

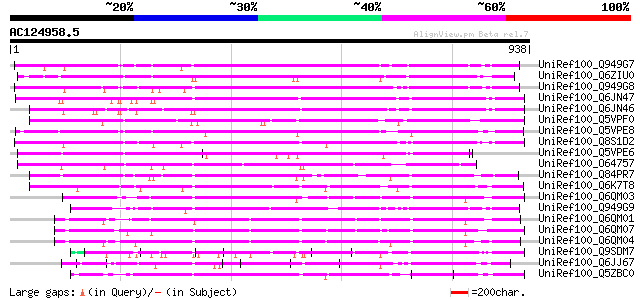
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124958.5 + phase: 0
(938 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q949G7 HcrVf3 protein [Malus floribunda] 574 e-162
UniRef100_Q6ZIU0 Putative HcrVf3 protein [Oryza sativa] 564 e-159
UniRef100_Q949G8 HcrVf2 protein [Malus floribunda] 554 e-156
UniRef100_Q6JN47 EIX receptor 1 [Lycopersicon esculentum] 548 e-154
UniRef100_Q6JN46 EIX receptor 2 [Lycopersicon esculentum] 546 e-153
UniRef100_Q5VPF0 Putative HcrVf3 protein [Oryza sativa] 530 e-149
UniRef100_Q5VPE8 Putative HcrVf3 protein [Oryza sativa] 515 e-144
UniRef100_Q8S1D2 HcrVf1 protein-like [Oryza sativa] 494 e-138
UniRef100_Q5VPE6 Putative HcrVf3 protein [Oryza sativa] 492 e-137
UniRef100_O64757 Putative disease resistance protein [Arabidopsi... 476 e-132
UniRef100_Q84PR7 Putative Cf2/Cf5 disease resistance protein hom... 475 e-132
UniRef100_Q6K7T8 Putative HcrVf2 protein [Oryza sativa] 474 e-132
UniRef100_Q6QM03 LLR protein WM1.1 [Aegilops tauschii] 429 e-118
UniRef100_Q949G9 HcrVf1 protein [Malus floribunda] 427 e-118
UniRef100_Q6QM01 LRR protein WM1.10 [Aegilops tauschii] 426 e-117
UniRef100_Q6QM07 LRR protein WM1.7 [Aegilops tauschii] 424 e-117
UniRef100_Q6QM04 LRR protein WM1.2 [Aegilops tauschii] 422 e-116
UniRef100_Q9SDM7 Hypothetical protein [Glycine max] 386 e-105
UniRef100_Q6JJ67 Putative disease resistance protein [Ipomoea tr... 373 e-101
UniRef100_Q5ZBC0 HcrVf1 protein-like [Oryza sativa] 373 e-101
>UniRef100_Q949G7 HcrVf3 protein [Malus floribunda]
Length = 915
Score = 574 bits (1480), Expect = e-162
Identities = 361/931 (38%), Positives = 530/931 (56%), Gaps = 47/931 (5%)
Query: 10 LIAKFIAILCLLMHGHVLCNG--GLNSQFIASEAEALLEFKEGLKDPSNLLSSW----KH 63
L+ +F+AI + LCNG G SE +ALL FK+ LKDP+N L+SW
Sbjct: 10 LLTRFLAIATITF-SIGLCNGNPGWPPLCKESERQALLMFKQDLKDPTNRLASWVAEEDS 68
Query: 64 GKDCCQWKGVGCNTTTGHVISLNLHCSNSLDKLQ----GHLNSSLLQLPYLSYLNLSGND 119
DCC W GV C+ TTGH+ L+L+ ++ L+ G +N SLL L +L++L+LS N
Sbjct: 69 DSDCCSWTGVVCDHTTGHIHELHLNNTDPFLDLKSSFGGKINPSLLSLKHLNFLDLSNNY 128
Query: 120 FMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFY--VNNLKWL 177
F + +P F + +L HL+L+++ F G + LGNLS L L+LS NS Y V NL+W+
Sbjct: 129 FYPTQIPSFFGSMTSLTHLNLAYSRFGGIIPHKLGNLSSLRYLNLSSNSIYLKVENLQWI 188
Query: 178 HGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSL 237
GLS LK LDLSGV LS+ +DW + +L SL L +S CQL+++P P P NF SL
Sbjct: 189 SGLSLLKHLDLSGVNLSKA-SDWLQ-VTNMLPSLVKLIMSDCQLYQIP--PLPTTNFTSL 244
Query: 238 VTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLN 297
V LDLS NNFN +P W+F + +L +++LS+ QG I + +T L +DLS N+
Sbjct: 245 VVLDLSFNNFNSLMPRWVF-SLKNLVSIHLSDCGFQGPIPSISQNITYLREIDLSDNNFT 303
Query: 298 GLIPN-FFDKLVN-----LVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSL 351
P+ F+ L + +L L +SG IP +L +SL++L +S+NQ NG+
Sbjct: 304 VQRPSEIFESLSRCGPDGIKSLSLRNTNVSGHIPMSL---RNLSSLEKLDISVNQFNGTF 360
Query: 352 ERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLE 411
I QL L L+++ N++E +S+V +N + LK N +TL S++WVPPFQLE
Sbjct: 361 TEVIGQLKMLTYLDISYNSLESAMSEVTFSNLTKLKNFVAKGNSLTLKTSRDWVPPFQLE 420
Query: 412 TIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRR 471
+ L + HLGP++P W++TQ + +S G+S +P WFW+L+ VEY+NLS N+L
Sbjct: 421 ILHLDSWHLGPKWPMWLRTQTQLKELSLSGTGISSTIPTWFWNLTSQVEYLNLSRNQLYG 480
Query: 472 CGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISH-VCEILCFNNSL 530
Q+ +DLS+N F+ LP +P +L LDLS + F ++ H C+ L
Sbjct: 481 QIQNIVAG-PSSVVDLSSNQFTGALPIVPTSLFFLDLSRSSFSESVFHFFCDRPDEPKQL 539
Query: 531 ENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSG 590
L+L N L+G +P+CW + ++ LNL NN G++P S G L+ L L + NN+L G
Sbjct: 540 SVLNLGNNLLTGKVPDCWMSWQHLRFLNLENNNLTGNVPMSMGYLQYLGSLHLRNNHLYG 599
Query: 591 KIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKS 650
++P +L+NC L++++L N G IP WIG + L VL L +N F+ +IP +C LKS
Sbjct: 600 ELPHSLQNCTWLSVVDLSENGFSGSIPIWIGKSLSGLNVLNLRSNKFEGDIPNEVCYLKS 659
Query: 651 LHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIPWK 710
L ILDL+ N+L+G IPRC F L+ +E Y S ++ K
Sbjct: 660 LQILDLAHNKLSGMIPRC-FHNLSALADFSESFYP-----TSYWGTNWSELSENAILVTK 713
Query: 711 GVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGE 770
G+ + +++ +K++DLS NF+ EIP E+ L+ L +LNLS N+ G IPS+IG
Sbjct: 714 GIEMEYSK---ILGFVKVMDLSCNFMYGEIPEELTGLLALQSLNLSNNRFTGRIPSNIGN 770
Query: 771 LESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNP 830
+ L LD S N L EIP SM N+ LS L+LSYN L+G+IP Q+QS D+ + GN
Sbjct: 771 MAWLESLDFSMNQLDGEIPPSMTNLTFLSHLNLSYNNLTGRIPESTQLQSLDQSSFVGN- 829
Query: 831 HLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWV 890
LCG PL K C N E DG G ++L E Y+S+ +GF TGFW+
Sbjct: 830 KLCGAPLNKNCSTNGVIPPPTV------EQDGGGGYRLL--EDEWFYVSLGVGFFTGFWI 881
Query: 891 FWGSLILIASWRHAYFRFISNMNDKIHVTVV 921
GSL++ W + ++ + K++ +V
Sbjct: 882 VLGSLLVNMPWSILLSQLLNRIVLKMYHVIV 912
>UniRef100_Q6ZIU0 Putative HcrVf3 protein [Oryza sativa]
Length = 940
Score = 564 bits (1453), Expect = e-159
Identities = 357/952 (37%), Positives = 521/952 (54%), Gaps = 93/952 (9%)
Query: 15 IAILCLLMHGHVLCNGGLNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVG 74
+A+LCL ++ + + I++E +AL+ F +KDP L SW HG++CC W GV
Sbjct: 12 LALLCLTINFREI------AACISTERDALVAFNTSIKDPDGRLHSW-HGENCCSWSGVS 64
Query: 75 CNTTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKN 134
C+ TGHVI L+L L G +N SL L L YLNLS +DF +P+F+ K
Sbjct: 65 CSKKTGHVIKLDL----GEYTLNGQINPSLSGLTRLVYLNLSQSDFGGVPIPEFIGCFKM 120
Query: 135 LKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYV---NNLKWLHGLSSLKILDLSGV 191
L++LDLSHA F G + LGNLS L LDLS + +V ++ +W+ L+SL+ LDLS +
Sbjct: 121 LRYLDLSHAGFGGTVPPQLGNLSRLSFLDLSSSGSHVITADDFQWVSKLTSLRYLDLSWL 180
Query: 192 VLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTI 251
L+ DW + + LH L+ LRL+ L + ++NF +L +DL N N ++
Sbjct: 181 YLA-ASVDWLQAVNM-LHLLEVLRLNDASLPATDLNSVSQINFTALKVIDLKNNELNSSL 238
Query: 252 PDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLV 311
PDW++ N L +L+LS+ L G+I + ++ L + L N LNG IP +L NLV
Sbjct: 239 PDWIW-NLSSLSDLDLSSCELSGRIPDELGKLAALQFIGLGNNKLNGAIPRSMSRLCNLV 297
Query: 312 ALDLSYNMLSGSIPSTL---------------------GQDHG----QNSLKELRLSINQ 346
+DLS N+LSG++ GQ G SL+ L LS N
Sbjct: 298 HIDLSRNILSGNLSEAARSMFPCMKKLQILNLADNKLTGQLSGWCEHMASLEVLDLSENS 357
Query: 347 LNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVP 406
L+G L SI +LSNL L+++ N + G +S++H N S L L L+ N + + +W P
Sbjct: 358 LSGVLPTSISRLSNLTYLDISFNKLIGELSELHFTNLSRLDALVLASNSFKVVVKHSWFP 417
Query: 407 PFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSS 466
PFQL +GL C +GPQFP W+Q+Q ID+ +AG+ +P+W W+ S + +N+S
Sbjct: 418 PFQLTKLGLHGCLVGPQFPTWLQSQTRIKMIDLGSAGIRGALPDWIWNFSSPMASLNVSM 477
Query: 467 NELR-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSN-------------- 511
N + + L TL++ +N +P +P ++R LDLS N
Sbjct: 478 NNITGELPASLVRSKMLITLNIRHNQLEGYIPDMPNSVRVLDLSHNNLSGSLPQSFGDKE 537
Query: 512 LFYGTISH------VCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFI 565
L Y ++SH + LC S+E +D+S NNLSG +PNCW ++M +++ + NNF
Sbjct: 538 LQYLSLSHNSLSGVIPAYLCDIISMELIDISNNNLSGELPNCWRMNSSMYVIDFSSNNFW 597
Query: 566 GSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQ 625
G IP + GSL +L L + N+LSG +P +L++C+ L +L++ N L G IP WIG +Q
Sbjct: 598 GEIPSTMGSLSSLTALHLSKNSLSGLLPTSLQSCKRLLVLDVGENNLSGYIPTWIGNGLQ 657
Query: 626 ILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCV-----FLALTTEESIN 680
L++LILG+N F IP+ L QL +L LDLS N+L+G+IPR + FL+ E +
Sbjct: 658 TLLLLILGSNQFSGEIPEELSQLHALQYLDLSNNKLSGSIPRSLGKLTSFLSRNLE--WD 715
Query: 681 EKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEI 740
+ +FM Y S K L ++G + F + +L IDLS N LT EI
Sbjct: 716 SSPFFQFMVYGVG-GAYFSVYKDTLQATFRGYRLTF----VISFLLTSIDLSENHLTGEI 770
Query: 741 PVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSW 800
P EIG L L++LNLSRN + GSIP +IG L L LDLS N+LS IP SM ++ LS+
Sbjct: 771 PSEIGNLYRLASLNLSRNHIEGSIPETIGNLAWLESLDLSWNDLSGPIPQSMKSLLFLSF 830
Query: 801 LDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHEN 860
L+LSYN LSGKIP GNQ+ +F+ + GN LCG PL ++C ++S D H H
Sbjct: 831 LNLSYNHLSGKIPYGNQLMTFEGDSFLGNEDLCGAPLTRSCHKDS---DKHKHHE----- 882
Query: 861 DGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNM 912
+ Y+ +GF+ GF + I A+ R AYF+F N+
Sbjct: 883 ----------IFDTLTYMFTLLGFAFGFCTVSTTFIFSAASRRAYFQFTDNI 924
>UniRef100_Q949G8 HcrVf2 protein [Malus floribunda]
Length = 980
Score = 554 bits (1427), Expect = e-156
Identities = 366/997 (36%), Positives = 526/997 (52%), Gaps = 114/997 (11%)
Query: 10 LIAKFIAILCLLMHGHVLCNG--GLNSQFIASEAEALLEFKEGLKDPSNLLSSW--KHGK 65
L+ +F+AI + LCNG G SE ALL FK+ LKDP N L+SW +
Sbjct: 10 LLTRFLAIATITF-SIGLCNGNPGWPPLCKVSERRALLMFKQDLKDPVNRLASWVAEEDS 68
Query: 66 DCCQWKGVGCNTTTGHVISLNLHCSNSLDKLQ----GHLNSSLLQLPYLSYLNLSGNDFM 121
DCC W GV C+ TGH+ L+L+ S S + G +N SLL L +L+YL+LS NDF
Sbjct: 69 DCCSWTGVVCDHVTGHIHELHLNSSYSDWEFNSFFGGKINPSLLSLKHLNYLDLSNNDFN 128
Query: 122 QSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFY-----VNNLKW 176
+ +P F + +L HL+L+++ G + LGNLS L L+LS SFY V NL+W
Sbjct: 129 GTQIPSFFGSMTSLTHLNLAYSELYGIIPHKLGNLSSLRYLNLS--SFYGSNLKVENLQW 186
Query: 177 LHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDS 236
+ GLS LK LDLS V LS+ +DW + +L SL L +S C+L ++P P P NF S
Sbjct: 187 ISGLSLLKHLDLSSVNLSKA-SDWLQ-VTNMLPSLVELDMSDCELDQIPPLPTP--NFTS 242
Query: 237 LVTLDLSGNNFNMTIPDWLF-----------------------ENCHHLQNLNLS----- 268
LV LDLS N+FN +P W+F +N L+ ++LS
Sbjct: 243 LVVLDLSRNSFNCLMPRWVFSLKNLVSLHLSFCGFQSPIPSISQNITSLREIDLSFNSIS 302
Query: 269 -------------------NNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVN 309
+N L GQ+ SI+ +T L L+L N N IP + L N
Sbjct: 303 LDPIPKLLFTQKILELSLESNQLTGQLPRSIQNMTGLTTLNLGGNEFNSTIPEWLYSLNN 362
Query: 310 LVAL------------------------DLSYNMLSGSIPSTLGQDHGQNSLKELRLSIN 345
L +L DLS N +SG IP +LG +SL++L +S N
Sbjct: 363 LESLLLFGNALRGEISSSIGNLKSLRHFDLSSNSISGPIPMSLGN---LSSLEKLYISEN 419
Query: 346 QLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWV 405
NG+ I QL L L+++ N++EG++S++ +N LK N TL S++WV
Sbjct: 420 HFNGTFTEVIGQLKMLTDLDISYNSLEGVVSEISFSNLIKLKHFVAKGNSFTLKTSRDWV 479
Query: 406 PPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLS 465
PPFQLE + L + HLGP++P W++TQ + +S G+S +P WFW+L+ +V+Y+NLS
Sbjct: 480 PPFQLEILKLDSWHLGPEWPMWLRTQTQLKELSLSGTGISSTIPTWFWNLTFHVQYLNLS 539
Query: 466 SNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISH-VCEIL 524
N+L Q+ +DLS+N F+ LP +P +L LDLS++ F G++ H C+
Sbjct: 540 HNQLYGQIQNIVAG-PSSAVDLSSNQFTGALPIVPTSLMWLDLSNSSFSGSVFHFFCDRP 598
Query: 525 CFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMY 584
L L L N L+G +P+CW + ++ LNL NN G++P S G L L L +
Sbjct: 599 DEPKQLGILRLGNNFLTGKVPDCWMSWPSLAFLNLENNNLTGNVPMSMGYLDWLESLHLR 658
Query: 585 NNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKT 644
NN+L G++P +L+NC L++++L N G IP WIG + L VL L +N F+ +IP
Sbjct: 659 NNHLYGELPHSLQNCTSLSVVDLSENGFSGSIPIWIGKSLSGLNVLNLRSNKFEGDIPNE 718
Query: 645 LCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHP 704
+C LKSL ILDL+ N+L+G IPRC S F + S T++
Sbjct: 719 VCYLKSLQILDLAHNKLSGMIPRCFHNLSAMANFSQSFSPTSFWGMVAS-----GLTENA 773
Query: 705 LLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSI 764
+L+ KG+ + + + +K +DLS NF+ EIP E+ L+ L LNLS N+ G I
Sbjct: 774 ILVT-KGMEMEYTK---ILGFVKGMDLSCNFMYGEIPEELTGLLALQYLNLSNNRFTGRI 829
Query: 765 PSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEV 824
PS IG + L LD S N L EIP SM + LS L+LSYN L+G+IP Q+QS D+
Sbjct: 830 PSKIGSMAQLESLDFSMNQLDGEIPPSMTILTFLSHLNLSYNNLTGRIPESTQLQSLDQS 889
Query: 825 FYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMAMGF 884
+ GN LCG PL K C N H DG G + +E Y+S+ +GF
Sbjct: 890 SFVGN-ELCGAPLNKNCSENGVIPPPTVEH------DGGGGYSL--VEDEWFYVSLGVGF 940
Query: 885 STGFWVFWGSLILIASWRHAYFRFISNMNDKIHVTVV 921
TGFW+ GSL++ W + ++ + K++ +V
Sbjct: 941 FTGFWIVLGSLLVNMPWSILLSQLLNRIVLKMYHVIV 977
>UniRef100_Q6JN47 EIX receptor 1 [Lycopersicon esculentum]
Length = 1031
Score = 548 bits (1412), Expect = e-154
Identities = 377/1041 (36%), Positives = 538/1041 (51%), Gaps = 144/1041 (13%)
Query: 11 IAKFIAILCLLMHGHVLCNGGLNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKD---C 67
+A+F+ L LL GG + + E +ALLEFK GL D + LS+W +D C
Sbjct: 9 LAQFLFTLSLLFLETSFGLGGNKTLCLDKERDALLEFKRGLTDSFDHLSTWGDEEDKQEC 68
Query: 68 CQWKGVGCNTTTGHVISLNLH----CSNSLD-----KLQGHLNSSLLQLPYLSYLNLSGN 118
C+WKG+ C+ TGHV ++LH CS +L G L+ SLL+L YL+YL+LS N
Sbjct: 69 CKWKGIECDRRTGHVTVIDLHNKFTCSAGASACFAPRLTGKLSPSLLELEYLNYLDLSVN 128
Query: 119 DFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLH 178
+F +S +P F+ + K L++L+LS + F G + NL+ L +LDL +N+ V +L+WL
Sbjct: 129 EFERSEIPRFIGSLKRLEYLNLSASFFSGVIPIQFQNLTSLRTLDLGENNLIVKDLRWLS 188
Query: 179 GLSS-------------------------LKILDLSGVVLSR------------------ 195
LSS LK LDLSG LS+
Sbjct: 189 HLSSLEFLSLSSSNFQVNNWFQEITKVPSLKELDLSGCGLSKLAPSQADLANSSFISLSV 248
Query: 196 ---CQND--------WFHDIRVILHSLDTL--RLSG---------CQLHKLPTS------ 227
C N+ W ++ L S+D L +LSG L L +
Sbjct: 249 LHLCCNEFSSSSEYSWVFNLTTSLTSIDLLYNQLSGQIDDRFGTLMYLEHLDLANNLKIE 308
Query: 228 ---PPPEMNFDSLVTLDLSGNNFNMTIPDW---------------LFENC---------- 259
P N L LD+S +P+ L EN
Sbjct: 309 GGVPSSFGNLTRLRHLDMSNTQTVQWLPELFLRLSGSRKSLEVLGLNENSLFGSIVNATR 368
Query: 260 -HHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYN 318
L+ L L N L G S +V+TL LDLS+N + G +P+ +L L L N
Sbjct: 369 FSSLKKLYLQKNMLNGSFMESAGQVSTLEYLDLSENQMRGALPDLA-LFPSLRELHLGSN 427
Query: 319 MLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDV 378
G IP +G+ + L+ L +S N+L G L S+ QLSNL + + N ++G I++
Sbjct: 428 QFRGRIPQGIGK---LSQLRILDVSSNRLEG-LPESMGQLSNLESFDASYNVLKGTITES 483
Query: 379 HLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHID 438
HL+N S+L LDLSFN + L S NW+PPFQL+ I L +C+LGP FPKW+Q Q N++ +D
Sbjct: 484 HLSNLSSLVDLDLSFNSLALKTSFNWLPPFQLQVISLPSCNLGPSFPKWLQNQNNYTVLD 543
Query: 439 ISNAGVSDYVPNWFWDLSPNVEYMNLSSNELR-RCGQDFSQKFKLKTLDLSNNSFSCPLP 497
IS A +SD +P+WF P+++ +NLS+N++ R + + +DLS N+FS LP
Sbjct: 544 ISLASISDTLPSWFSSFPPDLKILNLSNNQISGRVSDLIENTYGYRVIDLSYNNFSGALP 603
Query: 498 RLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIIL 557
+P N++ L N F+G+IS +C S +LDLS N SG +P+CW N T++ +L
Sbjct: 604 LVPTNVQIFYLHKNQFFGSISSICR---SRTSPTSLDLSHNQFSGELPDCWMNMTSLAVL 660
Query: 558 NLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIP 617
NLA NNF G IP S GSL NL L + N+LSG +P + CQ L +L+L N+L G IP
Sbjct: 661 NLAYNNFSGEIPHSLGSLTNLKALYIRQNSLSGMLP-SFSQCQGLQILDLGGNKLTGSIP 719
Query: 618 YWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEE 677
WIGTD+ L +L L N +IP +CQL+ L ILDLS N L+G IP C +
Sbjct: 720 GWIGTDLLNLRILSLRFNRLHGSIPSIICQLQFLQILDLSANGLSGKIPHCFNNFTLLYQ 779
Query: 678 SINEKSYMEFMT--IEESLP---IYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLS 732
N MEF+ P +Y+ LL+ WK + L+ LK IDLS
Sbjct: 780 DNNSGEPMEFIVQGFYGKFPRRYLYIG----DLLVQWKNQESEYKNPLLY---LKTIDLS 832
Query: 733 SNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSM 792
SN L +P EI + L +LNLSRN+L G++ IG++ L LD+SRN LS IP +
Sbjct: 833 SNELIGGVPKEIADMRGLKSLNLSRNELNGTVIEGIGQMRMLESLDMSRNQLSGVIPQDL 892
Query: 793 ANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPLRKA---CPRNSSFED 849
AN+ LS LDLS N LSG+IP Q+QSFD Y N LCGPPL++ P + +
Sbjct: 893 ANLTFLSVLDLSNNQLSGRIPSSTQLQSFDRSSYSDNAQLCGPPLQECPGYAPPSPLIDH 952
Query: 850 THCSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFI 909
++ +EH+ + ++ +E YISM + F FW G LI+ +SWR+AYF+F+
Sbjct: 953 GSNNNPQEHDEE----EEFPSLE---FYISMVLSFFVAFWGILGCLIVNSSWRNAYFKFL 1005
Query: 910 SNMNDKIHVTVVVALNKLRRK 930
++ + + V +L++K
Sbjct: 1006 TDTTSWLDMISRVWFARLKKK 1026
>UniRef100_Q6JN46 EIX receptor 2 [Lycopersicon esculentum]
Length = 1021
Score = 546 bits (1407), Expect = e-153
Identities = 372/1009 (36%), Positives = 530/1009 (51%), Gaps = 141/1009 (13%)
Query: 37 IASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSNSLDK- 95
I E ALLEFK GL D LS+W ++CC WKG+ C+ TGHVI L+LH +
Sbjct: 36 IEKERGALLEFKRGLNDDFGRLSTWGDEEECCNWKGIECDKRTGHVIVLDLHSEVTCPGH 95
Query: 96 ------LQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNL 149
L G ++ SLL+L YL++L+LS N F S +P F+ + K L++L+LS ++F G +
Sbjct: 96 ACFAPILTGKVSPSLLELEYLNFLDLSVNGFENSEIPRFIGSLKRLEYLNLSSSDFSGEI 155
Query: 150 -----------LDNLGN-------------LSLLESLDLSDNSFYVNNLKWLHGLS---S 182
+ +LGN LS LE L L N F N W ++ S
Sbjct: 156 PAQFQNLTSLRILDLGNNNLIVKDLVWLSHLSSLEFLRLGGNDFQARN--WFREITKVPS 213
Query: 183 LKILDLSGVVLSR---------------------CQND--------WFHDIRVILHSLDT 213
LK LDLS LS+ C N+ W + L S+D
Sbjct: 214 LKELDLSVCGLSKFVPSPADVANSSLISLSVLHLCCNEFSTSSEYSWLFNFSTSLTSID- 272
Query: 214 LRLSGCQLHKLPTS-----------------------PPPEMNFDSLVTLDLSGNNFNMT 250
LS QL + P N L LD+S
Sbjct: 273 --LSHNQLSRQIDDRFGSLMYLEHLNLANNFGAEGGVPSSFGNLTRLHYLDMSNTQTYQW 330
Query: 251 IPDWLFE---NCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKL 307
+P+ + L+ L L++N+L G I ++ R ++L L L KN LNG ++
Sbjct: 331 LPELFLRLSGSRKSLEVLGLNDNSLFGSI-VNVPRFSSLKKLYLQKNMLNGFFMERVGQV 389
Query: 308 VNLVALDLSYNMLSGSIPST----------LGQDH-------GQNSLKELRL---SINQL 347
+L LDLS N + G +P LG + G L +LR+ S N+L
Sbjct: 390 SSLEYLDLSDNQMRGPLPDLALFPSLRELHLGSNQFQGRIPQGIGKLSQLRIFDVSSNRL 449
Query: 348 NGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPP 407
G L S+ QLSNL + + N ++G I++ H +N S+L LDLSFN ++LN +WVPP
Sbjct: 450 EG-LPESMGQLSNLERFDASYNVLKGTITESHFSNLSSLVDLDLSFNLLSLNTRFDWVPP 508
Query: 408 FQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSN 467
FQL+ I L +C++GP FPKW+QTQ N++ +DIS A +SD +P+WF +L P ++ +NLS+N
Sbjct: 509 FQLQFIRLPSCNMGPSFPKWLQTQNNYTLLDISLANISDMLPSWFSNLPPELKILNLSNN 568
Query: 468 ELR-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCF 526
+ R + K +DLS+N+FS LP +P N++ L N F G+IS +C
Sbjct: 569 HISGRVSEFIVSKQDYMIIDLSSNNFSGHLPLVPANIQIFYLHKNHFSGSISSICRNTI- 627
Query: 527 NNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNN 586
+ ++DLS N SG +P+CW N +N+ +LNLA NNF G +P S GSL NL L + N
Sbjct: 628 -GAATSIDLSRNQFSGEVPDCWMNMSNLAVLNLAYNNFSGKVPQSLGSLTNLEALYIRQN 686
Query: 587 NLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLC 646
+ G +P + CQ+L +L++ N+L G IP WIGTD+ L +L L +N FD +IP +C
Sbjct: 687 SFRGMLP-SFSQCQLLQILDIGGNKLTGRIPAWIGTDLLQLRILSLRSNKFDGSIPSLIC 745
Query: 647 QLKSLHILDLSENQLTGAIPRCV--FLALTTEESINEKSYMEFMTIEESLP---IYLSRT 701
QL+ L ILDLSEN L+G IP+C+ F L E E M+F + +P +Y+
Sbjct: 746 QLQFLQILDLSENGLSGKIPQCLNNFTILRQENGSGES--MDFKVRYDYIPGSYLYIG-- 801
Query: 702 KHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLL 761
LLI WK + L+ LK+IDLSSN L IP EI ++ L +LNLSRN L
Sbjct: 802 --DLLIQWKNQESEYKNALLY---LKIIDLSSNKLVGGIPKEIAEMRGLRSLNLSRNDLN 856
Query: 762 GSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSF 821
G++ IG+++ L LDLSRN LS IP ++N+ LS LDLS N LSG+IP Q+QSF
Sbjct: 857 GTVVEGIGQMKLLESLDLSRNQLSGMIPQGLSNLTFLSVLDLSNNHLSGRIPSSTQLQSF 916
Query: 822 DEVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMA 881
D Y GN LCGPPL + + ++ +EH++D D+ +E Y+SM
Sbjct: 917 DRSSYSGNAQLCGPPLEECPGYAPPIDRGSNTNPQEHDDD----DEFSSLE---FYVSMV 969
Query: 882 MGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIHVTVVVALNKLRRK 930
+GF FW G LI+ SWR+AYF F+++M +H+T V +L+ K
Sbjct: 970 LGFFVTFWGILGCLIVNRSWRNAYFTFLTDMKSWLHMTSRVCFARLKGK 1018
>UniRef100_Q5VPF0 Putative HcrVf3 protein [Oryza sativa]
Length = 961
Score = 530 bits (1365), Expect = e-149
Identities = 364/972 (37%), Positives = 501/972 (51%), Gaps = 121/972 (12%)
Query: 37 IASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSN--SLD 94
I E +AL + K L+DP +LSSW G +CC W GV CN TGH+I LNL N D
Sbjct: 25 IGKERDALFDLKATLRDPGGMLSSWV-GLNCCNWYGVTCNNRTGHIIKLNLANYNISKED 83
Query: 95 KLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLG 154
L G ++ SL+ L +L YLNL NDF + +P F+ + KNL+HLDLS ANF G + LG
Sbjct: 84 ALTGDISPSLVHLTHLMYLNLRSNDFGGARIPAFIGSLKNLRHLDLSFANFGGKIPPQLG 143
Query: 155 NLSLLESLDLS---------DNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIR 205
NLS L LD+S +S V+NL W+ LSSL LD+S LS +DW +
Sbjct: 144 NLSKLNYLDISFPYNNFSSFTSSSSVDNLLWVSQLSSLVYLDMSLWNLS-VASDWLQSLN 202
Query: 206 VILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNL 265
+ L SL LRLSG L + + NF L +DLSGNNF+ P+WL + + L +
Sbjct: 203 M-LASLKVLRLSGTNLPPTNQNSLSQSNFTVLNEIDLSGNNFSSRFPNWL-ASIYTLSLI 260
Query: 266 NLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIP 325
NL L G I S+ +T L L L+ NSL G IP KL NL LDLS N L G I
Sbjct: 261 NLDYCELHGSIPESVGNLTALNTLYLADNSLIGAIP--ISKLCNLQILDLSNNNLIGDIA 318
Query: 326 ST---------------LGQDHGQNSLK----------ELRLSINQLNGSLERSIYQLSN 360
LG ++ SL + LS N L+G + +I QL+
Sbjct: 319 DLGKAMTRCMKGLSMIKLGNNNLSGSLSGWIGSFPNLFSVDLSKNSLSGHVHTNISQLTE 378
Query: 361 LVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHL 420
L+ L+L+ N++E ++S+ HL N + LK LDLS+N + +++ NW+PPFQL + L + L
Sbjct: 379 LIELDLSHNSLEDVLSEQHLTNLTKLKKLDLSYNSLRISVGANWLPPFQLYELLLGSSPL 438
Query: 421 GPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFW---------DLSPNV------------ 459
Q P+W+QTQ +D+ G +P+W W DLS N+
Sbjct: 439 QSQVPQWLQTQVGMQTLDLHRTGTLGQLPDWLWTSLTSLINLDLSDNLLTGMLPASLVHM 498
Query: 460 ---EYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPN--LRNLDLSSNLFY 514
+++ LSSN+L GQ L LDLSNNS S LP R + LSSN
Sbjct: 499 KSLQFLGLSSNQLE--GQIPDMPESLDLLDLSNNSLSGSLPNSVGGNKTRYILLSSNRLN 556
Query: 515 GTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGS 574
+I C L +DLS N+LSG +PNCW N T + +++ + NN G IP S GS
Sbjct: 557 RSIP---AYFCNMPWLSAIDLSNNSLSGELPNCWKNSTELFLVDFSYNNLEGHIPSSLGS 613
Query: 575 LKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGN 634
L L L + NN LSG +P +L +C +L L++ N L G IP WIG ++Q LM+L L +
Sbjct: 614 LTFLGSLHLNNNRLSGLLPSSLSSCGLLVFLDIGDNNLEGSIPEWIGDNMQYLMILRLRS 673
Query: 635 NSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESL 694
N F +IP L QL+ L +LDL+ N+L+G +P + I S M +
Sbjct: 674 NRFTGSIPSELSQLQGLQVLDLANNKLSGPLP----------QGIGNFSEMASQRSRHII 723
Query: 695 PIYLSRT--------KHPLLIPWKGVNVFFNEGRLFFEIL---KMIDLSSNFLTHEIPVE 743
P+ +S L I KG E RL+ +IL K IDLS+N+LT IP E
Sbjct: 724 PMQISGDSFGGSLYHNESLYITIKG------EERLYSKILYLMKSIDLSNNYLTGGIPAE 777
Query: 744 IGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDL 803
+G LV L LNLS+N L G IP +IG + SL LDLS N LS IP SM ++ LS L++
Sbjct: 778 VGDLVGLKNLNLSKNLLSGHIPETIGNMSSLESLDLSWNRLSGIIPESMTSLHLLSHLNM 837
Query: 804 SYNALSGKIPIGNQMQSF---DEVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHEN 860
SYN LSG +P G+Q+Q+ D Y GN +LC H + E
Sbjct: 838 SYNNLSGMVPQGSQLQTLGDEDPYIYAGNKYLC----------------IHLASGSCFEQ 881
Query: 861 DGNHGDKVLGMEINP--LYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIHV 918
NH D+ +++ LYI +GF GF W L+ + YF+F+ + +K+
Sbjct: 882 KDNHVDQAEHNDVHDIWLYIFSGLGFGVGFSSVWWLLVCSKAVGKRYFQFVDSTCEKVIH 941
Query: 919 TVVVALNKLRRK 930
+++ K+ +K
Sbjct: 942 WMILLEKKVNKK 953
>UniRef100_Q5VPE8 Putative HcrVf3 protein [Oryza sativa]
Length = 980
Score = 515 bits (1326), Expect = e-144
Identities = 333/987 (33%), Positives = 499/987 (49%), Gaps = 120/987 (12%)
Query: 11 IAKFIAILCLLMHGHVLCNGGLNSQF---IASEAEALLEFKEGL-KDPSNLLSSWKHGKD 66
+A I++LC H + N G + I SE +ALL FK GL D + L SW+ G D
Sbjct: 13 LAALISLLC-----HSIANAGKEAAAAVCITSERDALLAFKAGLCADSAGELPSWQ-GHD 66
Query: 67 CCQWKGVGCNTTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVP 126
CC W V CN TGHVI L++ G +NSSL L +L YLNLSGNDF +P
Sbjct: 67 CCSWGSVSCNKRTGHVIGLDI--GQYALSFTGEINSSLAALTHLRYLNLSGNDFGGVAIP 124
Query: 127 DFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKIL 186
DF+ + L+HLDLSHA F G + LGNLS+L L L+ ++ ++N W+ L +L+ L
Sbjct: 125 DFIGSFSKLRHLDLSHAGFAGLVPPQLGNLSMLSHLALNSSTIRMDNFHWVSRLRALRYL 184
Query: 187 DLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNN 246
DL + L C +DW I L L LRL+ L + +NF +L LDLS N
Sbjct: 185 DLGRLYLVAC-SDWLQAISS-LPLLQVLRLNDAFLPATSLNSVSYVNFTALTVLDLSNNE 242
Query: 247 FNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDK 306
N T+P W++ + H L L+LS+ L G + +I +++L+ L L N L G IP +
Sbjct: 243 LNSTLPRWIW-SLHSLSYLDLSSCQLSGSVPDNIGNLSSLSFLQLLDNHLEGEIPQHMSR 301
Query: 307 LVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLE-------------- 352
L +L +D+S N LSG+I + L+ L++ N L G+L
Sbjct: 302 LCSLNIIDMSRNNLSGNITAEKNLFSCMKELQVLKVGFNNLTGNLSGWLEHLTGLTTLDL 361
Query: 353 ----------RSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSK 402
I +LS L+ L+L+ N G +S+VHL N S L L L+ N + + +
Sbjct: 362 SKNSFTGQIPEDIGKLSQLIYLDLSYNAFGGRLSEVHLGNLSRLDFLSLASNKLKIVIEP 421
Query: 403 NWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYM 462
NW+P FQL +GL CH+GP P W+++Q ID+ + ++ +P+W W+ S ++ +
Sbjct: 422 NWMPTFQLTGLGLHGCHVGPHIPAWLRSQTKIKMIDLGSTKITGTLPDWLWNFSSSITTL 481
Query: 463 NLSSNELR-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISH-- 519
++SSN + L T ++ +N +P LP +++ LDLS N G++
Sbjct: 482 DISSNSITGHLPTSLVHMKMLSTFNMRSNVLEGGIPGLPASVKVLDLSKNFLSGSLPQSL 541
Query: 520 ------------------VCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAM 561
+ LC +S+E +DLS N SGV+P+CW N + + ++ +
Sbjct: 542 GAKYAYYIKLSDNQLNGTIPAYLCEMDSMELVDLSNNLFSGVLPDCWKNSSRLHTIDFSN 601
Query: 562 NNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIG 621
NN G IP + G + +L +L + N+LSG +P +L++C L +L+L SN L G +P W+G
Sbjct: 602 NNLHGEIPSTMGFITSLAILSLRENSLSGTLPSSLQSCNGLIILDLGSNSLSGSLPSWLG 661
Query: 622 TDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINE 681
+ L+ L L +N F IP++L QL +L LDL+ N+L+G +P+ FL T ++
Sbjct: 662 DSLGSLITLSLRSNQFSGEIPESLPQLHALQNLDLASNKLSGPVPQ--FLGNLTSMCVDH 719
Query: 682 KSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEI---------------- 725
+ ++IP + +GR + I
Sbjct: 720 --------------------GYAVMIPSAKFATVYTDGRTYLAIHVYTDKLESYSSTYDY 759
Query: 726 -LKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNL 784
L IDLS N T EIP EIG + L ALNLS N +LGSIP IG L L LDLS N+L
Sbjct: 760 PLNFIDLSRNQFTGEIPREIGAISFLLALNLSGNHILGSIPDEIGNLSHLEALDLSSNDL 819
Query: 785 SCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHL---CGPPLRKAC 841
S IP S+ ++ LS L+LSYN LSG IP +Q +F + Y GN L CG L + C
Sbjct: 820 SGSIPPSITDLINLSVLNLSYNDLSGVIPCSSQFSTFTDEPYLGNADLCGNCGASLSRIC 879
Query: 842 PRNSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLILIASW 901
++++ + +H+N + G Y+ +GF+ G V LI +
Sbjct: 880 SQHTT--------TRKHQNMIDRG----------TYLCTLLGFAYGLSVVSAILIFSRTA 921
Query: 902 RHAYFRFISNMNDKIHVTVVVALNKLR 928
R+AYF+F D+ V + LN+++
Sbjct: 922 RNAYFQFTDKTLDEFRAIVQIKLNRIK 948
>UniRef100_Q8S1D2 HcrVf1 protein-like [Oryza sativa]
Length = 953
Score = 494 bits (1271), Expect = e-138
Identities = 348/979 (35%), Positives = 508/979 (51%), Gaps = 94/979 (9%)
Query: 10 LIAKFIAILCLLMHGHVLCNGGLNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQ 69
LIA + + ++ N I SE AL+ FK GL DP NLLSSW+ G DCCQ
Sbjct: 10 LIALALLLFTPIISNEASANANSTGGCIPSERSALISFKSGLLDPGNLLSSWE-GDDCCQ 68
Query: 70 WKGVGCNTTTGHVISLNLH--CSNSLDK-------LQGHLNSSLLQLPYLSYLNLSGNDF 120
W GV CN TGH++ LNL N L L G + SLL L L +L+LS N+F
Sbjct: 69 WNGVWCNNETGHIVELNLPGGSCNILPPWVPLEPGLGGSIGPSLLGLKQLEHLDLSCNNF 128
Query: 121 MQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDN---SFYVNNLKWL 177
T+P+FL + NL+ LDLS + F G + LGNLS L L N S Y ++ WL
Sbjct: 129 -SGTLPEFLGSLHNLRSLDLSWSTFVGTVPPQLGNLSNLRYFSLGSNDNSSLYSTDVSWL 187
Query: 178 HGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSL 237
LSSL+ LD+S V LS DW + L SL LRL GCQL S P N SL
Sbjct: 188 SRLSSLEHLDMSLVNLSAVV-DWVSVVNK-LPSLRFLRLFGCQLSSTVDSVPNN-NLTSL 244
Query: 238 VTLDLSGNNFNMTI-PDWLFE-----------------------NCHHLQNLNLSNNNLQ 273
TLDLS NNFN I P+W ++ N + +++LS NNL
Sbjct: 245 ETLDLSLNNFNKRIAPNWFWDLTSLKLLDISDSGFYGPFPNEIGNMTSIVDIDLSGNNLV 304
Query: 274 GQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVN-----LVALDLSYNMLSGSIPSTL 328
G I ++++ + L +++ ++NG I F++L L L L L+GS+P+TL
Sbjct: 305 GMIPFNLKNLCNLEKFNVAGTNINGNITEIFNRLPRCSWNKLQVLFLPDCNLTGSLPTTL 364
Query: 329 GQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKV 388
++L L L N + G + I +LSNL +L L+ NN++G+I + HL+ +L +
Sbjct: 365 ---EPLSNLSMLELGNNNITGPIPLWIGELSNLTMLGLSSNNLDGVIHEGHLSGLESLDL 421
Query: 389 LDLSFN-HVTLNMSKNWVPPF-QLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSD 446
L LS N H+ + ++ WVPPF Q+ I L +C LGP+FP W++ + ++DISN +SD
Sbjct: 422 LILSDNNHIAIKVNSTWVPPFKQITDIELRSCQLGPKFPTWLRYLTDVYNLDISNTSISD 481
Query: 447 YVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNL 506
VP+WFW + +V ++N+ +N++ + + +DLS+N FS P+P+LP +L +L
Sbjct: 482 KVPDWFWKAASSVTHLNMRNNQIAGALPSTLEYMRTIVMDLSSNKFSGPIPKLPVSLTSL 541
Query: 507 DLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIG 566
D S N G + ++L +L L N+LSG IP+ ++ +L+++ N G
Sbjct: 542 DFSKNNLSGPLPSDIGA----SALVSLVLYGNSLSGSIPSYLCKMQSLELLDISRNKITG 597
Query: 567 SIPD-------SFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYW 619
I D + + N+ + + NNLSG+ P KNC+ L L+L N+ G +P W
Sbjct: 598 PISDCAIDSSSANYTCTNIINISLRKNNLSGQFPSFFKNCKNLVFLDLAENQFSGTLPAW 657
Query: 620 IGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCV--FLALTTEE 677
IG + L+ L L +NSF +IP L L L LDL+ N +G IP + F +T E+
Sbjct: 658 IGEKLPSLVFLRLRSNSFSGHIPIELTSLAGLQYLDLAHNNFSGCIPNSLAKFHRMTLEQ 717
Query: 678 SINEK---SYMEFMTIEES-LPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSS 733
++ + + I ++ + Y+ + + KG + G + + + IDLSS
Sbjct: 718 DKEDRFSGAIRHGIGINDNDMVNYIEN----ISVVTKGQERLYT-GEIVYMV--NIDLSS 770
Query: 734 NFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMA 793
N LT EIP EI LV L+ LNLS N L G IP IG L L LDLS N LS IP+S+A
Sbjct: 771 NNLTGEIPEEIISLVALTNLNLSWNSLSGQIPEKIGSLSQLESLDLSHNVLSGGIPSSIA 830
Query: 794 NIDRLSWLDLSYNALSGKIPIGNQMQSFDE--VFYKGNPHLCGPPLRKACPRNSSFEDTH 851
++ LS ++LSYN LSG+IP GNQ+ ++ Y GN LCG PL C N DT
Sbjct: 831 SLTYLSHMNLSYNNLSGRIPAGNQLDILEDPASMYVGNIDLCGHPLPNNCSING---DTK 887
Query: 852 CSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISN 911
D ++ M + SM +GF G + + ++ WR+ F F+
Sbjct: 888 IER-----------DDLVNMS---FHFSMIIGFMVGLLLVFYFMLFSRRWRNTCFVFVDG 933
Query: 912 MNDKIHVTVVVALNKLRRK 930
+ D+ +V V V +L R+
Sbjct: 934 LYDRTYVQVAVTCRRLWRR 952
>UniRef100_Q5VPE6 Putative HcrVf3 protein [Oryza sativa]
Length = 884
Score = 492 bits (1266), Expect = e-137
Identities = 314/874 (35%), Positives = 458/874 (51%), Gaps = 68/874 (7%)
Query: 15 IAILCLLMHGHVLCNGG--LNSQFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKG 72
I + LL+ ++ N G ++ I+SE +ALL FK G DP+ + G+DCC W G
Sbjct: 5 IMLAALLVLCQLIKNAGKITDAACISSERDALLAFKAGFADPAGGALRFWQGQDCCAWSG 64
Query: 73 VGCNTTTGHVISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTT 132
V C+ G V+SL++ + +G +NSSL L +L YLNLSGNDF +PDF+ +
Sbjct: 65 VSCSKKIGSVVSLDIGHYDLT--FRGEINSSLAVLTHLVYLNLSGNDFGGVAIPDFIGSF 122
Query: 133 KNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVN--NLKWLHGLSSLKILDLSG 190
+ L++LDLSHA F G + LGNLS+L LDLS S V + W+ L+SL LDLS
Sbjct: 123 EKLRYLDLSHAGFGGTVPPRLGNLSMLSHLDLSSPSHTVTVKSFNWVSRLTSLVYLDLSW 182
Query: 191 VVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMT 250
+ L+ +DW L L L L+ L + NF ++ LDL NNF+
Sbjct: 183 LYLA-ASSDWLQATNT-LPLLKVLCLNHAFLPATDLNALSHTNFTAIRVLDLKSNNFSSR 240
Query: 251 IPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNL 310
+PDW+ L L+LS+ L G + ++ +T+L+ L N+L G IP +L NL
Sbjct: 241 MPDWI-SKLSSLAYLDLSSCELSGSLPRNLGNLTSLSFFQLRANNLEGEIPGSMSRLCNL 299
Query: 311 VALDLSYNMLSGSIPSTLGQDHG-QNSLKELRLSINQLNGSLER---------------- 353
+DLS N SG I N LK L L++N L GSL
Sbjct: 300 RHIDLSGNHFSGDITRLANTLFPCMNQLKILDLALNNLTGSLSGWVRHIASVTTLDLSEN 359
Query: 354 --------SIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWV 405
I +LSNL L+L+ N+ +G +S++H AN S L +L L +V + +WV
Sbjct: 360 SLSGRVSDDIGKLSNLTYLDLSANSFQGTLSELHFANLSRLDMLILESIYVKIVTEADWV 419
Query: 406 PPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLS 465
PPFQL + L C +GP FP W+++Q I++S A + +P+W W+ S + +++S
Sbjct: 420 PPFQLRVLVLYGCQVGPHFPAWLKSQAKIEMIELSRAQIKSKLPDWLWNFSSTISALDVS 479
Query: 466 SNELR-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISH----- 519
N + + + L+ LD+S+N +P LP +++ LDLSSN YG +
Sbjct: 480 GNMINGKLPKSLKHMKALELLDMSSNQLEGCIPDLPSSVKVLDLSSNHLYGPLPQRLGAK 539
Query: 520 ---------------VCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNF 564
+ LC +E + LS NN SGV+PNCW G+ + +++ + NN
Sbjct: 540 EIYYLSLKDNFLSGSIPTYLCEMVWMEQVLLSLNNFSGVLPNCWRKGSALRVIDFSNNNI 599
Query: 565 IGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDI 624
G I + G L +L L+++ N LSG +P +LK C L L+L N L G IP WIG +
Sbjct: 600 HGEISSTMGHLTSLGSLLLHRNKLSGPLPTSLKLCNRLIFLDLSENNLSGTIPTWIGDSL 659
Query: 625 QILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCV--FLAL-----TTEE 677
Q L++L L +N+F IP+ L QL +L ILD+++N L+G +P+ + A+ ++
Sbjct: 660 QSLILLSLRSNNFSGKIPELLSQLHALQILDIADNNLSGPVPKSLGNLAAMQLGRHMIQQ 719
Query: 678 SINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLT 737
+ S + FM + + L + +N G F+ IDLS N L
Sbjct: 720 QFSTISDIHFMVYGAGGAVLYRLYAYLYLNSLLAGKLQYN-GTAFY-----IDLSGNQLA 773
Query: 738 HEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDR 797
EIP+EIG L L+ LNLS N + GSIP +G L SL VLDLSRN+LS IP ++
Sbjct: 774 GEIPIEIGFLSGLTGLNLSGNHIRGSIPEELGNLRSLEVLDLSRNDLSGPIPQCFLSLSG 833
Query: 798 LSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPH 831
LS L+LSYN LSG IP GN++ +F E Y GN H
Sbjct: 834 LSHLNLSYNDLSGAIPFGNELATFAESTYFGNAH 867
Score = 110 bits (276), Expect = 2e-22
Identities = 132/507 (26%), Positives = 213/507 (41%), Gaps = 83/507 (16%)
Query: 349 GSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPF 408
G + S+ L++LV LNL+ N+ G+ + +F L+ LDLS
Sbjct: 88 GEINSSLAVLTHLVYLNLSGNDFGGVAIPDFIGSFEKLRYLDLS---------------- 131
Query: 409 QLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFW-DLSPNVEYMNLSSN 467
+ G P + SH+D+S+ + V ++ W ++ Y++LS
Sbjct: 132 --------HAGFGGTVPPRLGNLSMLSHLDLSSPSHTVTVKSFNWVSRLTSLVYLDLSWL 183
Query: 468 ELRRCGQDFSQKFK----LKTLDLSNNSFSCPLPRLPPN---------LRNLDLSSNLFY 514
L D+ Q LK L L N++F LP N +R LDL SN F
Sbjct: 184 YLA-ASSDWLQATNTLPLLKVLCL-NHAF---LPATDLNALSHTNFTAIRVLDLKSNNFS 238
Query: 515 GTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGS 574
S + + + +SL LDLS LSG +P N T++ L NN G IP S
Sbjct: 239 ---SRMPDWISKLSSLAYLDLSSCELSGSLPRNLGNLTSLSFFQLRANNLEGEIPGSMSR 295
Query: 575 LKNLHMLIMYNNNLSG---KIPETLKNCQ-VLTLLNLKSNRLRGPIPYWIGTDIQILMVL 630
L NL + + N+ SG ++ TL C L +L+L N L G + W+ I + L
Sbjct: 296 LCNLRHIDLSGNHFSGDITRLANTLFPCMNQLKILDLALNNLTGSLSGWV-RHIASVTTL 354
Query: 631 ILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEES-INEKSYMEFMT 689
L NS + + +L +L LDLS N G + F L+ + I E Y++ +T
Sbjct: 355 DLSENSLSGRVSDDIGKLSNLTYLDLSANSFQGTLSELHFANLSRLDMLILESIYVKIVT 414
Query: 690 IEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVE 749
+ +P + L+++ L + P + +
Sbjct: 415 EADWVPPFQ---------------------------LRVLVLYGCQVGPHFPAWLKSQAK 447
Query: 750 LSALNLSRNQLLGSIPSSIGELES-LNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNAL 808
+ + LSR Q+ +P + S ++ LD+S N ++ ++P S+ ++ L LD+S N L
Sbjct: 448 IEMIELSRAQIKSKLPDWLWNFSSTISALDVSGNMINGKLPKSLKHMKALELLDMSSNQL 507
Query: 809 SGKIPIGNQMQSFDEVFYKGNPHLCGP 835
G IP + S +V + HL GP
Sbjct: 508 EGCIP---DLPSSVKVLDLSSNHLYGP 531
>UniRef100_O64757 Putative disease resistance protein [Arabidopsis thaliana]
Length = 905
Score = 476 bits (1225), Expect = e-132
Identities = 326/929 (35%), Positives = 466/929 (50%), Gaps = 133/929 (14%)
Query: 14 FIAILCLLMHGHVLCNGGLNS-QFIASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKG 72
FI+ L L++ L G S + I++E +ALL F+ L D S+ L SW G DCC W G
Sbjct: 10 FISFLILILLLKNLNYGSAASPKCISTERQALLTFRAALTDLSSRLFSWS-GPDCCNWPG 68
Query: 73 VGCNTTTGHVISLNLHCSNS--------LDKLQGHLNSSLLQLPYLSYLNLSGNDFMQST 124
V C+ T HV+ ++L + L+G ++ SL QL +LSYL+LS NDF +
Sbjct: 69 VLCDARTSHVVKIDLRNPSQDVRSDEYKRGSLRGKIHPSLTQLKFLSYLDLSSNDFNELE 128
Query: 125 VPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSF--------YVNNLKW 176
+P+F+ +L++L+LS ++F G + +LGNLS LESLDL SF +NL+W
Sbjct: 129 IPEFIGQIVSLRYLNLSSSSFSGEIPTSLGNLSKLESLDLYAESFGDSGTLSLRASNLRW 188
Query: 177 LHGL-SSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFD 235
L L SSLK L++ V LS W D I +L L L +L LP + +
Sbjct: 189 LSSLSSSLKYLNMGYVNLSGAGETWLQDFSRI-SALKELHLFNSELKNLPPTLSSSADLK 247
Query: 236 SLVTLDLSGNNFNMTIPDWL-----------------------FENCHHLQNLNLSNN-N 271
L LDLS N+ N IP+WL F+N L+ L+LSNN
Sbjct: 248 LLEVLDLSENSLNSPIPNWLFGLTNLRKLFLRWDFLQGSIPTGFKNLKLLETLDLSNNLA 307
Query: 272 LQGQI-----------------------------SYSIERVTTLAILDLSKNSLNGLIPN 302
LQG+I ++S + +L LDLS N L G +P
Sbjct: 308 LQGEIPSVLGDLPQLKFLDLSANELNGQIHGFLDAFSRNKGNSLVFLDLSSNKLAGTLPE 367
Query: 303 FFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLV 362
L NL LDLS N +GS+PS++G SLK+L LS N +NG++ S+ QL+ LV
Sbjct: 368 SLGSLRNLQTLDLSSNSFTGSVPSSIG---NMASLKKLDLSNNAMNGTIAESLGQLAELV 424
Query: 363 VLNLAVNNMEGIISDVHLANFSNLKVLDLS---FNHVTLNMSKNWVPPFQLETIGLANCH 419
LNL N G++ H N +LK + L+ + + + W+PPF+LE I + NC
Sbjct: 425 DLNLMANTWGGVLQKSHFVNLRSLKSIRLTTEPYRSLVFKLPSTWIPPFRLELIQIENCR 484
Query: 420 LGPQFPKWIQTQKNFSHIDISNAGVSDYVP-NWFWDLSPNVEYMNLSSNELR-RCGQDFS 477
+G FP W+Q Q + + + N G+ D +P +WF +S V Y+ L++N ++ R Q +
Sbjct: 485 IG-LFPMWLQVQTKLNFVTLRNTGIEDTIPDSWFSGISSKVTYLILANNRIKGRLPQKLA 543
Query: 478 QKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEIL--------CFNNS 529
KL T+DLS+N+F P N L L N F G++ ++L F+NS
Sbjct: 544 FP-KLNTIDLSSNNFEGTFPLWSTNATELRLYENNFSGSLPQNIDVLMPRMEKIYLFSNS 602
Query: 530 --------------LENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSL 575
L+ L L N+ SG P CW + ++++ NN G IP+S G L
Sbjct: 603 FTGNIPSSLCEVSGLQILSLRKNHFSGSFPKCWHRQFMLWGIDVSENNLSGEIPESLGML 662
Query: 576 KNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNN 635
+L +L++ N+L GKIPE+L+NC LT ++L N+L G +P W+G + L +L L +N
Sbjct: 663 PSLSVLLLNQNSLEGKIPESLRNCSGLTNIDLGGNKLTGKLPSWVG-KLSSLFMLRLQSN 721
Query: 636 SFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLP 695
SF IP LC + +L ILDLS N+++G IP+C+ N + + + I
Sbjct: 722 SFTGQIPDDLCNVPNLRILDLSGNKISGPIPKCISNLTAIARGTNNEVFQNLVFI----- 776
Query: 696 IYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNL 755
R + I I+LS N ++ EIP EI L+ L LNL
Sbjct: 777 --------------------VTRAREYEAIANSINLSGNNISGEIPREILGLLYLRILNL 816
Query: 756 SRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIG 815
SRN + GSIP I EL L LDLS+N S IP S A I L L+LS+N L G IP
Sbjct: 817 SRNSMAGSIPEKISELSRLETLDLSKNKFSGAIPQSFAAISSLQRLNLSFNKLEGSIP-- 874
Query: 816 NQMQSFDEVFYKGNPHLCGPPLRKACPRN 844
++ D Y GN LCG PL K CP++
Sbjct: 875 KLLKFQDPSIYIGNELLCGKPLPKKCPKD 903
>UniRef100_Q84PR7 Putative Cf2/Cf5 disease resistance protein homolog [Oryza sativa]
Length = 966
Score = 475 bits (1223), Expect = e-132
Identities = 342/965 (35%), Positives = 488/965 (50%), Gaps = 132/965 (13%)
Query: 37 IASEAEALLEFKEGLKDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSNSLDKL 96
IA E +ALL+ K GL+DPSN L+SW+ C +W+GV C+ GHV +L L + +
Sbjct: 44 IARERDALLDLKAGLQDPSNYLASWQGDNCCDEWEGVVCSKRNGHVATLTLEYAG----I 99
Query: 97 QGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNL 156
G ++ SLL L +L ++L+GNDF +P+ K+++HL L ANF G + +LGNL
Sbjct: 100 GGKISPSLLALRHLKSMSLAGNDFGGEPIPELFGELKSMRHLTLGDANFSGLVPPHLGNL 159
Query: 157 SLLESLDLSDNS---FYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDT 213
S L LDL+ Y NL WL L++L+ L L GV LS DW H + + L SL
Sbjct: 160 SRLIDLDLTSYKGPGLYSTNLAWLSRLANLQHLYLGGVNLSTAF-DWAHSLNM-LPSLQH 217
Query: 214 LRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTI-------PDWLFE--------- 257
L L C L PP MN SL +DLSGN F+ + P W F
Sbjct: 218 LSLRNCGLRNA-IPPPLHMNLTSLEVIDLSGNPFHSPVAVEKLFWPFWDFPRLETIYLES 276
Query: 258 ------------NCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFD 305
N L NL L+ N+L G + + +R++ L L L++N+++G I D
Sbjct: 277 CGLQGILPEYMGNSTSLVNLGLNFNDLTG-LPTTFKRLSNLKFLYLAQNNISGDIEKLLD 335
Query: 306 KLVN--LVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVV 363
KL + L L+L N L GS+P+ G+ SL LR+S N+++G + I +L+NL
Sbjct: 336 KLPDNGLYVLELYGNNLEGSLPAQKGR---LGSLYNLRISDNKISGDIPLWIGELTNLTS 392
Query: 364 LNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQ 423
L L NN G+I+ HLAN ++LK+L LS N + + NWVPPF+L GL +C LGP+
Sbjct: 393 LELDSNNFHGVITQFHLANLASLKILGLSHNTLAIVADHNWVPPFKLMIAGLKSCGLGPK 452
Query: 424 FPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRR-CGQDFSQKFKL 482
FP W+++Q + +DISN ++D +P+WFW N Y LS N++ ++K
Sbjct: 453 FPGWLRSQDTITMMDISNTSIADSIPDWFWTTFSNTRYFVLSGNQISGVLPAMMNEKMVA 512
Query: 483 KTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTI------SHVCEILCFNNS------- 529
+ +D SNN L ++P NL LDLS N G + + ++ F NS
Sbjct: 513 EVMDFSNNLLEGQLQKVPENLTYLDLSKNNLSGPLPLDFGAPFLESLILFENSLSGKIPQ 572
Query: 530 -------LENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLI 582
LE +DLS N L G PNC + ++ A N S D G +N+ ML
Sbjct: 573 SFCQLKYLEFVDLSANLLQGPFPNC-------LNISQAGNT---SRADLLGVHQNIIMLN 622
Query: 583 MYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIP 642
+ +NNLSG P L+ CQ L L+L NR G +P WI ++ L +
Sbjct: 623 LNDNNLSGMFPLFLQKCQNLIFLDLAFNRFSGSLPAWI-DELSALALF------------ 669
Query: 643 KTLCQLKSLHILDLSENQLTGAIPRCV--FLALTTEESINEK-SYMEF------------ 687
TL ++K L LDL+ N +GAIP + A++ + N+ SY+ +
Sbjct: 670 -TLTKMKELQYLDLAYNSFSGAIPWSLVNLTAMSHRPADNDSLSYIVYYGWSLSTSNVRV 728
Query: 688 --------MTIEESLPIY---LSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFL 736
EES P + S T LL+ KG + F G ++ + IDLS N L
Sbjct: 729 IMLANLGPYNFEESGPDFSHITSATNESLLVVTKGQQLEFRSGIIY---MVNIDLSCNNL 785
Query: 737 THEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANID 796
T IP +I L L LNLS N L G IP++IG L+S+ LDLS N L +IPTS++
Sbjct: 786 TGHIPEDISMLTALKNLNLSWNHLSGVIPTNIGALQSIESLDLSHNELFGQIPTSLSAPA 845
Query: 797 RLSWLDLSYNALSGKIPIGNQMQSFDE--VFYKGNPHLCGPPLRKACPRNSSFEDTHCSH 854
LS L+LSYN LSG+IP GNQ+++ D+ Y GNP LCGPPL + +CS
Sbjct: 846 SLSHLNLSYNNLSGQIPYGNQLRTLDDQASIYIGNPGLCGPPLSR-----------NCSE 894
Query: 855 SEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMND 914
S + D DK L + LY+ M +G+ G WV + + + WR F + D
Sbjct: 895 SSKLLPDAVDEDKSLSDGVF-LYLGMGIGWVVGLWVVLCTFLFMQRWRIICFLVSDRLYD 953
Query: 915 KIHVT 919
+I +
Sbjct: 954 RIRAS 958
>UniRef100_Q6K7T8 Putative HcrVf2 protein [Oryza sativa]
Length = 960
Score = 474 bits (1220), Expect = e-132
Identities = 336/957 (35%), Positives = 488/957 (50%), Gaps = 109/957 (11%)
Query: 37 IASEAEALLEFKEGL-KDPSNLLSSWKHGKDCCQWKGVGCNTTTGHVISLNLHCSNS-LD 94
+ SE ALL K G DP L+SW DCC+W GV C+ TGHV L LH + + +D
Sbjct: 37 VPSERAALLAIKAGFTSDPDGRLASWGAAADCCRWDGVVCDNATGHVTELRLHNARADID 96
Query: 95 K---LQGHLNSSLLQLPYLSYLNLSGNDFM------QSTVPDFLSTTKNLKHLDLSHANF 145
L G ++ SLL LP L+YL+LS N+ + S +P FL + +L++L+LS
Sbjct: 97 GGAGLGGEISRSLLGLPRLAYLDLSQNNLIGGDGVSPSPLPRFLGSLCDLRYLNLSFTGL 156
Query: 146 KGNLLDNLGNLSLLESLDLSDN--SFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHD 203
G + LGNL+ L LDLS N Y ++ WL G+SSL+ LD+S V L+ +
Sbjct: 157 AGEIPPQLGNLTRLRQLDLSSNVGGLYSGDISWLSGMSSLEYLDMSVVNLNASVG--WAG 214
Query: 204 IRVILHSLDTLRLSGCQLHKLPTSPPPEMNFD-------------------------SLV 238
+ L SL L LS C L P SPP N +L
Sbjct: 215 VVSNLPSLRVLALSDCGLTAAP-SPPARANLTRLQKLDLSTNVINTSSANSWFWDVPTLT 273
Query: 239 TLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNG 298
LDLSGN + PD L N +L+ LNL N++ G I +++R+ L ++DL+ NS+NG
Sbjct: 274 YLDLSGNALSGVFPDAL-GNMTNLRVLNLQGNDMVGMIPATLQRLCGLQVVDLTVNSVNG 332
Query: 299 LIPNFFDKLVNLV-----ALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLER 353
+ F +L V L LS +SG +P +G+ + L L LS N+L+G +
Sbjct: 333 DMAEFMRRLPRCVFGKLQVLQLSAVNMSGHLPKWIGE---MSELTILDLSFNKLSGEIPL 389
Query: 354 SIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETI 413
I LSNL L L N + G +S+ H A+ +L+ +DLS N++++ + +W PP +L
Sbjct: 390 GIGSLSNLTRLFLHNNLLNGSLSEEHFADLVSLEWIDLSLNNLSMEIKPSWKPPCKLVYA 449
Query: 414 GLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCG 473
+ +GP FP WI+ Q + ++DISNAG+ D +P WFW + Y+N+S N++
Sbjct: 450 YFPDVQMGPHFPAWIKHQPSIKYLDISNAGIVDELPPWFWKSYSDAVYLNISVNQISGVL 509
Query: 474 QDFSQKFKLKTLD--LSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLE 531
S KF L L +N+ + +P LP L LDLS N G L
Sbjct: 510 PP-SLKFMRSALAIYLGSNNLTGSVPLLPEKLLVLDLSRNSLSGPFPQEFGA----PELV 564
Query: 532 NLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIP-------DSFGSLKNLHMLIMY 584
LD+S N +SG++P N++ L+L+ NN G +P D G L LI+Y
Sbjct: 565 ELDVSSNMISGIVPETLCRFPNLLHLDLSNNNLTGHLPRCRNISSDGLG----LITLILY 620
Query: 585 NNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKT 644
NN +G+ P LK+C+ +T L+L N G +P WIG + L L + +N F +IP
Sbjct: 621 RNNFTGEFPVFLKHCKSMTFLDLAQNMFSGIVPEWIGRKLPSLTHLRMKSNRFSGSIPTQ 680
Query: 645 LCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLS----- 699
L +L L LDL++N+L+G+IP + + M MT + LP+ L+
Sbjct: 681 LTELPDLQFLDLADNRLSGSIPPSL-------------ANMTGMT-QNHLPLALNPLTGY 726
Query: 700 ------RTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSAL 753
R L + KG + + G ++ + +DLS N L IP E+ L L L
Sbjct: 727 GASGNDRIVDSLPMVTKGQDRSYTSGVIY---MVSLDLSDNVLDGSIPDELSSLTGLVNL 783
Query: 754 NLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIP 813
NLS N+L G+IP IG L+ L LDLS N LS EIP+S++++ LS L+LSYN LSG+IP
Sbjct: 784 NLSMNRLTGTIPRKIGALQKLESLDLSINVLSGEIPSSLSDLTSLSQLNLSYNNLSGRIP 843
Query: 814 IGNQMQSF--DEVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDKVLGM 871
GNQ+Q+ Y GN LCGPPL+K C S E S + HE G
Sbjct: 844 SGNQLQALANPAYIYIGNAGLCGPPLQKNC----SSEKNRTSQPDLHEGKGL-------S 892
Query: 872 EINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIHVTVVVALNKLR 928
+ Y+ +A+GF G W+ + SL+ + +WR YF+ I+ D ++V + V K R
Sbjct: 893 DTMSFYLGLALGFVVGLWMVFCSLLFVKTWRIVYFQAINKAYDTLYVFIGVRWAKFR 949
>UniRef100_Q6QM03 LLR protein WM1.1 [Aegilops tauschii]
Length = 1032
Score = 429 bits (1103), Expect = e-118
Identities = 298/853 (34%), Positives = 443/853 (50%), Gaps = 77/853 (9%)
Query: 96 LQGHLNSSL--LQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNL 153
L + N SL + L L L+L N F S + +LK+LDL + G D L
Sbjct: 235 LLDYANQSLQHVNLTKLEKLDLFNNYFEHSLASGWFWKATSLKYLDLGNNRLFGQFPDTL 294
Query: 154 GNLSLLESLDLSDN-SFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLD 212
GN++ L+ LD+S+N + ++ L L L+I+DLS ++ DI V++ SL
Sbjct: 295 GNMTNLQVLDISENWNPHMMMAGNLENLCGLEIIDLSYNYING-------DIAVLMESL- 346
Query: 213 TLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNL 272
P+ L +DL NNF T+P+ L + L+ L+LS NNL
Sbjct: 347 -----------------PQCTRKKLQEMDLRYNNFTGTLPN-LVSDFTRLRILSLSGNNL 388
Query: 273 QGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDH 332
G I + +T L L+L N L G IP + L L +L+LS N+L+GSIP+ G+
Sbjct: 389 VGSIPPWLVNLTRLTTLELFSNHLTGSIPPWLGNLTCLTSLELSDNLLTGSIPAEFGK-- 446
Query: 333 GQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLS 392
L L LS N LN S+ I L NL+ L+L+ N+ G+I++ HLAN ++LK +DLS
Sbjct: 447 -LMYLTILDLSSNHLNESVPAEIGSLVNLIFLDLSNNSFTGVITEEHLANLTSLKQIDLS 505
Query: 393 FNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWF 452
N+ + ++ +W P LE+ A+C +GP FP W+Q Q + +DIS + P+WF
Sbjct: 506 LNNFKIALNSDWRAPSTLESAWFASCQMGPLFPPWLQ-QLKITALDISTTSLKGEFPDWF 564
Query: 453 WDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNL 512
W NV Y+++S+N++ + L L +N + P+P LP N+ LD+S+N
Sbjct: 565 WSAFSNVTYLDISNNQISGNLPAHMDSMAFEKLYLRSNRLTGPIPTLPTNITLLDISNNT 624
Query: 513 FYGTI-----SHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGS 567
F TI + EILC ++ N + G IP +I L+L+ N G
Sbjct: 625 FSETIPSNLVAPRLEILCMHS---------NQIGGYIPESICKLEQLIYLDLSNNILEGE 675
Query: 568 IPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQIL 627
+P F + N+ LI+ NN+LSGKIP L+N L L+L N+ G +P WIG ++ L
Sbjct: 676 VPQCFDT-HNIENLILSNNSLSGKIPAFLQNNTSLEFLDLSWNKFSGRLPTWIG-NLVYL 733
Query: 628 MVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKS-YME 686
L+L +N F +NIP + +L L LDLS N +GAIPR + LT ++ E+S YM
Sbjct: 734 RFLVLSHNEFSDNIPVNITKLGHLQYLDLSHNNFSGAIPRHLS-NLTFMTTLQEESRYMV 792
Query: 687 FMTIEE---SLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVE 743
+ ++ + L + KG + ++ +F IDLS N LT +IP +
Sbjct: 793 EVEVDSMGGTTEFEADSLGQILSVNTKGQQLIYHRTLAYFV---SIDLSCNSLTGKIPTD 849
Query: 744 IGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDL 803
I L L LNLS NQL G IP+ IG ++SL LDLS+N L EIP+S+ N+ LS+LDL
Sbjct: 850 ITSLAALMNLNLSSNQLSGQIPNMIGAMQSLESLDLSQNKLYGEIPSSLTNLTSLSYLDL 909
Query: 804 SYNALSGKIPIGNQMQSFD----EVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHE 859
SYN+LSG+IP G Q+ + + + Y GN LCGPP+ K C N ++ S+E
Sbjct: 910 SYNSLSGRIPSGPQLDTLNMDNQTLMYIGNNGLCGPPVHKNCSGNDAYIHGDLESSKE-- 967
Query: 860 NDGNHGDKVLGMEINPL--YISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIH 917
E +PL Y + +GF G W+ + +L+ +WR AYFR + D+++
Sbjct: 968 ------------EFDPLTFYFGLVLGFVVGLWMVFCALLFKKTWRIAYFRLFDKVYDQVY 1015
Query: 918 VTVVVALNKLRRK 930
V VVV +K
Sbjct: 1016 VFVVVKWASFAKK 1028
>UniRef100_Q949G9 HcrVf1 protein [Malus floribunda]
Length = 1015
Score = 427 bits (1098), Expect = e-118
Identities = 293/844 (34%), Positives = 437/844 (51%), Gaps = 106/844 (12%)
Query: 110 LSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSF 169
L L+LS N F S +P ++ + KNL L L F+G + N++ L +DLS+NS
Sbjct: 243 LVVLDLSEN-FFNSLMPRWVFSLKNLVSLHLRFCGFQGPIPSISQNITSLREIDLSENSI 301
Query: 170 YVNNL-KWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSP 228
++ + KWL L + SL + +L+G +LP+S
Sbjct: 302 SLDPIPKWLFNQKDLAL------------------------SLKSNQLTG----QLPSS- 332
Query: 229 PPEMNFDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAI 288
N L L+L N FN TIP WL+ ++L++L LS N L+G+IS SI +T+L
Sbjct: 333 --FQNMTGLKVLNLESNYFNSTIPKWLY-GLNNLESLLLSYNALRGEISSSIGNMTSLVN 389
Query: 289 LDLSKNSLNGLIPNFFDKLVNLVALDLS------------------------------YN 318
L+L N L G IPN L L +DLS Y
Sbjct: 390 LNLENNQLQGKIPNSLGHLCKLKVVDLSENHFTVRRPSEIFESLSGCGPDGIKSLSLRYT 449
Query: 319 MLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDV 378
+SG IP +LG +SL++L +S N NG+ I QL L L+++ N EG++S++
Sbjct: 450 NISGPIPMSLGN---LSSLEKLDISGNHFNGTFTEVIGQLKMLTDLDISYNWFEGVVSEI 506
Query: 379 HLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHID 438
+N + LK N TL S++WVPPFQLET+ L + HLGP++P W++TQ +
Sbjct: 507 SFSNLTKLKHFVAKGNSFTLKTSRDWVPPFQLETLRLDSWHLGPKWPMWLRTQTQLKELS 566
Query: 439 ISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPR 498
+S G+S +P WFW+L+ +V Y+NLS N+L GQ + +DL +N F+ LP
Sbjct: 567 LSGTGISSTIPTWFWNLTFHVWYLNLSHNQL--YGQIQNIVAGRSVVDLGSNQFTGALPI 624
Query: 499 LPPNLRNLDLSSNLFYGTISH-VCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIIL 557
+P +L LDLS++ F G++ H C+ L L L N L+G +P+CW + + +
Sbjct: 625 VPTSLVWLDLSNSSFSGSVFHFFCDRPDETKLLYILHLGNNFLTGKVPDCWMSWPQLGFV 684
Query: 558 NLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIP 617
NL NN G++P S G L P +L+NC +L+ ++L N G IP
Sbjct: 685 NLENNNLTGNVPMSMGEL-----------------PHSLQNCTMLSFVDLSENGFSGSIP 727
Query: 618 YWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEE 677
WIG + L VL L +N F+ +IP +C L+SL ILDL+ N+L+G IPRC F L+
Sbjct: 728 IWIGKSLSWLYVLNLRSNKFEGDIPNEVCYLQSLQILDLAHNKLSGMIPRC-FHNLSALA 786
Query: 678 SINEKSYMEFMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLT 737
+ +E S+ F+T + ++ +L+ KG + +++ +K +DLS NF+
Sbjct: 787 NFSE-SFFPFITGNTDGEFW----ENAILVT-KGTEMEYSK---ILGFVKGMDLSCNFMY 837
Query: 738 HEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDR 797
EIP E+ L+ L +LNLS N+ G IPS IG + L LD S N L EIP SM N+
Sbjct: 838 GEIPKELTGLLALQSLNLSNNRFTGRIPSKIGNMAQLESLDFSMNQLDGEIPPSMTNLTF 897
Query: 798 LSWLDLSYNALSGKIPIGNQMQSFDEVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEE 857
LS L+LSYN L+G+I Q+QS D+ + GN LCG PL K C N H
Sbjct: 898 LSHLNLSYNNLTGRILESTQLQSLDQSSFVGN-ELCGAPLNKNCSENGVIPPPTVEH--- 953
Query: 858 HENDGNHGDKVLGMEINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIH 917
DG G ++L E Y+++ +GF TGFW+ GSL++ W + ++ + K++
Sbjct: 954 ---DGGGGYRLL--EDEWFYVTLGVGFFTGFWIVLGSLLVNMPWSILLSQLLNRIVLKMY 1008
Query: 918 VTVV 921
+V
Sbjct: 1009 HVIV 1012
>UniRef100_Q6QM01 LRR protein WM1.10 [Aegilops tauschii]
Length = 1060
Score = 426 bits (1096), Expect = e-117
Identities = 309/877 (35%), Positives = 441/877 (50%), Gaps = 95/877 (10%)
Query: 82 VISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLS 141
VI L+ +S ++ HLN L L L+LS N F S + +LK+L L
Sbjct: 231 VIDLSYCLLDSANQSLLHLN-----LTKLEKLDLSWNFFKHSLGSGWFWKVTSLKYLHLE 285
Query: 142 HANFKGNLLDNLGNLSLLESLDLSDNS----FYVNNLKWLHGLSSLKILDLSGVVLSRCQ 197
G D LGN++ L LD+S N N+K L SL+ILDLSG
Sbjct: 286 WNLLFGKFPDTLGNMTYLRVLDISYNGNPDMMMTGNIK---KLCSLEILDLSGN------ 336
Query: 198 NDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPDWLFE 257
R++G + L P+ +L LDLS NNF T+P+ +
Sbjct: 337 -----------------RING-DIESLFVESLPQCTRKNLQKLDLSYNNFTGTLPN-IVS 377
Query: 258 NCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSY 317
+ L L+LSNNNL G I + +T L LDL N LNG IP L L +LDLS
Sbjct: 378 DFSKLSILSLSNNNLVGPIPAQLGNLTCLTSLDLFWNHLNGSIPPELGALTTLTSLDLSM 437
Query: 318 NMLSGSIPSTLGQDH---------------------GQNSLKELRLSINQLNGSLERSIY 356
N L+GSIP+ LG SL L LS N LNGS+ I
Sbjct: 438 NDLTGSIPAELGNLRYLSELCLSDNNITAPIPPELMNSTSLTHLDLSSNHLNGSVPTEIG 497
Query: 357 QLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLA 416
L+NL+ L L+ N G+I++ + AN ++LK +DLSFN++ + ++ +W PF LE A
Sbjct: 498 SLNNLIYLYLSNNRFTGVITEENFANLTSLKDIDLSFNNLKIVLNSDWRAPFTLEFASFA 557
Query: 417 NCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDF 476
+C +GP FP +Q K + +DISN + +P+WFW N Y+++S+N++
Sbjct: 558 SCQMGPLFPPGLQRLKT-NALDISNTTLKGEIPDWFWSTFSNATYLDISNNQISGSLPAH 616
Query: 477 SQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLS 536
+ L L +N + P+P LP N+ LD+S+N F TI + LE L +
Sbjct: 617 MHSMAFEKLHLGSNRLTGPIPTLPTNITLLDISNNTFSETIPSNLGA----SRLEILSMH 672
Query: 537 FNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETL 596
N + G IP ++ L+L+ N G +P F K H LI+ NN+LSGKIP L
Sbjct: 673 SNQIGGYIPESICKLEQLLYLDLSNNILEGEVPHCFHFYKIEH-LILSNNSLSGKIPAFL 731
Query: 597 KNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDL 656
+N L L++ NR G +P WIG + L L+L +N F +NIP + +L L LDL
Sbjct: 732 QNNTGLQFLDVSWNRFSGRLPTWIGNLVN-LRFLVLSHNIFSDNIPVDITKLGHLQYLDL 790
Query: 657 SENQLTGAIPRCVFLALTTEESINEKSYMEFMTIEESL---PIYLS--RTKHPLLIPWKG 711
S N +G IP ++ T S + YM +T ++ PI++ R L + KG
Sbjct: 791 SRNNFSGGIPW--HMSNLTFMSTLQSMYMVEVTEYDTTRLGPIFIEADRLGQILSVNTKG 848
Query: 712 VNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGEL 771
+ ++ +F IDLS N LT EIP +I L L LNLS NQL G IPS IG +
Sbjct: 849 QQLIYHGTLAYFV---SIDLSCNSLTGEIPTDITSLAALMNLNLSSNQLSGQIPSMIGAM 905
Query: 772 ESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFD----EVFYK 827
+SL LDLS+N LS EIP+S++N+ LS+++LS N+LSG+IP G Q+ + + Y
Sbjct: 906 QSLVSLDLSQNKLSGEIPSSLSNLTSLSYMNLSCNSLSGRIPSGPQLDILNLDNQSLIYI 965
Query: 828 GNPHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDKVLGMEINPL--YISMAMGFS 885
GN LCGPP+ K C N + + S+E E +PL Y + +GF
Sbjct: 966 GNTGLCGPPVHKNCSGNDPYIHSDLESSKE--------------EFDPLTFYFGLVLGFV 1011
Query: 886 TGFWVFWGSLILIASWRHAYFRFISNMNDKIHVTVVV 922
G W+ + +L+ +WR AYFRF + D+++V VVV
Sbjct: 1012 VGLWMVFCALLFKKTWRIAYFRFFDKVYDQVYVFVVV 1048
>UniRef100_Q6QM07 LRR protein WM1.7 [Aegilops tauschii]
Length = 1102
Score = 424 bits (1089), Expect = e-117
Identities = 311/910 (34%), Positives = 456/910 (49%), Gaps = 103/910 (11%)
Query: 82 VISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLS 141
+I L + +S D+ HLN L L L+L+ NDF S + +LK+L+L
Sbjct: 231 IIDLTVCSLDSADQSLPHLN-----LTKLERLDLNNNDFEHSLTYGWFWKATSLKYLNLG 285
Query: 142 HANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKW-LHGLSSLKILDLSGVVLSRCQNDW 200
+ G D LGN++ L+ LD+S N + L L SL+I+DLS +N+
Sbjct: 286 YNGLFGQFPDTLGNMTNLQVLDISVNKITDMMMTGNLENLCSLEIIDLS-------RNEI 338
Query: 201 FHDIRVILHSL---------------------------DTLRLSGCQL--HKLPTSPPPE 231
DI V++ SL D RLS L + L PP+
Sbjct: 339 NTDISVMMKSLPQCTWKKLQELDLGGNKFRGTLPNFIGDFTRLSVLWLDYNNLVGPIPPQ 398
Query: 232 M-NFDSLVTLDLSGNNFNMTIPDWL-----------------------FENCHHLQNLNL 267
+ N L +LDL GN+ +IP L N +L L L
Sbjct: 399 LGNLTCLTSLDLGGNHLTGSIPTELGALTTLTYLDIGSNDLNGGVPAELGNLRYLTALYL 458
Query: 268 SNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPST 327
S+N + G I + + +L LDLS N + G IP L L L+L N L+GSIP
Sbjct: 459 SDNEIAGSIPPQLGNLRSLTALDLSDNEIAGSIPPQLGNLTGLTYLELRNNHLTGSIPRE 518
Query: 328 LGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLK 387
L SL L L N L GS+ I L NL L+L+ N+ G+I++ HLAN ++L+
Sbjct: 519 LMHS---TSLTILDLPGNHLIGSVPTEIGSLINLQFLDLSNNSFTGMITEEHLANLTSLQ 575
Query: 388 VLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDY 447
+DLS N++ + ++ +W PPF LE+ +C +GP FP W+Q K + +DIS+ G+
Sbjct: 576 KIDLSSNNLKIVLNSDWRPPFMLESASFGSCQMGPLFPPWLQQLKT-TQLDISHNGLKGE 634
Query: 448 VPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLD 507
P+WFW + YM++S+N++ + + L++N + P+P LP ++ LD
Sbjct: 635 FPDWFWSTFSHALYMDISNNQISGRLPAHLHGMAFEEVYLNSNQLTGPIPALPKSIHLLD 694
Query: 508 LSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGS 567
+S N F+GTI + L+ L + N +SG IP +I L+L+ N G
Sbjct: 695 ISKNQFFGTIPSILGA----PRLQMLSMHSNQISGYIPESICKLEPLIYLDLSNNILEGE 750
Query: 568 IPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQIL 627
I F + +L LI+ NN+LSGKIP +L+N L L+L N+ G +P WIGT + L
Sbjct: 751 IVKCF-DIYSLEHLILGNNSLSGKIPASLRNNACLKFLDLSWNKFSGGLPTWIGTLVH-L 808
Query: 628 MVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFLALTTEESINEKSYMEF 687
LIL +N F +NIP + +L L LDLS N +GAIP + +LT ++ E+S M
Sbjct: 809 RFLILSHNKFSDNIPVDITKLGYLQYLDLSSNNFSGAIPWHLS-SLTFMSTLQEES-MGL 866
Query: 688 MTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKL 747
+ I R L + KG + ++ +F IDLS N LT EIP +I L
Sbjct: 867 VGDVRGSEIVPDRLGQILSVNTKGQQLTYHRTLAYFV---SIDLSCNSLTGEIPTDITSL 923
Query: 748 VELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNA 807
L LNLS NQL G IPS IG ++SL LDLS+N LS EIP+S++N+ LS+++LS N+
Sbjct: 924 AALMNLNLSSNQLSGQIPSMIGAMQSLVSLDLSQNKLSGEIPSSLSNLTSLSYMNLSCNS 983
Query: 808 LSGKIPIGNQMQSFD----EVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGN 863
LSG+IP G Q+ + + + Y GN LCGPP+ K C N F
Sbjct: 984 LSGRIPSGRQLDTLNMDNPSLMYIGNNGLCGPPVHKNCSGNDPF---------------I 1028
Query: 864 HGD-KVLGMEINPL--YISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIHVTV 920
HGD + E++PL Y + +GF G W+ + +L+ +WR AYFR + D+++V V
Sbjct: 1029 HGDLRSSNQEVDPLTFYFGLVLGFVVGLWMVFCALLFKKTWRIAYFRLFDKVYDQVYVFV 1088
Query: 921 VVALNKLRRK 930
VV +K
Sbjct: 1089 VVKWASFAKK 1098
>UniRef100_Q6QM04 LRR protein WM1.2 [Aegilops tauschii]
Length = 1060
Score = 422 bits (1085), Expect = e-116
Identities = 305/861 (35%), Positives = 445/861 (51%), Gaps = 63/861 (7%)
Query: 82 VISLNLHCSNSLDKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLS 141
VI L+L +S ++ HLN L L L+LS N F S + +LK+L L
Sbjct: 231 VIDLSLCSLHSANQSLPHLN-----LTKLEKLDLSLNYFEHSLGSGWFWKAISLKYLALG 285
Query: 142 HANFKGNLLDNLGNLSLLESLDLSDN----SFYVNNLKWLHGLSSLKILDLSGVVLSRCQ 197
H + G D LGN++ L+ LD+S N + L L L SL+I+DL G
Sbjct: 286 HNSLFGQFPDTLGNMTSLQVLDVSYNWNPDMMMIGKL--LKNLCSLEIIDLDG------- 336
Query: 198 NDWFHDIRVILHSLDTLRLSGCQLHKLP------TSPPPEMNFDSLVTLDLSGNNFNMTI 251
N+ +I V++ S Q L T P +F SL TL LSGN+ I
Sbjct: 337 NEISGEIEVLMESWPQCTWKNLQELDLSSNTFTGTLPNFLGDFTSLRTLSLSGNSLAGPI 396
Query: 252 PDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLV 311
P L N L +L+LS+N+ G I + + L L+L N + G IP L L
Sbjct: 397 PPQL-GNLTCLTSLDLSSNHFTGSIRDELGNLRYLTALELQGNEITGSIPLQLGNLTCLT 455
Query: 312 ALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNM 371
++DL N L+GSIP+ +G+ L L LS N LNGS+ + L NL+ L+L N+
Sbjct: 456 SIDLGDNHLTGSIPAEVGK---LTYLTSLDLSSNHLNGSVPTEMGSLINLISLDLRNNSF 512
Query: 372 EGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQ 431
G+I+ H AN ++LK +DLS+N++ + ++ +W PF LE+ +C +GP FP W+Q
Sbjct: 513 TGVITGEHFANLTSLKQIDLSYNNLKMVLNSDWRAPFTLESASFGSCQMGPLFPPWLQQL 572
Query: 432 KNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNS 491
K + ++IS+ G+ P+WFW NV ++++S+N++ + L LS+N
Sbjct: 573 KT-TQLNISSNGLKGEFPDWFWSAFSNVTHLDISNNQINGSLPAHMDSMAFEELHLSSNR 631
Query: 492 FSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIPNCWTNG 551
+ P+P LP N+ LD+S+N F TI L+ L + NN+ G IP
Sbjct: 632 LAGPIPTLPINITLLDISNNTFSETIPSNLVA----PGLKVLCMQSNNIGGYIPESVCKL 687
Query: 552 TNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNR 611
+ L+L+ N G IP + N+ LI+ NN+LSGKIP L+N L L+L N
Sbjct: 688 EQLEYLDLSNNILEGKIPQC-PDIHNIKYLILSNNSLSGKIPAFLQNNTNLKFLDLSWNN 746
Query: 612 LRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCVFL 671
G +P WIG + L+ LIL +N F ++IP + +L L LDLS+N+ GAIP C
Sbjct: 747 FSGRLPTWIGK-LANLLFLILSHNKFSDSIPVNVTKLGHLQYLDLSDNRFFGAIP-CHLS 804
Query: 672 ALTTEESINEKSYME----FMTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILK 727
LT ++ E M+ ++ E + I LL+ KG ++ ++ +F
Sbjct: 805 NLTFMRTLQEDIDMDGPILYVFKEYATGIAPQELGQTLLVNTKGQHLIYHMTLAYFV--- 861
Query: 728 MIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCE 787
IDLS N LT EIP +I L L LNLS NQL G IP+ IG ++SL LDLS+N L E
Sbjct: 862 GIDLSHNSLTGEIPTDITSLDALVNLNLSSNQLSGEIPNMIGAMQSLESLDLSQNKLYGE 921
Query: 788 IPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSF----DEVFYKGNPHLCGPPLRKACPR 843
IP+S+ N+ LS+LDLSYN+LSG+IP G Q+ + + Y GN LCGPP+ K
Sbjct: 922 IPSSLTNLTSLSYLDLSYNSLSGRIPSGPQLDTLSAENQSLMYIGNSGLCGPPVHK---- 977
Query: 844 NSSFEDTHCSHSEEHENDGNHGDKVLGMEINPL--YISMAMGFSTGFWVFWGSLILIASW 901
+CS +E +D K E +PL Y + +GF G W+ + L+ +W
Sbjct: 978 -------NCSGNEPSIHDDLKSSK---KEFDPLNFYFGLVLGFVVGLWMVFCVLLFKRTW 1027
Query: 902 RHAYFRFISNMNDKIHVTVVV 922
R AYFR + D+++V VVV
Sbjct: 1028 RIAYFRLFDRVYDQVYVFVVV 1048
>UniRef100_Q9SDM7 Hypothetical protein [Glycine max]
Length = 578
Score = 386 bits (991), Expect = e-105
Identities = 229/603 (37%), Positives = 349/603 (56%), Gaps = 35/603 (5%)
Query: 337 LKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHV 396
L+ L L N L G + ++ LSNLV L+L+ N +EG I + + LK L LS+ ++
Sbjct: 1 LQVLNLGANSLTGDVPVTLGTLSNLVTLDLSSNLLEGSIKESNFVKLFTLKELRLSWTNL 60
Query: 397 TLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLS 456
L+++ W PPFQLE + L++ +GP+FP+W++ Q + + +S AG++D VP+WFW +
Sbjct: 61 FLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLKRQSSVKVLTMSKAGIADLVPSWFWIWT 120
Query: 457 PNVEYMNLSSNELRRCGQDFSQKF-KLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYG 515
+E+++LS+N LR D S F ++LS+N F LP + N+ L++++N G
Sbjct: 121 LQIEFLDLSNNLLRG---DLSNIFLNSSVINLSSNLFKGRLPSVSANVEVLNVANNSISG 177
Query: 516 TISHVCEILCFN----NSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDS 571
TIS LC N N L LD S N LSG + +CW + ++ +NL NN G IP+S
Sbjct: 178 TIS---PFLCGNPNATNKLSVLDFSNNVLSGDLGHCWVHWQALVHVNLGSNNLSGEIPNS 234
Query: 572 FGSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLI 631
G L L L++ +N SG IP TL+NC + +++ +N+L IP W+ ++Q LMVL
Sbjct: 235 MGYLSQLESLLLDDNRFSGYIPSTLQNCSTMKFIDMGNNQLSDTIPDWMW-EMQYLMVLR 293
Query: 632 LGNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCV----FLALTTEESINEKSYMEF 687
L +N+F+ +I + +CQL SL +LDL N L+G+IP C+ +A + N SY
Sbjct: 294 LRSNNFNGSIAQKMCQLSSLIVLDLGNNSLSGSIPNCLDDMKTMAGEDDFFANPSSY--- 350
Query: 688 MTIEESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKL 747
+ K L++ K + + + + +++MIDLSSN L+ IP EI KL
Sbjct: 351 ---SYGSDFSYNHYKETLVLVPKKDELEYRDNLI---LVRMIDLSSNKLSGAIPSEISKL 404
Query: 748 VELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNA 807
L LNLSRN L G IP+ +G+++ L LDLS NN+S +IP S++++ LS+L+LSY+
Sbjct: 405 FALRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNISGQIPQSLSDLSFLSFLNLSYHN 464
Query: 808 LSGKIPIGNQMQSFDEVFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDK 867
LSG+IP Q+QSFDE+ Y GNP LCGPP+ K C + S S H DGN
Sbjct: 465 LSGRIPTSTQLQSFDELSYTGNPELCGPPVTKNCTNKEWLRE---SASVGH-GDGNF--- 517
Query: 868 VLGMEINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIHVTVVVALNKL 927
+ YI M +GF+ GFW F + +WR AYF ++ ++ D I+V +V+ + +L
Sbjct: 518 ---FGTSEFYIGMGVGFAAGFWGFCSVVFFNRTWRLAYFHYLDHLRDLIYVMIVLKVRRL 574
Query: 928 RRK 930
K
Sbjct: 575 LGK 577
Score = 123 bits (309), Expect = 2e-26
Identities = 127/456 (27%), Positives = 196/456 (42%), Gaps = 83/456 (18%)
Query: 236 SLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNL----------QGQISYS------ 279
+LVTLDLS N +I + F L+ L LS NL Q+ Y
Sbjct: 24 NLVTLDLSSNLLEGSIKESNFVKLFTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFG 83
Query: 280 --------IERVTTLAILDLSKNSLNGLIPN-FFDKLVNLVALDLSYNMLSGSIPSTLGQ 330
++R +++ +L +SK + L+P+ F+ + + LDLS N+L G + +
Sbjct: 84 IGPKFPEWLKRQSSVKVLTMSKAGIADLVPSWFWIWTLQIEFLDLSNNLLRGDLSNIF-- 141
Query: 331 DHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEGIISDVHLAN---FSNLK 387
NS + LS N G L +N+ VLN+A N++ G IS N + L
Sbjct: 142 ---LNS-SVINLSSNLFKGRLPS---VSANVEVLNVANNSISGTISPFLCGNPNATNKLS 194
Query: 388 VLDLSFNHVTLNMSKNWVP------------------------PFQLETIGLANCHLGPQ 423
VLD S N ++ ++ WV QLE++ L +
Sbjct: 195 VLDFSNNVLSGDLGHCWVHWQALVHVNLGSNNLSGEIPNSMGYLSQLESLLLDDNRFSGY 254
Query: 424 FPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFSQKFKLK 483
P +Q ID+ N +SD +P+W W++ + S+N Q Q L
Sbjct: 255 IPSTLQNCSTMKFIDMGNNQLSDTIPDWMWEMQYLMVLRLRSNNFNGSIAQKMCQLSSLI 314
Query: 484 TLDLSNNSFSCPLPRLPPNLRNLDLSSNLF-------YG---TISHVCEILCF---NNSL 530
LDL NNS S +P +++ + + F YG + +H E L + L
Sbjct: 315 VLDLGNNSLSGSIPNCLDDMKTMAGEDDFFANPSSYSYGSDFSYNHYKETLVLVPKKDEL 374
Query: 531 E---------NLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHML 581
E +DLS N LSG IP+ + + LNL+ N+ G IP+ G +K L L
Sbjct: 375 EYRDNLILVRMIDLSSNKLSGAIPSEISKLFALRFLNLSRNHLSGEIPNDMGKMKLLESL 434
Query: 582 IMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIP 617
+ NN+SG+IP++L + L+ LNL + L G IP
Sbjct: 435 DLSLNNISGQIPQSLSDLSFLSFLNLSYHNLSGRIP 470
Score = 96.3 bits (238), Expect = 4e-18
Identities = 81/278 (29%), Positives = 133/278 (47%), Gaps = 39/278 (14%)
Query: 135 LKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLS 194
L H++L N G + +++G LS LESL L DN F L S++K +D+ LS
Sbjct: 217 LVHVNLGSNNLSGEIPNSMGYLSQLESLLLDDNRFSGYIPSTLQNCSTMKFIDMGNNQLS 276
Query: 195 RCQNDWFHDIR-VILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPD 253
DW +++ +++ L + +G K+ SL+ LDL N+ + +IP+
Sbjct: 277 DTIPDWMWEMQYLMVLRLRSNNFNGSIAQKM-------CQLSSLIVLDLGNNSLSGSIPN 329
Query: 254 WL------------------------FENCHHLQNLNLSNNNLQGQISYSIERVTTLAIL 289
L F H+ + L L + ++ Y + + + ++
Sbjct: 330 CLDDMKTMAGEDDFFANPSSYSYGSDFSYNHYKETLVLVPK--KDELEYR-DNLILVRMI 386
Query: 290 DLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNG 349
DLS N L+G IP+ KL L L+LS N LSG IP+ +G+ L+ L LS+N ++G
Sbjct: 387 DLSSNKLSGAIPSEISKLFALRFLNLSRNHLSGEIPNDMGK---MKLLESLDLSLNNISG 443
Query: 350 SLERSIYQLSNLVVLNLAVNNMEG-IISDVHLANFSNL 386
+ +S+ LS L LNL+ +N+ G I + L +F L
Sbjct: 444 QIPQSLSDLSFLSFLNLSYHNLSGRIPTSTQLQSFDEL 481
Score = 89.7 bits (221), Expect = 4e-16
Identities = 139/520 (26%), Positives = 207/520 (39%), Gaps = 134/520 (25%)
Query: 110 LSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLD-NLGNLSLLESLDLSDNS 168
L LNL N + VP L T NL LDLS +G++ + N L L+ L LS +
Sbjct: 1 LQVLNLGANS-LTGDVPVTLGTLSNLVTLDLSSNLLEGSIKESNFVKLFTLKELRLSWTN 59
Query: 169 FY--VNN----------------------LKWLHGLSSLKILDLSGVVLSRCQNDWFHDI 204
+ VN+ +WL SS+K+L +S ++ WF
Sbjct: 60 LFLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLKRQSSVKVLTMSKAGIADLVPSWFWIW 119
Query: 205 RVILHSLDTLR------LSGCQLHKLPTS----------PPPEMNFDSLVTLDLSGNNFN 248
+ + LD LS L+ + P N + L+++ N+ +
Sbjct: 120 TLQIEFLDLSNNLLRGDLSNIFLNSSVINLSSNLFKGRLPSVSANVE---VLNVANNSIS 176
Query: 249 MTIPDWLFEN---CHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFD 305
TI +L N + L L+ SNN L G + + L ++L N+L+G IPN
Sbjct: 177 GTISPFLCGNPNATNKLSVLDFSNNVLSGDLGHCWVHWQALVHVNLGSNNLSGEIPNSMG 236
Query: 306 KLVNLVALDLSYNMLSGSIPSTLGQ-------DHGQNSLKE--------------LRLSI 344
L L +L L N SG IPSTL D G N L + LRL
Sbjct: 237 YLSQLESLLLDDNRFSGYIPSTLQNCSTMKFIDMGNNQLSDTIPDWMWEMQYLMVLRLRS 296
Query: 345 NQLNGSLERSIYQLSNLVVLNLAVNNMEGII------------SDVHLANFSNLKV-LDL 391
N NGS+ + + QLS+L+VL+L N++ G I D AN S+ D
Sbjct: 297 NNFNGSIAQKMCQLSSLIVLDLGNNSLSGSIPNCLDDMKTMAGEDDFFANPSSYSYGSDF 356
Query: 392 SFNHVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKN-----FSHIDISNAGVSD 446
S+NH ET+ L PK + + ID+S+ +S
Sbjct: 357 SYNHYK-------------ETLVLV--------PKKDELEYRDNLILVRMIDLSSNKLSG 395
Query: 447 YVPNWFWDLSPNVEYMNLSSNELR-RCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRN 505
+P+ L + ++NLS N L D + L++LDLS N+ S ++P +L +
Sbjct: 396 AIPSEISKLFA-LRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNIS---GQIPQSLSD 451
Query: 506 LDLSSNLFYGTISHVCEILCFNNSLENLDLSFNNLSGVIP 545
L L L+LS++NLSG IP
Sbjct: 452 LSF---------------------LSFLNLSYHNLSGRIP 470
>UniRef100_Q6JJ67 Putative disease resistance protein [Ipomoea trifida]
Length = 476
Score = 373 bits (958), Expect = e-101
Identities = 218/495 (44%), Positives = 293/495 (59%), Gaps = 36/495 (7%)
Query: 418 CHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNELRRCGQDFS 477
C LGP+FPKW+QTQ FS +DIS+ G+SD +PNWFWDL VEY+ LS+N++ D S
Sbjct: 1 CKLGPKFPKWLQTQSGFSELDISSTGISDTMPNWFWDLCSKVEYLALSNNKIDGELPDLS 60
Query: 478 QKFKL-KTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNNSLENLDLS 536
KF + +DLS+N+F P+ LPP +++L LS+N F G+IS VC +L F + +DLS
Sbjct: 61 TKFGVFPEIDLSHNNFRGPIHSLPPKVKSLYLSNNSFVGSISFVCRVLKFMS----IDLS 116
Query: 537 FNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYNNNLSGKIPETL 596
N SG IP+CW + + + LNLA NNF G +P SFG L L L + NNN +G++P +L
Sbjct: 117 DNQFSGEIPDCWHHLSRLNNLNLANNNFSGKVPPSFGYLYYLKELQLRNNNFTGELPSSL 176
Query: 597 KNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDL 656
+NC +L +L+L N+L G +P W GT + L+++ L N F +P +LC L +H+LDL
Sbjct: 177 QNCTLLRILDLGRNQLTGRVPSWFGTSLVDLIIVNLRENQFHGELPLSLCHLNDIHVLDL 236
Query: 657 SENQLTGAIPRC----VFLALTTEE---SINEKSYMEFMTIEESLPIYLSRTKHPLLIPW 709
S+N+++G IP C +L+LT ++ K+Y F +S K +LI W
Sbjct: 237 SQNRISGKIPHCFSNFTYLSLTNSSLGTTVASKAYFVFQNDIDSY-------KSNILIQW 289
Query: 710 KGVNVFFNEGRLFFEILKMIDLSSNFLTHEIPVEIGKLVELSALNLSRNQLLGSIPSSIG 769
K N GRL +LK+IDLSSN L +IP E L L +LNLSRN L G I IG
Sbjct: 290 K-YNEREYSGRL--RLLKLIDLSSNLLGGDIPEEFSSLHGLISLNLSRNHLTGKIIREIG 346
Query: 770 ELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKIPIGNQMQSFDEVFYKGN 829
++E L LDLS N LS EIP S+ + L L+LS N LSGKIP QMQSF+ Y N
Sbjct: 347 QMEMLESLDLSYNQLSGEIPISLGRLSFLQILELSNNNLSGKIPSSTQMQSFNASSYAHN 406
Query: 830 PHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHGDKVLGMEINPLYISMAMGFSTGFW 889
LCG PL K CPRN + +E E+D + G+ YISM +GFS FW
Sbjct: 407 SGLCGDPLPK-CPRN-------VPNKDEDEDDDD------GLITQGFYISMVLGFSLSFW 452
Query: 890 VFWGSLILIASWRHA 904
F SWR+A
Sbjct: 453 GFLVIFFFKGSWRNA 467
Score = 114 bits (284), Expect = 2e-23
Identities = 120/430 (27%), Positives = 194/430 (44%), Gaps = 55/430 (12%)
Query: 175 KWLHGLSSLKILDLSGVVLSRCQNDWFHDI--RVILHSLDTLRLSGCQLHKLPTSPPPEM 232
KWL S LD+S +S +WF D+ +V +L ++ G +L L T
Sbjct: 9 KWLQTQSGFSELDISSTGISDTMPNWFWDLCSKVEYLALSNNKIDG-ELPDLST------ 61
Query: 233 NFDSLVTLDLSGNNFNMTIPDWLFENCHHL----QNLNLSNNNLQGQISYSIERVTTLAI 288
F +DLS NNF I H L ++L LSNN+ G IS+ + RV
Sbjct: 62 KFGVFPEIDLSHNNFRGPI--------HSLPPKVKSLYLSNNSFVGSISF-VCRVLKFMS 112
Query: 289 LDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLN 348
+DLS N +G IP+ + L L L+L+ N SG +P + G + LKEL+L N
Sbjct: 113 IDLSDNQFSGEIPDCWHHLSRLNNLNLANNNFSGKVPPSFGYLY---YLKELQLRNNNFT 169
Query: 349 GSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPF 408
G L S+ + L +L+L N + G + + +L +++L N + +
Sbjct: 170 GELPSSLQNCTLLRILDLGRNQLTGRVPSWFGTSLVDLIIVNLRENQFHGELPLSLCHLN 229
Query: 409 QLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNE 468
+ + L+ + + P NF+++ ++N+ + V + + + N + + SN
Sbjct: 230 DIHVLDLSQNRISGKIPHCF---SNFTYLSLTNSSLGTTVASKAYFVFQN-DIDSYKSNI 285
Query: 469 L---RRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILC 525
L + +++S + +L L+ +DLSSNL G I L
Sbjct: 286 LIQWKYNEREYSGRLRL--------------------LKLIDLSSNLLGGDIPEEFSSL- 324
Query: 526 FNNSLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSIPDSFGSLKNLHMLIMYN 585
+ L +L+LS N+L+G I + L+L+ N G IP S G L L +L + N
Sbjct: 325 --HGLISLNLSRNHLTGKIIREIGQMEMLESLDLSYNQLSGEIPISLGRLSFLQILELSN 382
Query: 586 NNLSGKIPET 595
NNLSGKIP +
Sbjct: 383 NNLSGKIPSS 392
Score = 94.4 bits (233), Expect = 1e-17
Identities = 84/295 (28%), Positives = 139/295 (46%), Gaps = 17/295 (5%)
Query: 94 DKLQGHLNSSLLQLPYLSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNL 153
++ G + L L+ LNL+ N+F P F LK L L + NF G L +L
Sbjct: 118 NQFSGEIPDCWHHLSRLNNLNLANNNFSGKVPPSF-GYLYYLKELQLRNNNFTGELPSSL 176
Query: 154 GNLSLLESLDLSDNSFYVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDT 213
N +LL LDL N W ++DL ++++ +N + ++ + L L+
Sbjct: 177 QNCTLLRILDLGRNQLTGRVPSWF----GTSLVDL--IIVNLRENQFHGELPLSLCHLND 230
Query: 214 LRLSGCQLHKLPTSPPPEMNFDSLVTLDLSGNNFNMTIPD---WLFENCHHLQNLNLSNN 270
+ + +++ P F + L L+ ++ T+ ++F+N N+
Sbjct: 231 IHVLDLSQNRISGKIPH--CFSNFTYLSLTNSSLGTTVASKAYFVFQNDIDSYKSNILIQ 288
Query: 271 NLQGQISYSIERVTTLAILDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQ 330
+ YS R+ L ++DLS N L G IP F L L++L+LS N L+G I +GQ
Sbjct: 289 WKYNEREYS-GRLRLLKLIDLSSNLLGGDIPEEFSSLHGLISLNLSRNHLTGKIIREIGQ 347
Query: 331 DHGQNSLKELRLSINQLNGSLERSIYQLSNLVVLNLAVNNMEG-IISDVHLANFS 384
L+ L LS NQL+G + S+ +LS L +L L+ NN+ G I S + +F+
Sbjct: 348 ---MEMLESLDLSYNQLSGEIPISLGRLSFLQILELSNNNLSGKIPSSTQMQSFN 399
Score = 93.2 bits (230), Expect = 3e-17
Identities = 101/415 (24%), Positives = 180/415 (43%), Gaps = 44/415 (10%)
Query: 121 MQSTVPD-FLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSFYVNNLKWLHG 179
+ T+P+ F +++L LS+ G L D + +DLS N+F +H
Sbjct: 27 ISDTMPNWFWDLCSKVEYLALSNNKIDGELPDLSTKFGVFPEIDLSHNNFRGP----IHS 82
Query: 180 L-SSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPPPEMNFDSLV 238
L +K L LS N + I + L + + P + L
Sbjct: 83 LPPKVKSLYLS-------NNSFVGSISFVCRVLKFMSIDLSDNQFSGEIPDCWHHLSRLN 135
Query: 239 TLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAILDLSKNSLNG 298
L+L+ NNF+ +P F ++L+ L L NNN G++ S++ T L ILDL +N L G
Sbjct: 136 NLNLANNNFSGKVPP-SFGYLYYLKELQLRNNNFTGELPSSLQNCTLLRILDLGRNQLTG 194
Query: 299 LIPNFF-DKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLNGSLERSIYQ 357
+P++F LV+L+ ++L N G +P +L N + L LS N+++G +
Sbjct: 195 RVPSWFGTSLVDLIIVNLRENQFHGELPLSLCH---LNDIHVLDLSQNRISGKIPHCFSN 251
Query: 358 LSNLVVLNLAV-------------NNMEGIISDV----------HLANFSNLKVLDLSFN 394
+ L + N ++ N+++ S++ + LK++DLS N
Sbjct: 252 FTYLSLTNSSLGTTVASKAYFVFQNDIDSYKSNILIQWKYNEREYSGRLRLLKLIDLSSN 311
Query: 395 HVTLNMSKNWVPPFQLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWD 454
+ ++ + + L ++ L+ HL + + I + +D+S +S +P
Sbjct: 312 LLGGDIPEEFSSLHGLISLNLSRNHLTGKIIREIGQMEMLESLDLSYNQLSGEIPISLGR 371
Query: 455 LSPNVEYMNLSSNELRRCGQDFSQKFKLKTLDLSNNSFSC--PLPRLPPNLRNLD 507
LS ++ + LS+N L +Q ++NS C PLP+ P N+ N D
Sbjct: 372 LS-FLQILELSNNNLSGKIPSSTQMQSFNASSYAHNSGLCGDPLPKCPRNVPNKD 425
>UniRef100_Q5ZBC0 HcrVf1 protein-like [Oryza sativa]
Length = 1128
Score = 373 bits (957), Expect = e-101
Identities = 278/845 (32%), Positives = 408/845 (47%), Gaps = 112/845 (13%)
Query: 110 LSYLNLSGNDFMQSTVPDFLSTTKNLKHLDLSHANFKGNLLDNLGNLSLLESLDLSDNSF 169
L Y N+SG T P F+ NL L L G L +G L L+ L LS+N+F
Sbjct: 367 LEYTNMSG------TFPTFIHKMSNLSVLLLFGNKLVGELPAGVGALGNLKILALSNNNF 420
Query: 170 YVNNLKWLHGLSSLKILDLSGVVLSRCQNDWFHDIRVILHSLDTLRLSGCQLHKLPTSPP 229
GL L+ + SLDTL L+ +K P
Sbjct: 421 --------RGLVPLETVS----------------------SLDTLYLNN---NKFNGFVP 447
Query: 230 PEMN-FDSLVTLDLSGNNFNMTIPDWLFENCHHLQNLNLSNNNLQGQISYSIERVTTLAI 288
E+ +L L L+ N F+ P W+ +L L+LS NNL G + I V L I
Sbjct: 448 LEVGAVSNLKKLFLAYNTFSGPAPSWI-GTLGNLTILDLSYNNLSGPVPLEIGAVN-LKI 505
Query: 289 LDLSKNSLNGLIPNFFDKLVNLVALDLSYNMLSGSIPSTLGQDHGQNSLKELRLSINQLN 348
L L+ N +G +P + +L L LSYN SG PS +G +L+ L LS N +
Sbjct: 506 LYLNNNKFSGFVPLGIGAVSHLKVLYLSYNNFSGPAPSWVG---ALGNLQILDLSHNSFS 562
Query: 349 GSLERSIYQLSNLVVLNLAVNNMEGIISDVHLANFSNLKVLDLSFNHVTLNMSKNWVPPF 408
G + I LSNL L+L+ N +G+IS H+ + S LK LDLS N + +++ N PPF
Sbjct: 563 GPVPPGIGSLSNLTTLDLSYNRFQGVISKDHVEHLSRLKYLDLSDNFLKIDIHTNSSPPF 622
Query: 409 QLETIGLANCHLGPQFPKWIQTQKNFSHIDISNAGVSDYVPNWFWDLSPNVEYMNLSSNE 468
+L +C LGP+FP W++ Q + + + N + D +P+WFW ++ S N+
Sbjct: 623 KLRNAAFRSCQLGPRFPLWLRWQTDIDVLVLENTKLDDVIPDWFWVTFSRASFLQASGNK 682
Query: 469 LRRCGQDFSQKFKLKTLDLSNNSFSCPLPRLPPNLRNLDLSSNLFYGTISHVCEILCFNN 528
L + + + L +N + +P+LP ++ L+LSSN G + + L
Sbjct: 683 LHGSLPPSLEHISVGRIYLGSNLLTGQVPQLPISMTRLNLSSNFLSGPLPSLKAPL---- 738
Query: 529 SLENLDLSFNNLSGVIPNCWTNGTNMIILNLAMNNFIGSI----------------PDSF 572
LE L L+ NN++G IP T + L+L+ N G + D F
Sbjct: 739 -LEELLLANNNITGSIPPSMCQLTGLKRLDLSGNKITGDLEQMQCWKQSDMTNTNSADKF 797
Query: 573 GSLKNLHMLIMYNNNLSGKIPETLKNCQVLTLLNLKSNRLRGPIPYWIGTDIQILMVLIL 632
GS ++ L + +N LSG P+ L+N L L+L NR G +P W+ + L +L L
Sbjct: 798 GS--SMLSLALNHNELSGIFPQFLQNASQLLFLDLSHNRFFGSLPKWLPERMPNLQILRL 855
Query: 633 GNNSFDENIPKTLCQLKSLHILDLSENQLTGAIPRCV--FLALTTEESINEKSYMEFMTI 690
+N F +IPK + L LH LD++ N ++G+IP + F A+T N + Y+
Sbjct: 856 RSNIFHGHIPKNIIYLGKLHFLDIAHNNISGSIPDSLANFKAMTVIAQ-NSEDYI----F 910
Query: 691 EESLPIYLSRTKHPLLIPWKGVNVFFNEGRLFFEILKMI---DLSSNFLTHEIPVEIGKL 747
EES+P+ + FEI + D S N LT IP EI L
Sbjct: 911 EESIPVITKDQQRDYT----------------FEIYNQVVNLDFSCNKLTGHIPEEIHLL 954
Query: 748 VELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNA 807
+ L+ LNLS NQ G+I IG+L+ L LDLS N LS EIP S++ + LS L+LSYN
Sbjct: 955 IGLTNLNLSSNQFSGTIHDQIGDLKQLESLDLSYNELSGEIPPSLSALTSLSHLNLSYNN 1014
Query: 808 LSGKIPIGNQMQSFDE--VFYKGNPHLCGPPLRKACPRNSSFEDTHCSHSEEHENDGNHG 865
LSG IP G+Q+Q+ D+ Y GNP LCGPPL K C N + + D +H
Sbjct: 1015 LSGTIPSGSQLQALDDQIYIYVGNPGLCGPPLLKNCSTNGT--------QQSFYEDRSH- 1065
Query: 866 DKVLGMEINPLYISMAMGFSTGFWVFWGSLILIASWRHAYFRFISNMNDKIHVTVVVALN 925
+ LY+ M++GF G W + ++++ +W AYFR I N+ DK +V V ++ +
Sbjct: 1066 -------MRSLYLGMSIGFVIGLWTVFCTMMMKRTWMMAYFRIIDNLYDKAYVQVAISWS 1118
Query: 926 KLRRK 930
+L RK
Sbjct: 1119 RLMRK 1123
Score = 49.3 bits (116), Expect = 5e-04
Identities = 33/77 (42%), Positives = 39/77 (49%), Gaps = 1/77 (1%)
Query: 726 LKMIDLSSNFLTH-EIPVEIGKLVELSALNLSRNQLLGSIPSSIGELESLNVLDLSRNNL 784
L+ +DLS NF IPV +G L LNLS G IPS IG + SL LD+S N
Sbjct: 123 LRHLDLSCNFFNGTSIPVFMGSFKNLRYLNLSWAGFGGKIPSQIGNISSLQYLDVSSNYF 182
Query: 785 SCEIPTSMANIDRLSWL 801
E T + LSWL
Sbjct: 183 FHEQNTFFMSSTDLSWL 199
Score = 40.0 bits (92), Expect = 0.33
Identities = 68/290 (23%), Positives = 115/290 (39%), Gaps = 36/290 (12%)
Query: 547 CWTNGTNMIILNLA-MNNFIGSIPDSFGSLKNLHMLIMYNNNLS---GKIPETLKNCQVL 602
C N++ LNL NNF D+ G + ++ +LS G++ +L L
Sbjct: 70 CSNRTGNIVALNLRNTNNFWYDFYDADG------LNLLRGGDLSLLGGELSSSLIALHHL 123
Query: 603 TLLNLKSNRLRGP-IPYWIGTDIQILMVLILGNNSFDENIPKTLCQLKSLHILDLSENQL 661
L+L N G IP ++G+ + L L L F IP + + SL LD+S N
Sbjct: 124 RHLDLSCNFFNGTSIPVFMGS-FKNLRYLNLSWAGFGGKIPSQIGNISSLQYLDVSSNYF 182
Query: 662 -------------TGAIPRCVFLALTTEESINEKSYMEFMTIEESLPIYLSRTKHPLLIP 708
+PR FL ++ S +++ + LP L +
Sbjct: 183 FHEQNTFFMSSTDLSWLPRLTFLRHVDMTDVDLSSVRDWVHMVNMLPAL-----QVLRLS 237
Query: 709 WKGVNVFFNE-GRLFFEILKMIDLSSNF-----LTHEIPVEIGKLVELSALNLSRNQLLG 762
G+N ++ L+++DLS N L H ++ L EL +
Sbjct: 238 ECGLNHTVSKLSHSNLTNLEVLDLSFNQFSYTPLRHNWFWDLTSLEELYLSEYAWFAPAE 297
Query: 763 SIPSSIGELESLNVLDLSRNNLSCEIPTSMANIDRLSWLDLSYNALSGKI 812
IP +G + +L VLDLS +++ P ++ N+ L L + N + +
Sbjct: 298 PIPDRLGNMSALRVLDLSYSSIVGLFPKTLENMCNLQVLLMDGNNIDADL 347
Score = 35.8 bits (81), Expect = 6.2
Identities = 29/75 (38%), Positives = 38/75 (50%), Gaps = 7/75 (9%)
Query: 749 ELSALNLSR----NQLLGSIPSSIGELESLNVLDLSRNNLS-CEIPTSMANIDRLSWLDL 803
+ LNL R + L G + SS+ L L LDLS N + IP M + L +L+L
Sbjct: 94 DADGLNLLRGGDLSLLGGELSSSLIALHHLRHLDLSCNFFNGTSIPVFMGSFKNLRYLNL 153
Query: 804 SYNALSGKIP--IGN 816
S+ GKIP IGN
Sbjct: 154 SWAGFGGKIPSQIGN 168
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,629,127,434
Number of Sequences: 2790947
Number of extensions: 72242139
Number of successful extensions: 315739
Number of sequences better than 10.0: 5034
Number of HSP's better than 10.0 without gapping: 2424
Number of HSP's successfully gapped in prelim test: 2668
Number of HSP's that attempted gapping in prelim test: 181459
Number of HSP's gapped (non-prelim): 39024
length of query: 938
length of database: 848,049,833
effective HSP length: 137
effective length of query: 801
effective length of database: 465,690,094
effective search space: 373017765294
effective search space used: 373017765294
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC124958.5


