM 186
Score = 45.8 bits (107), Expect = 0.009
Identities = 36/144 (25%), Positives = 62/144 (43%), Gaps = 16/144 (11%)
Query: 375 ASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFN 434
AS+H+ +L L S ++ + + DG++ + GT+ LIL +V++V + +
Sbjct: 169 ASHHMTGNLELLSGMRSMS-PVLIILADGNKRVAVSEGTVRLGSHLILKSVFYVKELESD 227
Query: 435 LMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYC 494
L+SV Q+ + +C + V QD+ T+ + G G N + H
Sbjct: 228 LISVGQMMDENHCVVQLADHFLVIQDRTTRMVTGIGKRENGSFCFRG---MENAAAVHTS 284
Query: 495 LSHPLSSLFSSKHCNKFDLWHLRL 518
+ P FDLWH RL
Sbjct: 285 VKAP------------FDLWHRRL 296
>UniRef100_Q9FL75 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1109
Score = 605 bits (1560), Expect = e-171
Identities = 294/543 (54%), Positives = 397/543 (72%), Gaps = 1/543 (0%)
Query: 764 VTSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALES 823
+ + FP +S + ++SH+AFL + EP +Y++AM WR+AM+ EI++L
Sbjct: 567 IFQETEFPYSK-MSCNRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRV 625
Query: 824 NNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVA 883
N T+ + LP GK A+G KW+YKIKY SDG+I+RYKARLV G Q +G+DY +TFAPVA
Sbjct: 626 NQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVA 685
Query: 884 KLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSI 943
K+ TVRL L +AA ++W ++Q DV+NAFL GDL EEVYMKLP GF VC+L+KS+
Sbjct: 686 KMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSL 745
Query: 944 YGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENA 1003
YGLKQA R WFSK S+ L Q GF QS+SDYSLF+YN D + VLVYVDD+II+G+ +A
Sbjct: 746 YGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDA 805
Query: 1004 ISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDF 1063
+++ K +L F +KDLG L Y LGIEVSR+ +G +L QRKY LDI+S+ G+ G RPS F
Sbjct: 806 VAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAF 865
Query: 1064 PMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDA 1123
P+EQ+ +L + LSD + YRR VGRL+YL VTRP++ Y+V+TL+QFMQ+P H++A
Sbjct: 866 PLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLAVTRPELSYSVHTLAQFMQNPRQDHWNA 925
Query: 1124 ATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKT 1183
A +V+RYLK + G+G+ LS++S++ + G+ DSD+A CP TRRS TGYF LG PISWKT
Sbjct: 926 AIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKT 985
Query: 1184 KKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPV 1243
KKQPT+SRSSAEAEYR++A L+ EL WLK +L DLG+ H Q + I+ DS++AI ++ NPV
Sbjct: 986 KKQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPV 1045
Query: 1244 FHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEI 1303
HERTKH+E+DCHF+R+ I G+IA S++ S QLADI TK LG + L KLG++++
Sbjct: 1046 QHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGILDV 1105
Query: 1304 SIP 1306
P
Sbjct: 1106 HAP 1108
Score = 119 bits (299), Expect = 5e-25
Identities = 59/121 (48%), Positives = 86/121 (70%), Gaps = 2/121 (1%)
Query: 548 QTYL-FYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMN 606
Q+YL F GEC+L+AAYLIN+ P+ +L+ KSP+++L + P+YS LRVFG LC+A N N
Sbjct: 466 QSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPNYSHLRVFGSLCYAHNQN 525
Query: 607 IQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNN 665
+ KF RS+ +FVGYP QKG+R++D++ ++ +VSRDV F ET FPY + F +
Sbjct: 526 HKGDKFVARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSCNRFTS 585
Query: 666 A 666
+
Sbjct: 586 S 586
Score = 105 bits (262), Expect = 9e-21
Identities = 62/218 (28%), Positives = 106/218 (48%), Gaps = 14/218 (6%)
Query: 26 MSDIPPSENSPLSSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVT 85
M+D PP S I V T +P +DN V+ P+L +NY W+
Sbjct: 1 MADAPPPP-------PSVIEVRRTISPYDLTA--ADNSGAVISHPILKTNNYEEWACGFK 51
Query: 86 MALRAKNKFGFVDGSLTIPKK-KEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQ 144
ALR++ KFGF+DG++ P D+ W N L+ SW+ ++ +E+ +I + + A
Sbjct: 52 TALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLTNISHRDVARD 111
Query: 145 IWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPC--- 201
+W ++ RF SN PK ++K ++ KQEGM + Y+ +L +WD +NS + C
Sbjct: 112 LWEQIRKRFFVSNGPKNQKMKADLATCKQEGMTMEGYYGKLNKIWDNINSYRPLRICASH 171
Query: 202 -ICGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILL 238
+ GN + + D ++ + L + R + + L
Sbjct: 172 HMTGNLELLSDMRSMSPVLIILADGNKRVAVSEGTVRL 209
Score = 45.8 bits (107), Expect = 0.009
Identities = 36/144 (25%), Positives = 62/144 (43%), Gaps = 16/144 (11%)
Query: 375 ASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFN 434
AS+H+ +L L S ++ + + DG++ + GT+ LIL +V++V + +
Sbjct: 169 ASHHMTGNLELLSDMRSMS-PVLIILADGNKRVAVSEGTVRLGSHLILKSVFYVKELESD 227
Query: 435 LMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYC 494
L+SV Q+ + +C + V QD+ T+ + G G N + H
Sbjct: 228 LISVGQMMDENHCVVQLADHFLVIQDRTTRMVTGIGKRENGSFCFRG---MENAAAVHTS 284
Query: 495 LSHPLSSLFSSKHCNKFDLWHLRL 518
+ P FDLWH RL
Sbjct: 285 VKAP------------FDLWHRRL 296
>UniRef100_O81617 F8M12.17 protein [Arabidopsis thaliana]
Length = 1633
Score = 603 bits (1554), Expect = e-170
Identities = 327/791 (41%), Positives = 476/791 (59%), Gaps = 73/791 (9%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAK-N 604
F+ + +CVLTAAYLIN+LP+P+L K+P ++LL P Y+ L+ CLC+A N
Sbjct: 673 FQSNVPLQYWSDCVLTAAYLINRLPSPLLDNKTPFELLLKKIPDYTLLK--SCLCYASTN 730
Query: 605 MNIQHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
++ ++KF R++P +F+GYP KGY++ D+++ I ++R+V FHET FP++
Sbjct: 731 VHDRNKFSPRARPCVFLGYPSGYKGYKVLDLESHSISITRNVVFHETKFPFK-------- 782
Query: 665 NAISINTQILDNEFDDLFTGS------PTH--PNIPPENSHNDNSNDTIVTISTPEDDAS 716
T E D+F S P H ++P ++ + N+ + S AS
Sbjct: 783 ------TSKFLKESVDMFPNSILPLPAPLHFVESMPLDDDLRADDNNA--STSNSASSAS 834
Query: 717 SNPPSFSAESLSNNPP-STEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKIN------ 769
S PP S + N + N +R P +L +Y C S ++
Sbjct: 835 SIPPLPSTVNTQNTDALDIDTNSVPIARPKRNAKAPAYLSEYHCNSVPFLSSLSPTTSTS 894
Query: 770 ----------------FPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDA 813
+P+ +S L+ +++ EPK+++QAMKS +W A
Sbjct: 895 IETPSSSIPPKKITTPYPMSTAISYDKLTPLFHSYICAYNVETEPKAFTQAMKSEKWTRA 954
Query: 814 MAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGI 873
+E+ ALE N TWI+ L EGK+ +GCKW++ IKY+ DGSI+RYKARLVA+G++Q +GI
Sbjct: 955 ANEELHALEQNKTWIVESLTEGKNVVGCKWVFTIKYNPDGSIERYKARLVAQGFTQQEGI 1014
Query: 874 DYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGF----- 928
DY +TF+PVAK +V+LLL +AA W L Q DV+NAFL G+L EE+YM LP G+
Sbjct: 1015 DYMETFSPVAKFGSVKLLLGLAAATGWSLTQMDVSNAFLHGELDEEIYMSLPQGYTPPTG 1074
Query: 929 -SHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFV 987
S KP VC+L KS+YGLKQASRQW+ + S+ + F QS +D ++F + I V
Sbjct: 1075 ISLPSKP-VCRLLKSLYGLKQASRQWYKRLSSVFLGANFIQSPADNTMFVKVSCTSIIVV 1133
Query: 988 LVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTL 1047
LVYVDD++I N+ +A+ +K+ L F IKDLG + LG+E++RS +GI +CQRKY
Sbjct: 1134 LVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVCQRKYAQ 1193
Query: 1048 DILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVN 1107
++L D G++GC+PS PM+ +L L GT L + T YR VGRLLYL +TRPDI +AV+
Sbjct: 1194 NLLEDVGLSGCKPSSIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDITFAVH 1253
Query: 1108 TLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRST 1167
TLSQF+ +P H AA +VLRYLKG+ G+ D+DW C +RRS
Sbjct: 1254 TLSQFLSAPTDIHMQAAHKVLRYLKGNPGQ----------------DADWGTCKDSRRSV 1297
Query: 1168 TGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPIT 1227
TG+ LG++ I+WK+KKQ +SRSS E+EYRSLA + E+ WL+ LL DL + P
Sbjct: 1298 TGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAK 1357
Query: 1228 IYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLG 1287
++CD+++A+H+A NPVFHERTKHIEIDCH VR++IK+G + ++ + +QLADI TKPL
Sbjct: 1358 LFCDNKSALHLATNPVFHERTKHIEIDCHTVRDQIKAGKLKTLHVPTGNQLADILTKPLH 1417
Query: 1288 GDAYKRILGKL 1298
+ +L ++
Sbjct: 1418 PGPFHSLLKRI 1428
Score = 235 bits (600), Expect = 6e-60
Identities = 163/527 (30%), Positives = 258/527 (48%), Gaps = 108/527 (20%)
Query: 32 SENSPLSSIS---SPITVHSTTN------PDPCVIHHSDNPSTVLVTPLL-TGDNYGSWS 81
S N+PL+S S P +V + +P +H SD+ VLV+ L T ++ SW
Sbjct: 3 SMNNPLNSSSLRPPPASVPEQSRYQADQYENPHHLHTSDHAGLVLVSERLNTASDFHSWR 62
Query: 82 RAVTMALRAKNKFGFVDGSLTIPK-KKEDILTWQRCNDLVASWILNSVSTEIRPSILYAE 140
R++ MAL +NK GF+DG++ P D W RCND V++W++NSVS +I S+L+
Sbjct: 63 RSIWMALNVRNKLGFIDGTIVKPPLDHRDYGAWSRCNDTVSTWLMNSVSKKIGQSLLFIP 122
Query: 141 TAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNP 200
TAE IW ++ RF Q +AP++Y ++Q +S ++Q M +S Y+T+L++LW+E + +
Sbjct: 123 TAEGIWKNMLSRFKQDDAPRVYDIEQRLSKIEQGSMDISAYYTELQTLWEEHKNYVDLPV 182
Query: 201 CICGNAK---SILDQQNQDRA--MEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQE 255
C CG + ++ ++ Q R+ +FL G+++ + R IL++ P +I+ +NIV Q+
Sbjct: 183 CTCGRCECDAAVKWERLQQRSHVTKFLMGLNESYEQTRRHILMLKPIRTIEEAFNIVTQD 242
Query: 256 EKQQEINFRPVPAVESAALQTSKAPYRTPGKRQRPFCEHCNRHGHTITTCYQIHGFP--- 312
E+Q+ I RP P V++ + + P C +C + GHT+ CY+I G+P
Sbjct: 243 ERQKAI--RPTPKVDN------------QDQLKLPLCTNCGKVGHTVQKCYKIIGYPPGY 288
Query: 313 -SNPSKPQKKTETSPSTSANQLSSAQYHKLLTLLAKEDNVGSSVN--LAGTAFT------ 363
+ S Q + +T P Q S + + + L + N V A + +T
Sbjct: 289 KAATSYRQPQIQTQPRMQMPQQSQPRMQQPIQHLISQFNAQVRVQEPAATSIYTSSPTAT 348
Query: 364 --------------CIPF-----------------------------SWIIDSGASNHIC 380
IPF +WIIDSGAS+H+C
Sbjct: 349 ITEHGLMAQTSTSGTIPFPSTSLKYENNNLTFQNHTLSSLQNVLSSDAWIIDSGASSHVC 408
Query: 381 TSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSV-- 438
+ L++F V+ ++V P+G++ + H GTI + +LIL NV VP FKFNL+SV
Sbjct: 409 SDLTMFRELIHVSG-VTVTLPNGTRVAITHTGTICITSTLILHNVLLVPDFKFNLISVCC 467
Query: 439 TQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVS 485
+LT L MIGRG N LY L S
Sbjct: 468 LELTRGL--------------------MIGRGKTYNNLYILETQRTS 494
>UniRef100_Q710T7 Gag-pol polyprotein [Populus deltoides]
Length = 1382
Score = 581 bits (1498), Expect = e-164
Identities = 330/749 (44%), Positives = 457/749 (60%), Gaps = 37/749 (4%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQ-HKFD 612
F GE VLTA LIN +P+ SP + L G P YSS RVFGC F + +++ +K
Sbjct: 665 FWGEAVLTAVSLINTIPSSHSSGLSPFEKLYGHVPDYSSFRVFGCTYFVLHPHVERNKLS 724
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQ 672
RS +F+GY +KGYR +D T+++YVS V F E + P+ I P ++++ +
Sbjct: 725 SRSAICVFLGYGEGKKGYRCFDPITQKLYVSHHVVFLEHI-PFFSI--PSTTHSLTKSDL 781
Query: 673 ILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPP 732
I + F + +G+ T P + +HN T+++ TPE SS P S+E + P
Sbjct: 782 IHIDPFSE-DSGNDTSPYVRSICTHNSAGTGTLLS-GTPEASFSSTAPQASSEIVDPPP- 838
Query: 733 STEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHRAFLINI 792
R S RIR L D+ + S S+S +FL I
Sbjct: 839 ---------RQSIRIRKST-KLPDF-------------------AYSCYSSSFTSFLAYI 869
Query: 793 IENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSD 852
EP SY +A+ + AM +E+ AL +TW L PLP GKS +GC+W+YKIK +SD
Sbjct: 870 HCLFEPSSYKEAILDPLGQQAMDEELSALHKTDTWDLVPLPPGKSVVGCRWVYKIKTNSD 929
Query: 853 GSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFL 912
GSI+RYKARLVAKGYSQ G+DY +TFAP+AK+ T+R L+++A+I+ W + Q DV NAFL
Sbjct: 930 GSIERYKARLVAKGYSQQYGMDYEETFAPIAKMTTIRTLIAVASIRQWHISQLDVKNAFL 989
Query: 913 QGDLSEEVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISD 972
GDL EEVYM PPG SH VCKL K++YGLKQA R WF KFS + GF S D
Sbjct: 990 NGDLQEEVYMAPPPGISHDSG-YVCKLKKALYGLKQAPRAWFEKFSIVISSLGFVSSSHD 1048
Query: 973 YSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVS 1032
+LF D I + +YVDD+IITG++ + IS +K LA+ F +KDLG L Y LGIEV+
Sbjct: 1049 SALFIKCTDAGRIILSLYVDDMIITGDDIDGISVLKTELARRFEMKDLGYLRYFLGIEVA 1108
Query: 1033 RSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRL 1092
S +G L Q KY +IL + +T + D P+E + R +DG PL DPT+YR VG L
Sbjct: 1109 YSPRGYLLSQSKYVANILERARLTDNKTVDTPIEVNARYSSSDGLPLIDPTLYRTIVGSL 1168
Query: 1093 LYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGY 1152
+YLT+T PDI YAV+ +SQF+ SP + H+ A ++LRYL+G+V + L LS++SS+ L Y
Sbjct: 1169 VYLTITHPDIAYAVHVVSQFVASPTTIHWAAVLRILRYLRGTVFQSLLLSSTSSLELRAY 1228
Query: 1153 ADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLK 1212
+D+D PT R+S TG+ LG + ISWK+KKQ +S+SS EAEY ++A+ + E+ W +
Sbjct: 1229 SDADHGSDPTDRKSVTGFCIFLGDSLISWKSKKQSIVSQSSTEAEYCAMASTTKEIVWSR 1288
Query: 1213 YLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYI 1272
+LL+D+GI +YCD+Q++I IA N VFHERTKHIEIDCH R +K G IA ++
Sbjct: 1289 WLLADMGISFSHLTPMYCDNQSSIQIAHNSVFHERTKHIEIDCHLTRHHLKHGTIALPFV 1348
Query: 1273 RSSDQLADIFTKPLGGDAYKRILGKLGVI 1301
SS Q+AD FTK + ++GKL ++
Sbjct: 1349 PSSLQIADFFTKAHSISRFCFLVGKLSML 1377
Score = 185 bits (470), Expect = 7e-45
Identities = 141/486 (29%), Positives = 224/486 (46%), Gaps = 47/486 (9%)
Query: 68 VTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKED-----ILTWQRCNDLVAS 122
V+ L G NY WS + L+ K +G+V G+ +PK E+ I TW+ N + +
Sbjct: 12 VSVRLDGKNYSYWSYVMRNFLKGKKMWGYVSGTYVVPKNTEEGDTVSIDTWEANNAKIIT 71
Query: 123 WILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYF 182
WI N V I + ETA+++W LQ F+QSN K YQL+ I AL Q+ M + ++
Sbjct: 72 WINNYVEHSIGTQLAKYETAKEVWDHLQRLFTQSNFAKQYQLENDIRALHQKNMSIQEFY 131
Query: 183 TQLKSLWDELNSIALVNPCICGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPF 242
+ + LWD+L V CG + ++++ Q R ++FL + F +R IL P
Sbjct: 132 SAMTDLWDQLALTESVELKACG---AYIERREQQRLVQFLTALRSDFEGLRGSILHRSPL 188
Query: 243 PSIQRIYN-IVRQEEKQQEINFRPV-PAVESAALQTSKAPYRTPGKRQRPF-------CE 293
PS+ + + ++ +E + Q + + + A + L P+ + +P+ C
Sbjct: 189 PSVDSVVSELLAEEIRLQSYSEKGILSASNPSVLAVPSKPF--SNHQNKPYTRVGFDECS 246
Query: 294 HCNRHGHTITTCYQI-----------------HGFPSNPSKPQKKTETSPS----TSANQ 332
C + GH C ++ H P P T S T N
Sbjct: 247 FCKQKGHWKAQCPKLRQQNQAWKSGSQSQSNAHRSPQGYKPPHHNTAAVASPGSITDPNT 306
Query: 333 LSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCIPFSWIIDSGASNHICTSLSLFSSYYPV 392
L+ Q+ K L+L + + S L ++ W++DSGAS+H+ S F+S P+
Sbjct: 307 LAE-QFQKFLSLQPQAMSASSIGQLPHSSSGISHSEWVLDSGASHHMSPDSSSFTSVSPL 365
Query: 393 NNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFS 452
++ I V DG+ + +G++ + L L NVY +P K NL S+ Q+ +S + +FS
Sbjct: 366 SS-IPVMTADGTPMPLAGVGSV-VTLHLSLPNVYLIPKLKLNLASIGQICDSGDYLVMFS 423
Query: 453 SSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYCLSHPLSSLFSSKHCNKFD 512
S C QD ++K+IG G N LY L++ V LS SL SS F
Sbjct: 424 GSFCCVQDLQSQKLIGTGRRENGLYILDELKVPVVVAATTVDLSFFRLSLSSS----SFY 479
Query: 513 LWHLRL 518
LWH RL
Sbjct: 480 LWHSRL 485
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,239,518,096
Number of Sequences: 2790947
Number of extensions: 96811719
Number of successful extensions: 278488
Number of sequences better than 10.0: 1732
Number of HSP's better than 10.0 without gapping: 1450
Number of HSP's successfully gapped in prelim test: 291
Number of HSP's that attempted gapping in prelim test: 270810
Number of HSP's gapped (non-prelim): 4071
length of query: 1309
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1170
effective length of database: 460,108,200
effective search space: 538326594000
effective search space used: 538326594000
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 81 (35.8 bits)
Medicago: description of AC124966.5


