

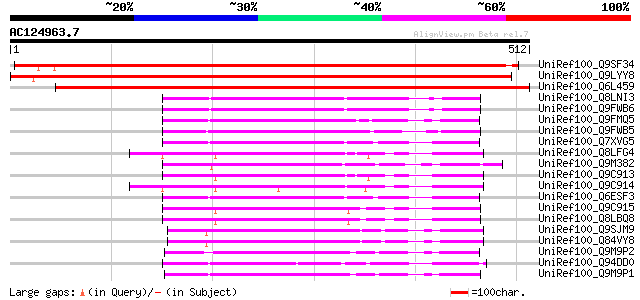
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.7 + phase: 0
(512 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SF34 Hypothetical protein F11F8_28 [Arabidopsis thal... 604 e-171
UniRef100_Q9LYY8 Hypothetical protein F9G14_280 [Arabidopsis tha... 598 e-169
UniRef100_Q6L459 Hypothetical protein PGEC589.7 [Solanum demissum] 567 e-160
UniRef100_Q8LNI3 Hypothetical protein OSJNBb0028C01.43 [Oryza sa... 151 4e-35
UniRef100_Q9FWB6 Putative alpha/beta hydrolase [Oryza sativa] 151 4e-35
UniRef100_Q9FMQ5 Gb|AAF31728.1 [Arabidopsis thaliana] 151 5e-35
UniRef100_Q9FWB5 Putative alpha/beta hydrolase [Oryza sativa] 144 8e-33
UniRef100_Q7XVG5 OSJNBa0073L04.5 protein [Oryza sativa] 143 1e-32
UniRef100_Q8LFG4 Hypothetical protein [Arabidopsis thaliana] 138 4e-31
UniRef100_Q9M382 Hypothetical protein F24B22.200 [Arabidopsis th... 136 1e-30
UniRef100_Q9C913 Hypothetical protein F1O17.5 [Arabidopsis thali... 136 2e-30
UniRef100_Q9C914 Hypothetical protein F1O17.4 [Arabidopsis thali... 131 4e-29
UniRef100_Q6ESF3 Hydrolase, alpha/beta fold protein-like [Oryza ... 131 4e-29
UniRef100_Q9C915 Hypothetical protein F1O17.3 [Arabidopsis thali... 130 9e-29
UniRef100_Q8LBQ8 Hypothetical protein [Arabidopsis thaliana] 130 9e-29
UniRef100_Q9SJM9 Expressed protein [Arabidopsis thaliana] 126 2e-27
UniRef100_Q84VY8 At2g36290 [Arabidopsis thaliana] 126 2e-27
UniRef100_Q9M9P2 T17B22.7 protein [Arabidopsis thaliana] 121 5e-26
UniRef100_Q94DD0 P0683F02.22 protein [Oryza sativa] 116 1e-24
UniRef100_Q9M9P1 T17B22.8 protein [Arabidopsis thaliana] 115 3e-24
>UniRef100_Q9SF34 Hypothetical protein F11F8_28 [Arabidopsis thaliana]
Length = 527
Score = 604 bits (1558), Expect = e-171
Identities = 288/506 (56%), Positives = 376/506 (73%), Gaps = 13/506 (2%)
Query: 5 RGTTWWSEELASLMENSLPESST----TTSFEDVRKSSSSEE---VEVGESLKEHAVGFL 57
RG W +ELASLM+ L + T E KSS E E ESLK+ GF+
Sbjct: 6 RGVPTWQDELASLMDGGLQYDGSPIDLTADTESRSKSSGFESGSGSESVESLKDQVTGFM 65
Query: 58 MAWCEILMELGRGCRDILQQNFFNEDSYLVQKLRGPCSKLSKRLSFLNDFLPEDRDPLLA 117
+W E+LM+L GC+D++QQ +DS++V+KLR P +K+SK+LSFLN++LPEDRDP+ A
Sbjct: 66 KSWGEMLMDLAIGCKDVVQQMVVTDDSFVVRKLRKPAAKVSKKLSFLNEYLPEDRDPVHA 125
Query: 118 WSIVFTVFLLAFAAISVDSNHQTSTKAAM-VRMHPPIASRILLPDGRYMAYQDQGVPPGR 176
W ++F VFLLA A+S S+H S +R+HP ASR+ LPDGRY+AYQ+ GV R
Sbjct: 126 WPVIFFVFLLALTALSFSSDHDRSVPLLKKIRLHPTSASRVQLPDGRYLAYQELGVSADR 185
Query: 177 ARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVTYDLPGFGESDPHPSRNFNSSAMDM 236
AR SL+ PHSFLSSRLAGIPGVK SLL+DYGVRLV+YDLPGFGESDPH +RN +SSA DM
Sbjct: 186 ARHSLIVPHSFLSSRLAGIPGVKESLLKDYGVRLVSYDLPGFGESDPHRARNLSSSASDM 245
Query: 237 LHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYESHMTKDEMKR 296
+ L A+ + DKFW+L +SSG +HAWA+++Y P++IAG AM+APM++PYE MTK+EM +
Sbjct: 246 IDLAAALGIVDKFWLLGYSSGSVHAWAAMRYFPDQIAGVAMVAPMINPYEPSMTKEEMAK 305
Query: 297 TWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDEILVDEPAF 356
TWE+W +RK+MY LA R+P LL F YR+SFL E +DK S+SLG+KD+++ +P F
Sbjct: 306 TWEQWQRKRKFMYFLARRWPSLLPFSYRRSFLSGNLEPLDKWMSVSLGEKDKLVTADPVF 365
Query: 357 EEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQA 416
E+ +QR++EESVRQG KPF+EEA LQVS W F++ E H+ KKC+T G+L WL SMY ++
Sbjct: 366 EDLYQRNVEESVRQGTAKPFVEEAALQVSNWGFSLPEFHMQKKCRTNGVLSWLMSMYSES 425
Query: 417 ECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFFFCDECHKQ 476
ECEL G+ IHIWQG+DDR+ PPS+T+YI RV+PEA +H+LPNEGHFSYF+ CDECH Q
Sbjct: 426 ECELIGFRKPIHIWQGMDDRVTPPSVTDYISRVIPEATVHRLPNEGHFSYFYLCDECHTQ 485
Query: 477 IFSTLFGTPQGPFERQEETMLEKNTE 502
IFS +FG P+GP E +LE+ TE
Sbjct: 486 IFSAIFGEPKGPVE-----LLEQRTE 506
>UniRef100_Q9LYY8 Hypothetical protein F9G14_280 [Arabidopsis thaliana]
Length = 514
Score = 598 bits (1541), Expect = e-169
Identities = 283/501 (56%), Positives = 373/501 (73%), Gaps = 6/501 (1%)
Query: 1 MAEARGTTWWSEELASLMENSL-----PESSTTTSFEDVRKSSSSEEVEVGESLKEHAVG 55
M E RG W EELASL++ L P T + S+ E E+LK+ G
Sbjct: 1 MEEIRGVPTWQEELASLVDAGLRYDGAPIDLTAATKRSGFVSADGSGSEPKETLKDQVTG 60
Query: 56 FLMAWCEILMELGRGCRDILQQNFFNEDSYLVQKLRGPCSKLSKRLSFLNDFLPEDRDPL 115
F+ +W E+L+EL +GC+DI+QQ +DS+LV+KLR P +K+SK+LSFLN+FLPEDRDP+
Sbjct: 61 FMKSWGEMLLELAKGCKDIVQQTVVTDDSFLVRKLRKPAAKVSKKLSFLNEFLPEDRDPI 120
Query: 116 LAWSIVFTVFLLAFAAISVD-SNHQTSTKAAMVRMHPPIASRILLPDGRYMAYQDQGVPP 174
AW ++F VFLLA AA+S N + T +R+HP A+R+ LPDGRY+AYQ+ GV
Sbjct: 121 HAWPVIFFVFLLALAALSFSPENDRPVTVITKLRLHPTGATRVQLPDGRYIAYQELGVSA 180
Query: 175 GRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVTYDLPGFGESDPHPSRNFNSSAM 234
RAR+SLV PHSFLSSRLAGIPGVK SLL +YGVRLV+YDLPGFGESDPH RN +SSA
Sbjct: 181 ERARYSLVMPHSFLSSRLAGIPGVKKSLLVEYGVRLVSYDLPGFGESDPHRGRNLSSSAS 240
Query: 235 DMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYESHMTKDEM 294
DM++L A+ + +KFW+L +S+G IH WA +KY PE+IAGAAM+AP+++PYE M K+E+
Sbjct: 241 DMINLAAAIGIDEKFWLLGYSTGSIHTWAGMKYFPEKIAGAAMVAPVINPYEPSMVKEEV 300
Query: 295 KRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDEILVDEP 354
+TWE+WL +RK+MY LA RFP LL FFYR+SFL +++D+ +LSLG+KD++L+ +P
Sbjct: 301 VKTWEQWLTKRKFMYFLARRFPILLPFFYRRSFLSGNLDQLDQWMALSLGEKDKLLIKDP 360
Query: 355 AFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYG 414
F+E +QR++EESVRQG KPF+EEA+LQVS W F + E KKC T G+L WL SMY
Sbjct: 361 TFQEVYQRNVEESVRQGITKPFVEEAVLQVSNWGFTLSEFRTQKKCATNGVLSWLMSMYS 420
Query: 415 QAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFFFCDECH 474
+AECEL G+ IHIWQG++DR+ PPSM++YI R++PEA +HK+ NEGHFS+F+FCDECH
Sbjct: 421 EAECELIGFRKPIHIWQGMEDRVAPPSMSDYISRMIPEATVHKIRNEGHFSFFYFCDECH 480
Query: 475 KQIFSTLFGTPQGPFERQEET 495
+QIF LFG P+G ER +ET
Sbjct: 481 RQIFYALFGEPKGQLERVKET 501
>UniRef100_Q6L459 Hypothetical protein PGEC589.7 [Solanum demissum]
Length = 547
Score = 567 bits (1462), Expect = e-160
Identities = 267/468 (57%), Positives = 354/468 (75%), Gaps = 1/468 (0%)
Query: 46 GESLKEHAVGFLMAWCEILMELGRGCRDILQQNFFNEDSYLVQKLRGPCSKLSKRLSFLN 105
GE+ K HAV F + E+ +E G+G RD+L+Q+ EDS LV+K+ C K+ +RLSFLN
Sbjct: 51 GENWKFHAVEFAKGFVEMSVEFGKGVRDVLKQSVIREDSILVRKVGPLCCKVCRRLSFLN 110
Query: 106 DFLPEDRDPLLAWSIVFTVFLLAFAA-ISVDSNHQTSTKAAMVRMHPPIASRILLPDGRY 164
++LPEDRDP AWS++F V LA A I+ + N +T V +HPP ASRI LPDGR+
Sbjct: 111 EYLPEDRDPAHAWSVIFFVLFLASAVLIASNDNIYPTTSVKKVCIHPPSASRISLPDGRH 170
Query: 165 MAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVTYDLPGFGESDPH 224
+AYQ QGVP ARFS++APHSF+SSRLAGIPG+K SLL++YG+RLVTYDLPGFGESDPH
Sbjct: 171 LAYQQQGVPAELARFSMIAPHSFVSSRLAGIPGIKTSLLQEYGIRLVTYDLPGFGESDPH 230
Query: 225 PSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSP 284
PSRN SSAMDMLHL AVNVTDKFWV+ S GC+HAWA+L+YIP+RIAGA M+APMVSP
Sbjct: 231 PSRNLESSAMDMLHLSYAVNVTDKFWVVGFSDGCMHAWAALRYIPDRIAGAVMVAPMVSP 290
Query: 285 YESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLG 344
YE MTK+E + W+KW ++K MY LA +FP+LL + YR+SFL H +I+ + +LSLG
Sbjct: 291 YEPRMTKEEKSKMWKKWTTKKKNMYILARKFPRLLPYLYRRSFLSGVHGQIETRLALSLG 350
Query: 345 KKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGG 404
+D+ L++ P FE++WQRD+EESVRQ N KPF+EEA+LQVS W F+ +L V +K G
Sbjct: 351 IRDKALLEHPLFEKFWQRDVEESVRQKNAKPFLEEAVLQVSNWGFSPADLKVQRKRPGKG 410
Query: 405 LLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHF 464
++ W+KS++GQ E L G+LG+IH+WQG++D +VPPS +++++RVLP+A++H+L EGHF
Sbjct: 411 IMHWIKSLFGQTEEILTGFLGQIHVWQGMEDMVVPPSTSDFLQRVLPDAMVHRLLYEGHF 470
Query: 465 SYFFFCDECHKQIFSTLFGTPQGPFERQEETMLEKNTEDALVSVSDIE 512
+YF+FCDECH+ IFST+FG PQGP E E + + + D E
Sbjct: 471 TYFYFCDECHRHIFSTVFGNPQGPLTPAPEPEPEPEPDQSPIQNDDGE 518
>UniRef100_Q8LNI3 Hypothetical protein OSJNBb0028C01.43 [Oryza sativa]
Length = 332
Score = 151 bits (382), Expect = 4e-35
Identities = 93/315 (29%), Positives = 159/315 (49%), Gaps = 30/315 (9%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP+ A R+ L DGR++AY++ GVP AR+ +V H F SRL + + E+ GV
Sbjct: 30 PPVTAPRVRLSDGRHLAYEESGVPKEAARYKIVFSHGFTGSRLDSLRA-SPEVAEELGVY 88
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
+V +D G+GESDP+P+R S+A+DM L DA+ + DKF+V+ S G W +L+YIP
Sbjct: 89 MVAFDRAGYGESDPNPNRTVKSAALDMAELADALGLGDKFYVVGVSLGSHAVWGALRYIP 148
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLP 329
ERIAGAAM+AP+V+ + ++ + + ++ +++ P +L ++ +S+LP
Sbjct: 149 ERIAGAAMMAPVVNYWWPGFPAEDAAAAYGRQSYGDQWALRVSHHAPAILHWWMDQSWLP 208
Query: 330 EKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDF 389
+ + K+D + + Q+ E + +QG + + + + +W+F
Sbjct: 209 T--STVVDNTTFLPNKRDADIRRTLTADGTLQKKKEMATQQGINESYYRDMTVMFGKWEF 266
Query: 390 NIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERV 449
+ L + C +HIWQG +D +VP ++ ++
Sbjct: 267 DPMAL-------------------PEPPCP-------VHIWQGDEDGLVPVALQRHVAGK 300
Query: 450 LPEAVIHKLPNEGHF 464
L H+LP GHF
Sbjct: 301 LGWVSYHELPGTGHF 315
>UniRef100_Q9FWB6 Putative alpha/beta hydrolase [Oryza sativa]
Length = 354
Score = 151 bits (382), Expect = 4e-35
Identities = 93/315 (29%), Positives = 159/315 (49%), Gaps = 30/315 (9%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP+ A R+ L DGR++AY++ GVP AR+ +V H F SRL + + E+ GV
Sbjct: 52 PPVTAPRVRLSDGRHLAYEESGVPKEAARYKIVFSHGFTGSRLDSLRA-SPEVAEELGVY 110
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
+V +D G+GESDP+P+R S+A+DM L DA+ + DKF+V+ S G W +L+YIP
Sbjct: 111 MVAFDRAGYGESDPNPNRTVKSAALDMAELADALGLGDKFYVVGVSLGSHAVWGALRYIP 170
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLP 329
ERIAGAAM+AP+V+ + ++ + + ++ +++ P +L ++ +S+LP
Sbjct: 171 ERIAGAAMMAPVVNYWWPGFPAEDAAAAYGRQSYGDQWALRVSHHAPAILHWWMDQSWLP 230
Query: 330 EKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDF 389
+ + K+D + + Q+ E + +QG + + + + +W+F
Sbjct: 231 T--STVVDNTTFLPNKRDADIRRTLTADGTLQKKKEMATQQGINESYYRDMTVMFGKWEF 288
Query: 390 NIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERV 449
+ L + C +HIWQG +D +VP ++ ++
Sbjct: 289 DPMAL-------------------PEPPCP-------VHIWQGDEDGLVPVALQRHVAGK 322
Query: 450 LPEAVIHKLPNEGHF 464
L H+LP GHF
Sbjct: 323 LGWVSYHELPGTGHF 337
>UniRef100_Q9FMQ5 Gb|AAF31728.1 [Arabidopsis thaliana]
Length = 340
Score = 151 bits (381), Expect = 5e-35
Identities = 91/314 (28%), Positives = 159/314 (49%), Gaps = 28/314 (8%)
Query: 151 PPIAS-RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP+ S RI L DGRY+AY++ GV A + ++ H F SS+ P + ++E+ G+
Sbjct: 35 PPVTSPRIKLSDGRYLAYRESGVDRDNANYKIIVVHGFNSSKDTEFP-IPKDVIEELGIY 93
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
V YD G+GESDPHPSR S A D+ L D + + KF+VL S G ++ LKYIP
Sbjct: 94 FVFYDRAGYGESDPHPSRTVKSEAYDIQELADKLKIGPKFYVLGISLGAYSVYSCLKYIP 153
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLP 329
R+AGA ++ P V+ + + + ++++ + E + ++ + +A+ P LL ++ + P
Sbjct: 154 HRLAGAVLMVPFVNYWWTKVPQEKLSKALELMPKKDQWTFKVAHYVPWLLYWWLTQKLFP 213
Query: 330 EKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDF 389
S KD +++ + E + LE+ +QG+ + + + + W+F
Sbjct: 214 SSSMVTGNNALCS--DKDLVVIKKKM--ENPRPGLEKVRQQGDHECLHRDMIAGFATWEF 269
Query: 390 NIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERV 449
+ E L++ + + E G +H+WQG++DR++P + YI
Sbjct: 270 DPTE---------------LENPFAEGE-------GSVHVWQGMEDRIIPYEINRYISEK 307
Query: 450 LPEAVIHKLPNEGH 463
LP H++ GH
Sbjct: 308 LPWIKYHEVLGYGH 321
>UniRef100_Q9FWB5 Putative alpha/beta hydrolase [Oryza sativa]
Length = 353
Score = 144 bits (362), Expect = 8e-33
Identities = 95/316 (30%), Positives = 157/316 (49%), Gaps = 31/316 (9%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP+ A+R+ + DGR++AY + GV ARF +V H F R+ P +LL++ GV
Sbjct: 47 PPVTAARVRVRDGRFLAYAESGVRREAARFKVVYSHGFSGGRMDS-PRASQALLKELGVY 105
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
+V +D G+GESDP P R+ S+AMD+ L DA+ + KF ++C S GC AWAS KYIP
Sbjct: 106 MVAFDRAGYGESDPDPRRSLRSAAMDIQDLADALQLGPKFHLICSSLGCHAAWASFKYIP 165
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLP 329
R+AGAAM+AP+++ + + ++ + + ++ +AY P LL ++ +++LP
Sbjct: 166 HRLAGAAMMAPVINYRWPGLPRGLARQLYRRQPVGDQWSLRVAYYAPWLLHWWMNQTWLP 225
Query: 330 EKHERIDK-QFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWD 388
F +L +K+ ++ +Q+ + +QG F + + RW
Sbjct: 226 TSTVISGSGSFPNALDEKNRLMALSTGL---FQKKARMATQQGVQDSFYRDMAVMFGRW- 281
Query: 389 FNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIER 448
+ AE E + +H++QG +D +VP + +I R
Sbjct: 282 ----------------------PEFEPAELEKPPF--PVHLFQGDEDGVVPVQLQRHICR 317
Query: 449 VLPEAVIHKLPNEGHF 464
L H+L GHF
Sbjct: 318 RLGWISYHELAGVGHF 333
>UniRef100_Q7XVG5 OSJNBa0073L04.5 protein [Oryza sativa]
Length = 344
Score = 143 bits (360), Expect = 1e-32
Identities = 85/314 (27%), Positives = 156/314 (49%), Gaps = 30/314 (9%)
Query: 151 PPIA-SRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP+ +R L DGR++AY + GVP +A++ ++ H F S R +P + L ++ G+
Sbjct: 35 PPVTGTRTQLKDGRHLAYLESGVPKDQAKYKIIFVHGFDSCRYDALP-ISPELAQELGIY 93
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
+++D PG+ ESDP+P+ S A+D+ L D + + KF+++ S G W+ LK+I
Sbjct: 94 QLSFDRPGYAESDPNPASTEKSIALDVEELADNLQLGPKFYLMGFSMGGEIMWSCLKHIS 153
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLP 329
R+AG A+L P+ + + S + + W + LP+ K+ +++ P L ++ + P
Sbjct: 154 HRLAGVAILGPVGNYWWSGLPSNVSWHAWNQQLPQDKWAVWVSHHLPWLTYWWNSQKLFP 213
Query: 330 EKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDF 389
+ L ++D++++ + AF Y + + +QG + + +W +
Sbjct: 214 A--SSVIAYNPALLSEEDKLIMPKFAFRTY----MPQIRQQGEYSCLHRDMTVGFGKWSW 267
Query: 390 NIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERV 449
+ EL E AG G++H+W G +D +VP S++ Y+
Sbjct: 268 SPLEL----------------------EDPFAGGKGKVHLWHGAEDLIVPVSLSRYLSEK 305
Query: 450 LPEAVIHKLPNEGH 463
LP V H+LP GH
Sbjct: 306 LPWVVYHELPKSGH 319
>UniRef100_Q8LFG4 Hypothetical protein [Arabidopsis thaliana]
Length = 372
Score = 138 bits (347), Expect = 4e-31
Identities = 100/363 (27%), Positives = 171/363 (46%), Gaps = 46/363 (12%)
Query: 119 SIVFTVFLLAFAAISVDSNHQTSTKAAMVRM-----HPPI-ASRILLPDGRYMAYQDQGV 172
S +F ++ I V +Q+ K ++ PPI A RI L DGRY+AY++ G+
Sbjct: 18 SFLFPSVVIVIVGIIVAFTYQSKLKPPPPKLCGSSGGPPITAPRIKLQDGRYLAYKEHGL 77
Query: 173 PPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYGVRLVTYDLPGFGESDPHPSRNFN 230
P +A +V H R + S L+E+ GV +V++D PG+ ESDPHPSR
Sbjct: 78 PREKANRKIVFIHGSDCCRHDAVFATLLSPDLVEELGVYMVSFDRPGYCESDPHPSRTPR 137
Query: 231 SSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYESHMT 290
S D+ L D +++ KF+VL +S G AW LKYIP R+AG ++AP+V+ Y ++
Sbjct: 138 SLVSDIEELADQLSLGSKFYVLGYSMGGQAAWGCLKYIPHRLAGVTLVAPVVNYYWKNLP 197
Query: 291 KDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDEIL 350
+ + R + +A+ P L+ ++ + + P ++ SL L + D+ +
Sbjct: 198 LNVSTEGFNFQQKRDQLAVRVAHYTPWLIYWWNTQKWFPGS-SIANRDHSL-LAQPDKDI 255
Query: 351 VD------EPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGG 404
+ +P + E Q+ + ES+ + + ++ W+F+ +L
Sbjct: 256 ISKLGSSRKPHWAEVRQQGIHESINR--------DMIVGFGNWEFDPLDL---------- 297
Query: 405 LLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHF 464
E G +H+WQG +D +VP + Y+ LP H++P GHF
Sbjct: 298 ------------ENPFLNKEGSVHLWQGDEDMLVPAKLQRYLAHQLPWVHYHEVPRSGHF 345
Query: 465 SYF 467
++
Sbjct: 346 FHY 348
>UniRef100_Q9M382 Hypothetical protein F24B22.200 [Arabidopsis thaliana]
Length = 342
Score = 136 bits (343), Expect = 1e-30
Identities = 95/339 (28%), Positives = 156/339 (45%), Gaps = 32/339 (9%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPG--VKASLLEDYG 207
PPI A RI L DGRY+AY++ GV A F +V H+F + R + V+ LE G
Sbjct: 31 PPITAPRIRLSDGRYLAYEEHGVSRQNATFKIVFIHAFSTFRRDAVIANRVRPGFLEKNG 90
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKY 267
+ +V+YD PG+GESDPH SRN + A D+ L D + + KF+V+ +S G W LKY
Sbjct: 91 IYVVSYDRPGYGESDPHSSRNEKTLAHDVEQLADQLQLGSKFYVVGYSMGGQAVWGVLKY 150
Query: 268 IPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSF 327
IP R+AGA +L P+ + + W K ++ + + P LL ++ +
Sbjct: 151 IPHRLAGATLLCPVTNSWWPSFPDSLTWELWNKQSKSERFSMLITHHTPWLLYWWNNQKL 210
Query: 328 LPEKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRW 387
+ + +D L+ + A ++ ++ +QG + + ++ +W
Sbjct: 211 F--STTAVMQSSPNMFSPQDLALLPKLAARVSYK---NQTTQQGTHESLDRDLIVGFGKW 265
Query: 388 DFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIE 447
F+ + +++ + + E G +H+WQG DDR+VP + I
Sbjct: 266 SFD---------------PMKIENPFPKGE-------GSVHMWQGDDDRLVPIQLQRIIA 303
Query: 448 RVLPEAVIHKLPNEGHFSYFFFCDECHKQIFSTLFGTPQ 486
+ L H++P GH F D + I L PQ
Sbjct: 304 QKLTWIKYHEIPGAGHI--FPMADGMAETILKELLPIPQ 340
>UniRef100_Q9C913 Hypothetical protein F1O17.5 [Arabidopsis thaliana]
Length = 372
Score = 136 bits (342), Expect = 2e-30
Identities = 94/326 (28%), Positives = 157/326 (47%), Gaps = 41/326 (12%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYG 207
PPI A RI L DGRY+AY++ G+P +A +V H R + S L+E+ G
Sbjct: 55 PPITAPRIKLQDGRYLAYKEHGLPREKANRKIVFIHGSDCCRHDAVFATLLSPDLVEELG 114
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKY 267
V +V++D PG+ ESDPHPSR S D+ L D +++ KF+VL +S G AW LKY
Sbjct: 115 VYMVSFDRPGYCESDPHPSRTPRSLVSDIEELADQLSLGSKFYVLGYSMGGQAAWGCLKY 174
Query: 268 IPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSF 327
IP R+AG ++AP+V+ Y ++ + + R + +A+ P L+ ++ + +
Sbjct: 175 IPHRLAGVTLVAPVVNYYWKNLPLNVSTEGFNFQQKRDQLAVRVAHYTPWLIYWWNTQKW 234
Query: 328 LPEKHERIDKQFSLSLGKKDEILVD------EPAFEEYWQRDLEESVRQGNLKPFIEEAL 381
P ++ SL L + D+ ++ +P + E Q+ + ES+ + + +
Sbjct: 235 FPGS-SIANRDHSL-LAQPDKDIISKLGSSRKPHWAEVRQQGIHESINR--------DMI 284
Query: 382 LQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPS 441
+ W+F +L E G +H+WQG +D +VP
Sbjct: 285 VGFGNWEFGPLDL----------------------ENPFLNKEGSVHLWQGDEDMLVPAK 322
Query: 442 MTEYIERVLPEAVIHKLPNEGHFSYF 467
+ Y+ LP H++P GHF ++
Sbjct: 323 LQRYLAHQLPWVHYHEVPRSGHFFHY 348
>UniRef100_Q9C914 Hypothetical protein F1O17.4 [Arabidopsis thaliana]
Length = 371
Score = 131 bits (330), Expect = 4e-29
Identities = 99/363 (27%), Positives = 169/363 (46%), Gaps = 45/363 (12%)
Query: 119 SIVFTVFLLAFAAISVDSNHQTSTKAAMVRM-----HPPI-ASRILLPDGRYMAYQDQGV 172
S +F ++ I V +Q+ K ++ PPI A RI L DGRY+AY++ G+
Sbjct: 18 SFLFPSVVIVIVGIIVALTYQSKLKPPQPKLCGSSSGPPITAPRIKLQDGRYLAYKEHGL 77
Query: 173 PPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYGVRLVTYDLPGFGESDPHPSRNFN 230
P +A +V H R + S L+E+ GV +V++D PG+ ESDPHPSR
Sbjct: 78 PREKANRKIVFIHGSDCCRHDAVFATLLSPDLVEELGVYMVSFDRPGYCESDPHPSRTPR 137
Query: 231 SSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWA--SLKYIPERIAGAAMLAPMVSPYESH 288
S D+ L D +++ KF+V+ S G AW +LKYIP R+AG ++AP+V+ Y +
Sbjct: 138 SLVSDIEELDDQLSLGSKFYVIGKSMGGQAAWGCLNLKYIPHRLAGVTLVAPVVNYYWRN 197
Query: 289 MTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDE 348
+ + + R ++ +A+ P L+ ++ + + P ++ LS +D
Sbjct: 198 LPLNVSTEGFNFQQKRDQWAVRVAHYAPWLIYWWNTQKWFPGS-SIANRDSLLSQSDRDI 256
Query: 349 I----LVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGG 404
I +P + E Q+ + ES+ + + ++ W+F+ +L
Sbjct: 257 ISKRGYTRKPHWAEVRQQGIHESINR--------DMIVGFGNWEFDPLDL---------- 298
Query: 405 LLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHF 464
+ G +H+WQG +D +VP + Y+ LP H++P GHF
Sbjct: 299 ------------DNPFLNNEGFVHLWQGDEDMLVPVKLQRYLAHQLPWVHYHEVPRSGHF 346
Query: 465 SYF 467
+F
Sbjct: 347 FHF 349
>UniRef100_Q6ESF3 Hydrolase, alpha/beta fold protein-like [Oryza sativa]
Length = 372
Score = 131 bits (330), Expect = 4e-29
Identities = 85/314 (27%), Positives = 149/314 (47%), Gaps = 30/314 (9%)
Query: 151 PPIAS-RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP+ R+ L DGR++AY + GVP +A+ ++ H F S R + V L E+ GV
Sbjct: 67 PPVTGPRLQLKDGRHLAYHEYGVPKDQAKHKIIFVHGFDSCRYDALQ-VSPELAEELGVY 125
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
+V++D PG+GESDPHP R +S A D+ L D + + KF+++ +S G W+ LK IP
Sbjct: 126 MVSFDRPGYGESDPHPGRTEDSIAFDIEGLADGLQLGPKFYLIGYSMGGEIMWSCLKNIP 185
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLP 329
R+AG ++L P+ + + S + W LP+ ++ +A+ P L ++ + P
Sbjct: 186 HRLAGVSILGPVGNYWWSGYPSNVSTEAWYVQLPQDQWAVRVAHHAPWLAYWWNTQKLFP 245
Query: 330 EKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDF 389
+ L ++D ++ + A+ Y + +QG + + L+ +W +
Sbjct: 246 A--SSVISFNPAILSREDLTVIPKFAYRTY----AGQVRQQGEHESLHRDMLVGFGKWGW 299
Query: 390 NIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERV 449
+ E+ E +H+W G +D +VP ++ +I +
Sbjct: 300 SPLEM----------------------ENPFPAGEAAVHLWHGAEDLIVPVQLSRHIAQR 337
Query: 450 LPEAVIHKLPNEGH 463
LP H+LP GH
Sbjct: 338 LPWVRYHELPTAGH 351
>UniRef100_Q9C915 Hypothetical protein F1O17.3 [Arabidopsis thaliana]
Length = 346
Score = 130 bits (327), Expect = 9e-29
Identities = 93/326 (28%), Positives = 154/326 (46%), Gaps = 47/326 (14%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYG 207
PPI A RI L DGR++AY++ G+P +A+ +V H S R + S L+++ G
Sbjct: 38 PPITAPRIKLRDGRHLAYKEYGLPREKAKHKIVFIHGSDSCRHDAVFATLLSPDLVQERG 97
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKY 267
V +V++D PG+GESDP P R S A+D+ L D +++ KF+V+ S G AW LKY
Sbjct: 98 VYMVSFDKPGYGESDPDPIRTPKSLALDIEELADQLSLGSKFYVIGKSMGGQAAWGCLKY 157
Query: 268 IPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSF 327
P R+AG ++AP+V+ Y ++ + + R ++ +A+ P L+ ++ +++
Sbjct: 158 TPHRLAGVTLVAPVVNYYWRNLPLNISTEGFNLQQKRDQWAVRVAHYAPWLIYWWNTQNW 217
Query: 328 LPEKH---------ERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIE 378
P + DK L LG +P E Q+ + ES+ +
Sbjct: 218 FPGSSVVNRDGGVLSQPDKDIILKLGSS-----RKPHLAEVRQQGIHESINR-------- 264
Query: 379 EALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMV 438
+ ++ W+F+ EL E G +H+WQG +D +V
Sbjct: 265 DMIVGFGNWEFDPLEL----------------------ENPFLNREGSVHLWQGDEDMLV 302
Query: 439 PPSMTEYIERVLPEAVIHKLPNEGHF 464
P ++ YI LP H++ GHF
Sbjct: 303 PVTLQRYIADKLPWLHYHEVAGGGHF 328
>UniRef100_Q8LBQ8 Hypothetical protein [Arabidopsis thaliana]
Length = 346
Score = 130 bits (327), Expect = 9e-29
Identities = 93/326 (28%), Positives = 154/326 (46%), Gaps = 47/326 (14%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYG 207
PPI A RI L DGR++AY++ G+P +A+ +V H S R + S L+++ G
Sbjct: 38 PPITAPRIKLRDGRHLAYKEYGLPREKAKHKIVFIHGSDSCRHDAVFATLLSPDLVQERG 97
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKY 267
V +V++D PG+GESDP P R S A+D+ L D +++ KF+V+ S G AW LKY
Sbjct: 98 VYMVSFDKPGYGESDPDPIRTPKSLALDIEELADQLSLGSKFYVIGKSMGGQAAWGCLKY 157
Query: 268 IPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSF 327
P R+AG ++AP+V+ Y ++ + + R ++ +A+ P L+ ++ +++
Sbjct: 158 TPHRLAGVTLVAPVVNYYWRNLPLNISTEGFNLQQKRDQWAVRVAHYAPWLIYWWNTQNW 217
Query: 328 LPEKH---------ERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIE 378
P + DK L LG +P E Q+ + ES+ +
Sbjct: 218 FPGSSVVNRDGGVLSQPDKDIILKLGSS-----RKPHLAEVRQQGIHESINR-------- 264
Query: 379 EALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMV 438
+ ++ W+F+ EL E G +H+WQG +D +V
Sbjct: 265 DMIVGFGNWEFDPLEL----------------------ENPFLNREGSVHLWQGDEDMLV 302
Query: 439 PPSMTEYIERVLPEAVIHKLPNEGHF 464
P ++ YI LP H++ GHF
Sbjct: 303 PVTLQRYIADKLPWLHYHEVAGGGHF 328
>UniRef100_Q9SJM9 Expressed protein [Arabidopsis thaliana]
Length = 364
Score = 126 bits (316), Expect = 2e-27
Identities = 88/314 (28%), Positives = 148/314 (47%), Gaps = 28/314 (8%)
Query: 156 RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRL--AGIPGVKASLLEDYGVRLVTY 213
RI L DGR +AY++ GVP A ++ H S R A + + E GV +V++
Sbjct: 57 RIKLRDGRQLAYKEHGVPRDEATHKIIVVHGSDSCRHDNAFAALLSPDIKEGLGVYMVSF 116
Query: 214 DLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIA 273
D PG+ ESDP P+R S A+D+ L D +++ KF+V+ +S G WA LKYIP R+A
Sbjct: 117 DRPGYAESDPDPNRTPKSLALDIEELADQLSLGSKFYVIGYSMGGQATWACLKYIPHRLA 176
Query: 274 GAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHE 333
G ++AP+V+ + + + + + ++ +A+ P L ++ +S+ P
Sbjct: 177 GVTLVAPVVNYWWKNFPSEISTEAFNQQGRNDQWAVRVAHYAPWLTHWWNSQSWFPGSSV 236
Query: 334 RIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEE 393
LS K EI+ A + + + QG + + ++ W+F+ E
Sbjct: 237 VARNLGMLSKADK-EIMFKLGAARSQHEAQIRQ---QGTHETLHRDMIVGFGTWEFDPME 292
Query: 394 LHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEA 453
L++++ E G +H+WQG DD +VP ++ YI + LP
Sbjct: 293 ---------------LENLFPNNE-------GSVHLWQGDDDVLVPVTLQRYIAKKLPWI 330
Query: 454 VIHKLPNEGHFSYF 467
H++P GH F
Sbjct: 331 HYHEIPGAGHLFPF 344
>UniRef100_Q84VY8 At2g36290 [Arabidopsis thaliana]
Length = 390
Score = 126 bits (316), Expect = 2e-27
Identities = 88/314 (28%), Positives = 148/314 (47%), Gaps = 28/314 (8%)
Query: 156 RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRL--AGIPGVKASLLEDYGVRLVTY 213
RI L DGR +AY++ GVP A ++ H S R A + + E GV +V++
Sbjct: 83 RIKLRDGRQLAYKEHGVPRDEATHKIIVVHGSDSCRHDNAFAALLSPDIREGLGVYMVSF 142
Query: 214 DLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIA 273
D PG+ ESDP P+R S A+D+ L D +++ KF+V+ +S G WA LKYIP R+A
Sbjct: 143 DRPGYAESDPDPNRTPKSLALDIEELADQLSLGSKFYVIGYSMGGQATWACLKYIPHRLA 202
Query: 274 GAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHE 333
G ++AP+V+ + + + + + ++ +A+ P L ++ +S+ P
Sbjct: 203 GVTLVAPVVNYWWKNFPSEISTEAFNQQGRNDQWAVRVAHYAPWLTHWWNSQSWFPGSSV 262
Query: 334 RIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEE 393
LS K EI+ A + + + QG + + ++ W+F+ E
Sbjct: 263 VARNLGMLSRADK-EIMFKLGAARSQHEAQIRQ---QGTHETLHRDMIVGFGTWEFDPME 318
Query: 394 LHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEA 453
L++++ E G +H+WQG DD +VP ++ YI + LP
Sbjct: 319 ---------------LENLFPNNE-------GSVHLWQGDDDVLVPVTLQRYIAKKLPWI 356
Query: 454 VIHKLPNEGHFSYF 467
H++P GH F
Sbjct: 357 HYHEIPGAGHLFPF 370
>UniRef100_Q9M9P2 T17B22.7 protein [Arabidopsis thaliana]
Length = 326
Score = 121 bits (303), Expect = 5e-26
Identities = 82/312 (26%), Positives = 150/312 (47%), Gaps = 38/312 (12%)
Query: 153 IASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVT 212
I+ RI L DGRY+AY++ G P +A+ ++ H F SS+L +++++ + +
Sbjct: 34 ISPRIKLNDGRYLAYKELGFPKDKAKNKIIILHGFGSSKL--------EMIDEFEIYFLL 85
Query: 213 YDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERI 272
+D G+GESDPHPSR + D+ L D + + KF VL S G + LKYIP R+
Sbjct: 86 FDRAGYGESDPHPSRTLKTDTYDIEELADKLQIGPKFHVLGMSLGAYPVYGCLKYIPHRL 145
Query: 273 AGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFY-RKSFLPEK 331
+GA ++ P+++ + S + + ++K + ++ +A+ FP LL ++ +K F P
Sbjct: 146 SGATLVVPILNFWWSCLPLNLSISAFKKLPIQNQWTLGVAHYFPWLLYWWMTQKWFSPFS 205
Query: 332 HERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNI 391
+ ++ ++D L D+ Y + E ++RQG + + W+F+
Sbjct: 206 QNPRE-----TMTERDIELADKHTKHAYIK---ESALRQGEYVSMQRDIIAGYENWEFDP 257
Query: 392 EELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLP 451
E L + + G +HIW L+D+ + + Y+ LP
Sbjct: 258 TE---------------LSNPFSDDN------KGSVHIWCALEDKQISHEVLLYLCDKLP 296
Query: 452 EAVIHKLPNEGH 463
+H++P+ GH
Sbjct: 297 WIKLHEVPDAGH 308
>UniRef100_Q94DD0 P0683F02.22 protein [Oryza sativa]
Length = 347
Score = 116 bits (291), Expect = 1e-24
Identities = 90/323 (27%), Positives = 149/323 (45%), Gaps = 36/323 (11%)
Query: 151 PPIAS-RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP++S R+ L DGR++AY++ GV A++ ++ H F S++ + P V L E+ G+
Sbjct: 37 PPVSSPRVQLKDGRHLAYREAGVGREIAKYKIIFSHGFASTKESDFP-VSQELAEELGIY 95
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
L+ +D G+G+SD +P R S A D+ L D + + +KF+V+ S G AW+ L YIP
Sbjct: 96 LLYFDRAGYGDSDANPKRGLKSDATDVEELADKLQLGEKFYVVGTSMGGYVAWSCLNYIP 155
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLL-SFFYRKSFL 328
R+AG A++ P V+ Y M + K + + +A+ P L ++F +K F
Sbjct: 156 YRLAGVALVVPAVN-YWWPMPASVSASAYRKLDVGDRRTFWIAHHMPWLFYAWFNQKWF- 213
Query: 329 PEKHERIDKQFSLSLGKKD-EILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRW 387
+ I + + +KD EIL + + Q D + +QG +A + W
Sbjct: 214 --RISPIVEGKPEAFTEKDWEILAE---IQRTGQLDRGRATKQGAYHSLCRDATILFGAW 268
Query: 388 DFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIE 447
+F+ + E G + IWQG +D++V Y+
Sbjct: 269 EFDPTAI----------------------ENPFPNGDGVVSIWQGREDKIVRVEAQRYVA 306
Query: 448 RVLPEAVIHKLPNEGHFSYFFFC 470
LP H+ P GH F C
Sbjct: 307 EKLPWVRYHEHPEGGH---LFMC 326
>UniRef100_Q9M9P1 T17B22.8 protein [Arabidopsis thaliana]
Length = 333
Score = 115 bits (288), Expect = 3e-24
Identities = 75/312 (24%), Positives = 152/312 (48%), Gaps = 30/312 (9%)
Query: 153 IASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVT 212
I+ RI L DGR++AY++ G P +A+ ++ H +S+ + + +++++ + +
Sbjct: 34 ISPRIKLNDGRHLAYKELGFPKDKAKNKIIIVHGNGNSKDVDLY-ITQEMIDEFKIYFLF 92
Query: 213 YDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERI 272
+D G+GESDP+P+R + D+ L D + V KF V+ S G + LKYIP R+
Sbjct: 93 FDRAGYGESDPNPTRTLKTDTYDIEELADKLQVGPKFHVIGMSLGAYPVYGCLKYIPNRL 152
Query: 273 AGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKH 332
+GA+++ P+V+ + S + ++ + +K + +A+ P LL ++ + + P
Sbjct: 153 SGASLVVPLVNFWWSRVPQNLLNAAMKKLPIGFQLTLRVAHYSPWLLYWWMTQKWFPNSR 212
Query: 333 ERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIE 392
D ++ ++D L ++ Y + E ++RQG ++ + W+F+
Sbjct: 213 NPKD-----TMTERDLELAEKHTKHSYIK---ESALRQGGYVTTQQDIIAGYGNWEFDPT 264
Query: 393 ELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPE 452
E LK+ + + G +H+W L+D+ + + YI LP
Sbjct: 265 E---------------LKNPFSDSN------KGSVHMWCALEDKQISRDVLLYICDKLPW 303
Query: 453 AVIHKLPNEGHF 464
+H++P+ GH+
Sbjct: 304 IKLHEVPDGGHY 315
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 876,784,407
Number of Sequences: 2790947
Number of extensions: 37157468
Number of successful extensions: 97299
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 140
Number of HSP's that attempted gapping in prelim test: 97142
Number of HSP's gapped (non-prelim): 214
length of query: 512
length of database: 848,049,833
effective HSP length: 132
effective length of query: 380
effective length of database: 479,644,829
effective search space: 182265035020
effective search space used: 182265035020
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC124963.7


