

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124951.5 + phase: 0
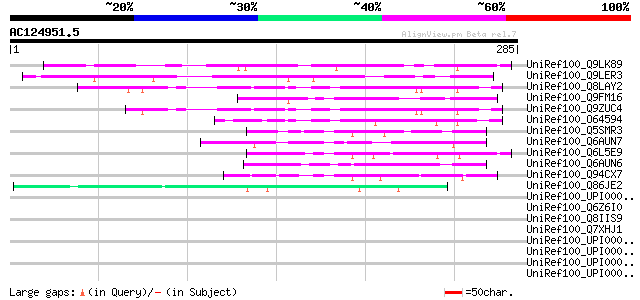
(285 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LK89 Arabidopsis thaliana genomic DNA, chromosome 3,... 113 5e-24
UniRef100_Q9LER3 Hypothetical protein T9L3_30 [Arabidopsis thali... 74 6e-12
UniRef100_Q8LAY2 Hypothetical protein [Arabidopsis thaliana] 73 1e-11
UniRef100_Q9FM16 Arabidopsis thaliana genomic DNA, chromosome 5,... 69 1e-10
UniRef100_Q9ZUC4 Hypothetical protein F5O8.26 [Arabidopsis thali... 67 4e-10
UniRef100_O64594 F17O7.4 [Arabidopsis thaliana] 65 3e-09
UniRef100_Q5SMR3 Hypothetical protein P0445H04.42 [Oryza sativa] 62 1e-08
UniRef100_Q6AUN7 Hypothetical protein OSJNBa0040E06.3 [Oryza sat... 61 4e-08
UniRef100_Q6L5E9 Hypothetical protein OJ1126_B10.13 [Oryza sativa] 61 4e-08
UniRef100_Q6AUN6 Hypothetical protein OSJNBa0040E06.4 [Oryza sat... 52 2e-05
UniRef100_Q94CX7 Hypothetical protein P0004A09.33 [Oryza sativa] 51 4e-05
UniRef100_Q86JE2 Similar to Dictyostelium discoideum (Slime mold... 47 6e-04
UniRef100_UPI000035F4C0 UPI000035F4C0 UniRef100 entry 46 0.001
UniRef100_Q6Z6I0 Hypothetical protein P0030G11.22 [Oryza sativa] 46 0.001
UniRef100_Q8IIS9 Hypothetical protein [Plasmodium falciparum] 45 0.002
UniRef100_Q7XHJ1 Pheromone receptor-like protein [Quercus robur] 44 0.005
UniRef100_UPI000036550F UPI000036550F UniRef100 entry 44 0.006
UniRef100_UPI000036CC67 UPI000036CC67 UniRef100 entry 43 0.008
UniRef100_UPI000036CC66 UPI000036CC66 UniRef100 entry 43 0.008
UniRef100_UPI0000465A1A UPI0000465A1A UniRef100 entry 43 0.008
>UniRef100_Q9LK89 Arabidopsis thaliana genomic DNA, chromosome 3, TAC clone: K16N12
[Arabidopsis thaliana]
Length = 242
Score = 113 bits (283), Expect = 5e-24
Identities = 93/280 (33%), Positives = 136/280 (48%), Gaps = 68/280 (24%)
Query: 20 PSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEF-VAFRSHRRNAADVSPIFNRDERRNS 78
PSFS YS + + ++A++V + + + +FEF A S + V P+FN
Sbjct: 12 PSFSTYSGDRLVEIAERVCYKEKS----DAEFEFSTAATSSSGDGGLVFPVFN------- 60
Query: 79 DAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEK------WTSS- 131
KNL+ GD P E + LK LF+ ++ ++SS
Sbjct: 61 ---------KNLISGDVSP-------------EKVIPLKDLFLRERNDQQPPQQTYSSSS 98
Query: 132 ---EVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT--SSSR 186
E +D+ DSIP+E YC WTP S+ SP C+KS STGSSS + S+ R
Sbjct: 99 DEEEEDDEFDSIPSEIYCPWTPARSTADMSPSGG-------CRKSKSTGSSSTSTWSTKR 151
Query: 187 WKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEV 246
W+ L+RSKSDGK+SL + P +++ +S KK+ + TVSA E
Sbjct: 152 WRLRDFLKRSKSDGKQSLKFLNPTNRDDDDESSKSK-----KKVSVS-----VTVSAHEK 201
Query: 247 FYGR----KKETRVKSYLPYKKELIGFSVGFNANVGRGFP 282
FY R K+E + KSYLPYK++L+G + G+ FP
Sbjct: 202 FYLRNKAIKEEDKRKSYLPYKQDLVGLFSNIH-RYGKTFP 240
>UniRef100_Q9LER3 Hypothetical protein T9L3_30 [Arabidopsis thaliana]
Length = 246
Score = 73.6 bits (179), Expect = 6e-12
Identities = 78/279 (27%), Positives = 124/279 (43%), Gaps = 57/279 (20%)
Query: 8 EETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHS-----QND-DFEFVAFRSHRR 61
++ SST+ PSFS Y+S ++ + A +V E+++ +S +ND +FEF
Sbjct: 5 QDLSSTVS--FSPSFSFYASGDLVETAVRVIRESESYYSVKVDGENDKEFEFETLPLGDE 62
Query: 62 NAADVSPIFNRDERRNSD--AMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKL 119
+ S + +R + D D ++ KNL G +S+
Sbjct: 63 SLFHFSTMTTTFKRSSDDDDVADDRVTSKNLFYGG-------------------WSVDPP 103
Query: 120 FIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSL---IASPKTSPMNSTIK--CKKSN 174
+ + E +SS+ +D + P++ YCFW+P S K S IK ++S+
Sbjct: 104 SLTSPSESLSSSDSDDSENISPSKFYCFWSPIRSPARDDSLKSKGSSRRCRIKEFLRRSH 163
Query: 175 STGSSSNTSSS-RWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPAT 233
S G+ S S++ R F LLRRS SDG E +++ GNG++V K
Sbjct: 164 SDGAVSTVSATKRCSFKDLLRRSHSDG----------GGEGSSISTSGNGSLVVK----G 209
Query: 234 EKKTPATVSAMEVFYGRKKETRVKSYLPYKKELIGFSVG 272
KT A Y + R KSYLPY+++LIG G
Sbjct: 210 RNKTAA--------YKPDVDKRRKSYLPYRQDLIGVFAG 240
>UniRef100_Q8LAY2 Hypothetical protein [Arabidopsis thaliana]
Length = 297
Score = 72.8 bits (177), Expect = 1e-11
Identities = 76/275 (27%), Positives = 114/275 (40%), Gaps = 78/275 (28%)
Query: 39 NENDNSHSQNDDFEFVAFRSHRRN-AAD----------VSPIFNRD---ERRNSDAMDIS 84
NE+D + ++F F AD V P+FNRD E N D + +
Sbjct: 55 NEDDEEEDEEEEFSFACVNGEGSPITADEAFEDGQIRPVFPLFNRDLLFEYENEDDKNDN 114
Query: 85 ISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLDSIPAES 144
+S+ + + +P+L +KLF+ + E ED + P
Sbjct: 115 VSVTD----ENRPRL-----------------RKLFVEDRNGNGDGEETEDS-EKEPLGP 152
Query: 145 YCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESL 204
YC WT + + ASP+T C+KSNSTG S W+F L+ RS SDG+++
Sbjct: 153 YCSWTGGTVAE-ASPET--------CRKSNSTGFSK-----LWRFRDLVLRSNSDGRDAF 198
Query: 205 NMVTPAKKENLKLNSGGNGNVVG----KKI--------------PATEKKTPATVSAMEV 246
+ + + +S + + KK+ T+KKT T SA E
Sbjct: 199 VFLNNSNDKTRTRSSSSSSSTAAEENDKKVITEKKKGKEKTSTSSETKKKTTTTKSAHEK 258
Query: 247 FYGR----KKETRVKSYLPYKKELIGFSVGFNANV 277
Y R K+E + +SYLPYK+ VGF NV
Sbjct: 259 LYMRNRAMKEEVKHRSYLPYKQ------VGFFTNV 287
>UniRef100_Q9FM16 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MQB2
[Arabidopsis thaliana]
Length = 268
Score = 68.9 bits (167), Expect = 1e-10
Identities = 51/152 (33%), Positives = 78/152 (50%), Gaps = 27/152 (17%)
Query: 129 TSSEVEDDLDSIPAESYCFWTPKSSSL----IASPKTSPMNSTIKCKKSNSTGSSSNTSS 184
+SSE E+DL +P E+YC W PK S+ + +SP +S I KS+S G S
Sbjct: 129 SSSEAEEDLTGVPPETYCVWKPKQSNSGDDDLQRLSSSPSHSKI---KSHSAG-----FS 180
Query: 185 SRWKFLSLLR-RSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSA 243
RWK +LL RS S+G + L P KK + ++ E++ P+ V
Sbjct: 181 KRWKLRNLLYVRSSSEGNDKLVFPAPVKKNDETVSD-----------QREEEEPPSKVDG 229
Query: 244 MEVFYGR-KKETRVKSYLPYKKELIGFSVGFN 274
E GR ++ET+ ++Y+PY+K++IG N
Sbjct: 230 EE--EGREREETKRQTYVPYRKDMIGILKNVN 259
>UniRef100_Q9ZUC4 Hypothetical protein F5O8.26 [Arabidopsis thaliana]
Length = 295
Score = 67.4 bits (163), Expect = 4e-10
Identities = 68/237 (28%), Positives = 102/237 (42%), Gaps = 67/237 (28%)
Query: 66 VSPIFNRD---ERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
V P+FNRD E N D + ++S+ + + +P+L +KLF+
Sbjct: 91 VFPLFNRDLLFEYENEDDKNDNVSVTD----ENRPRL-----------------RKLFVE 129
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT 182
+ E E + P YC WT + + ASP+T C+KSNSTG S
Sbjct: 130 DRNGNGDGEETEGS-EKEPLGPYCSWTGGTVAE-ASPET--------CRKSNSTGFSK-- 177
Query: 183 SSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVG----KKI-------- 230
W+F L+ RS SDG+++ + + + +S + + KK+
Sbjct: 178 ---LWRFRDLVLRSNSDGRDAFVFLNNSNDKTRTRSSSSSSSTAAEENDKKVITEKKKGK 234
Query: 231 ------PATEKKTPATVSAMEVFYGR----KKETRVKSYLPYKKELIGFSVGFNANV 277
T+KKT T SA E Y R K+E + +SYLPYK+ VGF NV
Sbjct: 235 EKTSTSSETKKKTTTTKSAHEKLYMRNRAMKEEVKHRSYLPYKQ------VGFFTNV 285
>UniRef100_O64594 F17O7.4 [Arabidopsis thaliana]
Length = 272
Score = 64.7 bits (156), Expect = 3e-09
Identities = 59/175 (33%), Positives = 84/175 (47%), Gaps = 39/175 (22%)
Query: 116 LKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNS 175
LKKLF+ E T+ E E++ D++ YC WT ++ ASP+T C+KSNS
Sbjct: 114 LKKLFV----ESTTTEEEEEESDTVGP--YCSWTNRTVEQ-ASPET--------CRKSNS 158
Query: 176 TGSSSNTSSSRWKFLSLLRRSKSDGKESL-------NMVTPAKKENLKLNSGGNGNVVGK 228
TG S W+F L+ RS SDGK++ + + +L+ + GK
Sbjct: 159 TGFSK-----LWRFRDLVLRSNSDGKDAFVFLSNGSSSTSSTSSTAARLSGVVKSSEKGK 213
Query: 229 KIPATEKKTP--ATVSAMEVFYGR----KKETRVKSYLPYKKELIGFSVGFNANV 277
+ T+KK T SA E Y R ++E + +SYLPYK VGF NV
Sbjct: 214 EKTKTDKKKDKMRTKSAHEKLYMRNRAMREEGKRRSYLPYK------HVGFFTNV 262
>UniRef100_Q5SMR3 Hypothetical protein P0445H04.42 [Oryza sativa]
Length = 304
Score = 62.4 bits (150), Expect = 1e-08
Identities = 51/146 (34%), Positives = 68/146 (45%), Gaps = 30/146 (20%)
Query: 134 EDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLS-- 191
E +L+ +P ESYC W P P T+P S +C+KS STG S RW+ +S
Sbjct: 123 EGELEGVPPESYCLWAPG-----GQPSTTPA-SPRRCRKSGSTG-----SVLRWRRISER 171
Query: 192 LLRRSKSDGKESLNMVTP---------AKKENLKLNSGGNGNVVGKKIPATEKKTPATVS 242
L+RRS SDGKE + K+N N GG+ VGK + +
Sbjct: 172 LVRRSHSDGKEKFVFLNAPGGGAPSPHPPKDNDDANGGGS---VGKGDAGRHGWSYYSKG 228
Query: 243 AMEVFYGRKKETRVKSYLPYKKELIG 268
G R +SYLPYK+EL+G
Sbjct: 229 G-----GGGSGGRRRSYLPYKQELVG 249
>UniRef100_Q6AUN7 Hypothetical protein OSJNBa0040E06.3 [Oryza sativa]
Length = 231
Score = 60.8 bits (146), Expect = 4e-08
Identities = 50/166 (30%), Positives = 72/166 (43%), Gaps = 27/166 (16%)
Query: 108 DAAEILYSLKKLFIGNEKEKWTSSEVEDD---LDSIPAESYCFWTPKSSSLIASPKTSPM 164
D A + L +L + S E D+ LD +PAE+YC W+P SP +
Sbjct: 73 DTATVRVPLGQLLLEERASAPPSGEQADEDGVLDGVPAETYCLWSP------GSPAPAVS 126
Query: 165 NSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
NS +C+KS STG S RW+ L+ RS SDGKE + L+SG +
Sbjct: 127 NSPARCQKSGSTG-----SVLRWR-QRLIGRSHSDGKEKF----------VFLSSGSDVR 170
Query: 225 VVGKKIPATEKKTPATVSAMEVFY--GRKKETRVKSYLPYKKELIG 268
G+ + + G R S+LPYK++L+G
Sbjct: 171 SKGRTTTTSRGDAGGRGGGWRYYASGGGNGGGRRPSFLPYKQDLVG 216
>UniRef100_Q6L5E9 Hypothetical protein OJ1126_B10.13 [Oryza sativa]
Length = 306
Score = 60.8 bits (146), Expect = 4e-08
Identities = 57/172 (33%), Positives = 80/172 (46%), Gaps = 34/172 (19%)
Query: 134 EDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLS-- 191
E LD P ESYC WTP + + AS SP +KS STG S +RW+ +S
Sbjct: 143 EGGLDGAPPESYCLWTPGAGAGSASASASPRPP----RKSGSTG-----SMARWRRISEL 193
Query: 192 LLRRSKSDGKES---LNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPA-------TV 241
++ RS SDGKE L + P+ KEN + K PA+ +KT TV
Sbjct: 194 VVGRSHSDGKEKFLFLPIPPPSSKEN-DVEHFKPKPKPPKPTPASGRKTAQAAAAEIDTV 252
Query: 242 SAM-EVFYGRK----------KETRVKSYLPYKKELIGFSVGFNANVGRGFP 282
+A+ + YG K T +++LPY++EL+G N + R P
Sbjct: 253 AAIHRIAYGAKGGGATGTSAGGGTPRRTFLPYREELVGLFANVN-GISRSHP 303
>UniRef100_Q6AUN6 Hypothetical protein OSJNBa0040E06.4 [Oryza sativa]
Length = 229
Score = 51.6 bits (122), Expect = 2e-05
Identities = 41/137 (29%), Positives = 64/137 (45%), Gaps = 15/137 (10%)
Query: 132 EVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLS 191
+++ DLD PAES + P +L + SP +C+KS STGSS RW+ S
Sbjct: 93 DLDLDLDGEPAESTFLYCPLCPALPVAAAASPA----RCRKSGSTGSSL----LRWRQRS 144
Query: 192 LLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFYGRK 251
+ RS SDGKE + + + + GG G VG + + G
Sbjct: 145 I-GRSHSDGKEKFVFLNASSSSSGSEHKGGRGGEVGHDGALSYYANGGSRG------GGG 197
Query: 252 KETRVKSYLPYKKELIG 268
R +S+LPY+++++G
Sbjct: 198 GGGRRRSFLPYRQDIVG 214
>UniRef100_Q94CX7 Hypothetical protein P0004A09.33 [Oryza sativa]
Length = 248
Score = 50.8 bits (120), Expect = 4e-05
Identities = 47/163 (28%), Positives = 76/163 (45%), Gaps = 25/163 (15%)
Query: 121 IGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSS 180
+G+ +S+++ + LD + +SYC W P SS +SP P +KS STG
Sbjct: 92 VGSTSSSSSSTDIAE-LDGVSPDSYCVWVP-GSSPASSPSRPP-------RKSGSTG--- 139
Query: 181 NTSSSRWKFLS--LLRRSKSDGKESLNMV----TPAKKENLKLNSGGNGNVVGKKIPATE 234
S +RW+ +S ++ RS SDGKE + +PA+ + G K+ TE
Sbjct: 140 --SIARWRRISELVVGRSHSDGKEKFRFLSAPSSPARDHPKPKPTTKGGAAAATKL-HTE 196
Query: 235 KKTPATVSAMEVFYGRKKE---TRVKSYLPYKKELIGFSVGFN 274
T A + K TR +++LPY+++L+G N
Sbjct: 197 LDTIAAGHRLSYSPNHKAHGGATR-RTFLPYRQDLMGIFANVN 238
>UniRef100_Q86JE2 Similar to Dictyostelium discoideum (Slime mold). MYOM protein
[Dictyostelium discoideum]
Length = 1144
Score = 47.0 bits (110), Expect = 6e-04
Identities = 62/263 (23%), Positives = 99/263 (37%), Gaps = 24/263 (9%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
N +ST DS + P+ S SSN +N ++N Q ++ E + ++
Sbjct: 98 NSTTPTSATSTPDSMITPTKSKGSSNKLNSKE----DKNLKKQKQKEEKERLEMEKQKQK 153
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
I + ER + K I DEK +L ++ + E L K+
Sbjct: 154 EEKEQKIKDEKERLEMEKQKQKEE-KEQKIKDEKERLEMEKQKQKEEKEKLKEEKEKSKR 212
Query: 123 NEKEKWTSSE---VEDDLDSIPAE----------SYCFWTPKSSSLIASPKTSPMNSTIK 169
+KEK E + D DS P + S F KS+SLI TSP+
Sbjct: 213 EQKEKEKEKEDITEQKDKDSTPTKVQLTREPSVISKIFSVKKSNSLIIERSTSPVQIDSS 272
Query: 170 CKKSNSTGSSSNTSSSRWKFLSLLRR---SKSDGKESLNMVTPAKKENLKL---NSGGNG 223
KS +++N SSS + R S D N T + EN + N+ NG
Sbjct: 273 PSKSFIESNNNNNSSSNNLANNTTDRNSISSEDANIIQNNTTDSNIENNETTNNNNNSNG 332
Query: 224 NVVGKKIPATEKKTPATVSAMEV 246
+ P + ++S+ ++
Sbjct: 333 SRTSTPTPINSEGIEKSISSSDL 355
>UniRef100_UPI000035F4C0 UPI000035F4C0 UniRef100 entry
Length = 665
Score = 45.8 bits (107), Expect = 0.001
Identities = 48/223 (21%), Positives = 83/223 (36%), Gaps = 3/223 (1%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
+ N +SS+ S + S SS++ N + + + +S S N + S N
Sbjct: 44 SSSSNSSSSSSSSSNSSSNSSSSSSSSSNSSSSSSNSSSSSSSSSNSSSNSSSSSSSSSN 103
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
++ S + + +S + S S + N +S ++ I S
Sbjct: 104 SSSSSSNSSSNSSSSSSSSSNSSSSSSSSSNSSSSNSNSSSNSSSSSSSISNSSSSSSSN 163
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT 182
N +S S + S + SS+ S +S NS+ S+S+ SSSN+
Sbjct: 164 NSSNSSSSISNSSSSHSSSSSSSSSSSSSSSNSSRSSSSSISNSSSSSSSSSSSSSSSNS 223
Query: 183 SSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNV 225
SSS S R S S S + + N +S + N+
Sbjct: 224 SSSS---SSSSRSSSSSSSRSSSRSSSGSNSNSSNSSSSSSNI 263
Score = 43.1 bits (100), Expect = 0.008
Identities = 49/223 (21%), Positives = 89/223 (38%), Gaps = 7/223 (3%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
N N +SS+ S S S SSN+ + + + + +S S N S +
Sbjct: 89 NSSSNSSSSSSSSSNSSSSSSNSSSNSSSSSSSSSNSSSSSSSSSNSSSS--NSNSSSNS 146
Query: 63 AADVSPIFNRDERRNSD-AMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFI 121
++ S I N +S+ + + S S+ N + +S + S +
Sbjct: 147 SSSSSSISNSSSSSSSNNSSNSSSSISNSSSSHSSSSSSSSSSSSSSSNSSRSSSSSISN 206
Query: 122 GNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSN 181
+ +SS S + S + +SSS +S +S +S SNS+ SSSN
Sbjct: 207 SSSSSSSSSSSSSSSNSSSSSSS----SSRSSSSSSSRSSSRSSSGSNSNSSNSSSSSSN 262
Query: 182 TSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
SSS + S ++S + S++ + + N+ +S N +
Sbjct: 263 ISSSNSRSSSNSSSNRSCSRSSISNSSSSSNINISSSSSSNSS 305
Score = 39.3 bits (90), Expect = 0.12
Identities = 47/234 (20%), Positives = 90/234 (38%), Gaps = 13/234 (5%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
+ N +SS+ S S S SS++ N + ++ N +S+S + S N
Sbjct: 20 SSSSNSRSSSSNSSSSSSSSSNSSSSSSNSSSSSSSSSNSSSNSSSSSSSSSNSSSSSSN 79
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
++ S + + NS + S S + + + +S+++ S
Sbjct: 80 SSSSSSS-SSNSSSNSSSSSSSSSNSSSSSSNSSSNSSSSSSSSSNSSSSSSSSSNSSSS 138
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGS---- 178
N SS + + + S + SSS I++ +S +S+ S+S+ S
Sbjct: 139 NSNSSSNSSSSSSSISNSSSSSSSNNSSNSSSSISNSSSSHSSSSSSSSSSSSSSSNSSR 198
Query: 179 --------SSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
SS++SSS S S S S + + + + + + +SG N N
Sbjct: 199 SSSSSISNSSSSSSSSSSSSSSSNSSSSSSSSSRSSSSSSSRSSSRSSSGSNSN 252
Score = 38.5 bits (88), Expect = 0.20
Identities = 51/243 (20%), Positives = 100/243 (40%), Gaps = 9/243 (3%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
+ + +SS+ +S S S SS++ + + ++ NS+S N S
Sbjct: 210 SSSSSSSSSSSSNSSSSSSSSSRSSSSSSSRSSSRSSSGSNSNSSNSSSSSSNISSSNSR 269
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
++ S NR R+S + S S N+ I + +S ++ I S + I
Sbjct: 270 SSSNSSS-NRSCSRSS--ISNSSSSSNINISSS----SSSNSSSSSSSSISSSSSNINIS 322
Query: 123 NEKEKWTSSEVEDDLDSIPAESYC--FWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSS 180
+ +SS + S + S + +SSS +S +S +S SNS+ SSS
Sbjct: 323 SSSSSNSSSSSSSSISSSSSNSSSNSSSSSRSSSSSSSRSSSRSSSGSNRNSSNSSSSSS 382
Query: 181 NTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPAT 240
N SSS + S ++S + S++ + + + +S N + ++ + +
Sbjct: 383 NISSSNSRSSSNSSSNRSCSRSSISNSSSSSSSSSISSSSSNSSSNSSSNSSSSSSSSSN 442
Query: 241 VSA 243
+S+
Sbjct: 443 ISS 445
Score = 35.4 bits (80), Expect = 1.7
Identities = 46/241 (19%), Positives = 84/241 (34%), Gaps = 7/241 (2%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
+ + SS+ S S S S+++ N + ++ N +S S N + S N
Sbjct: 65 SSSSSSSNSSSSSSNSSSSSSSSSNSSSNSSSSSSSSSNSSSSSSNSSSNSSSSSSSSSN 124
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
++ S + NS++ S S + + + +S + S
Sbjct: 125 SSSSSSSSSNSSSSNSNSSSNSSSSSSSISNSSSSSSSNNSSNSSSSISNSSS------- 177
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT 182
+ +SS S + S SSS +S +S +S S+S+ SSS++
Sbjct: 178 SHSSSSSSSSSSSSSSSNSSRSSSSSISNSSSSSSSSSSSSSSSNSSSSSSSSSRSSSSS 237
Query: 183 SSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVS 242
SS S S S S + + NS N + I + + +S
Sbjct: 238 SSRSSSRSSSGSNSNSSNSSSSSSNISSSNSRSSSNSSSNRSCSRSSISNSSSSSNINIS 297
Query: 243 A 243
+
Sbjct: 298 S 298
Score = 34.3 bits (77), Expect = 3.8
Identities = 37/180 (20%), Positives = 74/180 (40%), Gaps = 5/180 (2%)
Query: 7 NEETSSTMDSFLCPSFSVYSSN-NINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAAD 65
N +SS+ +S S S+ SS+ NIN + +N + +S S + + ++
Sbjct: 295 NISSSSSSNSSSSSSSSISSSSSNINISSSSSSNSSSSSSSSISSSSSNSSSNSSSSSRS 354
Query: 66 VSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEK 125
S +R R+S + + S + ++ R+S + S + I N
Sbjct: 355 SSSSSSRSSSRSSSGSNRNSSNSS----SSSSNISSSNSRSSSNSSSNRSCSRSSISNSS 410
Query: 126 EKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSS 185
+SS + + + S + SSS ++ + C S+S+ SSS++S++
Sbjct: 411 SSSSSSSISSSSSNSSSNSSSNSSSSSSSSSNISSSNSRTAQHSCSSSSSSSSSSSSSTA 470
Score = 34.3 bits (77), Expect = 3.8
Identities = 43/205 (20%), Positives = 81/205 (38%), Gaps = 9/205 (4%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQ----NDDFEFVAFRS 58
N + + +SS + SS+NIN + +N + +S S + + + S
Sbjct: 267 NSRSSSNSSSNRSCSRSSISNSSSSSNINISSSSSSNSSSSSSSSISSSSSNINISSSSS 326
Query: 59 HRRNAADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKK 118
+++ S I + +S++ S S + +G R +S+++ S
Sbjct: 327 SNSSSSSSSSISSSSSNSSSNSSSSSRSSSSSSSRSSSRSSSGSNRNSSNSSS---SSSN 383
Query: 119 LFIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGS 178
+ N + SS S + S + SSS+ +S S NS+ S+S+ S
Sbjct: 384 ISSSNSRSSSNSSSNRSCSRSSISNSSS--SSSSSSISSSSSNSSSNSSSNSSSSSSSSS 441
Query: 179 SSNTSSSRWKFLSLLRRSKSDGKES 203
+ ++S+SR S S S S
Sbjct: 442 NISSSNSRTAQHSCSSSSSSSSSSS 466
Score = 33.9 bits (76), Expect = 5.0
Identities = 44/199 (22%), Positives = 82/199 (41%), Gaps = 9/199 (4%)
Query: 7 NEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAADV 66
N SS+ S + S S SSN+ ++ + ++ +++S S N + + S +++
Sbjct: 252 NSSNSSSSSSNISSSNSRSSSNSSSNRSCSRSSISNSSSSSNINIS--SSSSSNSSSSSS 309
Query: 67 SPIFNRDERRN----SDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
S I + N S + S S ++ N S ++ S + G
Sbjct: 310 SSISSSSSNINISSSSSSNSSSSSSSSISSSSSNSSSNSSSSSRSSSSSSSRSSSRSSSG 369
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSS---SLIASPKTSPMNSTIKCKKSNSTGSS 179
+ + SS ++ S + S + S S I++ +S +S+I SNS+ +S
Sbjct: 370 SNRNSSNSSSSSSNISSSNSRSSSNSSSNRSCSRSSISNSSSSSSSSSISSSSSNSSSNS 429
Query: 180 SNTSSSRWKFLSLLRRSKS 198
S+ SSS S + S S
Sbjct: 430 SSNSSSSSSSSSNISSSNS 448
>UniRef100_Q6Z6I0 Hypothetical protein P0030G11.22 [Oryza sativa]
Length = 255
Score = 45.8 bits (107), Expect = 0.001
Identities = 45/155 (29%), Positives = 65/155 (41%), Gaps = 35/155 (22%)
Query: 142 AESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLL---RRSKS 198
A++YC WTP+S+ SP + KS STG S++SS RW+ L+ RS+S
Sbjct: 104 ADTYCAWTPRSAP--GSPGRD------RFPKSASTGGESSSSSRRWRLRDLVGAGGRSRS 155
Query: 199 DGKE---------------SLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSA 243
DGK+ L P ++ K S KK TE +A
Sbjct: 156 DGKDKFAFLHHHAAAPPSSKLKTPPPPQQPQQKKQSAVKTKPAAKKGVVTEMD---MATA 212
Query: 244 MEVFY-----GRKKETRVKSYLPYKKELIG-FSVG 272
+FY G + + SYL Y+ G F++G
Sbjct: 213 HRLFYSKASAGGDRRPQQASYLTYRPAFSGLFALG 247
>UniRef100_Q8IIS9 Hypothetical protein [Plasmodium falciparum]
Length = 3334
Score = 45.1 bits (105), Expect = 0.002
Identities = 58/264 (21%), Positives = 104/264 (38%), Gaps = 24/264 (9%)
Query: 25 YSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAADVSPIFNRDERRNSDAMDIS 84
Y+ NN + + N N+ S+ +N+ V+F + N + N + N++ + +
Sbjct: 2950 YNHNNNMNYHENKMN-NNLSNMRNNLLRNVSFNMNNNNNNN-----NNNNNNNNNNNNNN 3003
Query: 85 ISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLDSIPAES 144
IS N++ + N G +N D + + L + N+ S + DD+ E+
Sbjct: 3004 ISFGNMMNNNNMNLRNMGNMKNMDQNKNMKKLSLNLMKNKNNLQQSKMMNDDMKGTNMEN 3063
Query: 145 YCFWTPKSSSLI-ASPKTSPMNSTIKC-KKSNSTGSSSNTSSSRWKFLS--LLRRSKSDG 200
F PK SL + S ++ C +N+ +SS TS+S + LS LL + K +G
Sbjct: 3064 NYFDIPKRESLFNKNSSFSKEDANNACTNNNNNNNTSSGTSNSFFSNLSSRLLNKEKKEG 3123
Query: 201 ------KESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFYGR---- 250
S N+ N+ N + E+K + V +
Sbjct: 3124 GGGNIFSRSFNLKKNNNISNISNNDLDKKDKTNMNTNKNEEKDNTFTNKDNVLNTKVDNT 3183
Query: 251 ----KKETRVKSYLPYKKELIGFS 270
K T V+ Y+P E++ S
Sbjct: 3184 NEEPKDYTFVEEYIPKVNEILELS 3207
Score = 33.1 bits (74), Expect = 8.5
Identities = 43/213 (20%), Positives = 79/213 (36%), Gaps = 20/213 (9%)
Query: 25 YSSNNINDVAQQVTNENDNS-----HSQNDDFEFVAFRSHRRNAADVSPIFNRDERRNSD 79
Y +NNIN N+N+++ +S ND + +++N+ D S + N N +
Sbjct: 60 YDNNNININNNNNNNDNNDNKDELLNSSNDLINEKKNKKNKKNSYDDSILCNNRGSINDE 119
Query: 80 AMDISISL-------KNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSE 132
+DI + N + GD ++ ++ IL + + L + +
Sbjct: 120 NLDIPNNCDTSHNNNNNSIHGDNNVFEKRNNSQDVHSSSILKNSEDLILNKSSNNSNNEN 179
Query: 133 VEDDLDSIP-AESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLS 191
V + D E+ C + P M++ ++ SSSN+S S
Sbjct: 180 VYSNEDMYSYKENLCEQQNNDETNNMFPSKINMDNCNINNNYSNNNSSSNSSRS------ 233
Query: 192 LLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
SK K+ N ++N N+ N N
Sbjct: 234 -YNESKKSSKDENNNNDNDNRDNNNNNNNNNNN 265
>UniRef100_Q7XHJ1 Pheromone receptor-like protein [Quercus robur]
Length = 191
Score = 43.9 bits (102), Expect = 0.005
Identities = 40/126 (31%), Positives = 63/126 (49%), Gaps = 22/126 (17%)
Query: 158 SPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLS-LLRRSKSDG----KESLNMVTPAKK 212
S K S +NS + + SSS+ SS +W LL RS S+G K+ T K
Sbjct: 52 STKQSSLNSKAPISAATYSSSSSSKSSKKWSLKDFLLFRSASEGRATDKDPFRKYTAVYK 111
Query: 213 --ENLKL----NSGGNGNVVGKKIPATEKKTPATVSAMEVFYGRKK----ETRVKSYLPY 262
E+ K+ ++ G+G+V G K ++ P VSA E+ Y K + + K++LPY
Sbjct: 112 RHEDTKIASFRSTDGSGSVTGSK-----RRGP--VSAHELHYTTNKAASEDLKKKTFLPY 164
Query: 263 KKELIG 268
K+ ++G
Sbjct: 165 KQGILG 170
>UniRef100_UPI000036550F UPI000036550F UniRef100 entry
Length = 615
Score = 43.5 bits (101), Expect = 0.006
Identities = 48/217 (22%), Positives = 83/217 (38%), Gaps = 7/217 (3%)
Query: 10 TSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAADVSPI 69
+SS +S S S SS+N + ++ N +S S N + S+ +++ S
Sbjct: 92 SSSISNSSNSSSSSNSSSSNSSSSCSYSSSSNSSSGSSNS-----SSNSNSSSSSSSSYS 146
Query: 70 FNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKW- 128
+ +S IS S + + N NS ++ S I + +
Sbjct: 147 SSSSSYSSSSNSSISSSSSSYISNSSSGSSNSSSSSNSSSSSSYSSSSNSSISSSSSSYI 206
Query: 129 -TSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRW 187
SS S + SY + SSS +S +S NS+ ++S+ +SSN+S S
Sbjct: 207 SNSSSSSSSSSSSSSSSYSSSSNSSSSNSSSSSSSSSNSSNSSSSNSSSSNSSNSSISSS 266
Query: 188 KFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
+ S S S N + + +S G+ N
Sbjct: 267 SISNSSNSSSSSNSSSSNSSSSCSYSSSSNSSSGSSN 303
Score = 41.2 bits (95), Expect = 0.031
Identities = 51/222 (22%), Positives = 84/222 (36%), Gaps = 16/222 (7%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
N N +SS+ S S S YSS++ + ++ + NS + +++ S
Sbjct: 303 NSSSNSNSSSSSSSSYSSSSSSYSSSSNSSISSSSNISSSNSSISSSSSRYISNSS---- 358
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
S N NS + S N I +S ++ YS
Sbjct: 359 ----SGSSNSSSSSNSSSSSSYSSSSNSSISSSSSSYISNSSSSSSSS---YSSSSSSSS 411
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT 182
+ +SS ++ S SY ++SSL S +S N++ S+S+ SS+N+
Sbjct: 412 SNSSSSSSSSSNNNNKS---SSYIVPPLRNSSLCHSFSSS--NNSRSSNSSSSSSSSNNS 466
Query: 183 SSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
SSSR S R S S S N + + NS + +
Sbjct: 467 SSSRSNNSSSSRSSNSRSSNSSNSSSGCSSSSSSSNSSSSSS 508
Score = 40.0 bits (92), Expect = 0.070
Identities = 51/234 (21%), Positives = 86/234 (35%), Gaps = 12/234 (5%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
+ N SS+ S S S YSSN + + + +++S S ++ + S N
Sbjct: 25 SSSSNSSNSSSSSSKSNSSSSSYSSNRSSSNSSSSNSSSNSSSSSSNSSNSSSSNSSSSN 84
Query: 63 AAD-------VSPIFNRDERRNSDAMDISISLK-----NLLIGDEKPKLNGGGRRNSDAA 110
+++ +S N NS + + S S N G N +S ++
Sbjct: 85 SSNSSISSSSISNSSNSSSSSNSSSSNSSSSCSYSSSSNSSSGSSNSSSNSNSSSSSSSS 144
Query: 111 EILYSLKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKC 170
S N +SS + S + S SSS +S S ++S+
Sbjct: 145 YSSSSSSYSSSSNSSISSSSSSYISNSSSGSSNSSSSSNSSSSSSYSSSSNSSISSSSSS 204
Query: 171 KKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
SNS+ SSS++SSS S S S S + + + NS + +
Sbjct: 205 YISNSSSSSSSSSSSSSSSYSSSSNSSSSNSSSSSSSSSNSSNSSSSNSSSSNS 258
Score = 37.4 bits (85), Expect = 0.45
Identities = 50/222 (22%), Positives = 85/222 (37%), Gaps = 15/222 (6%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTN--ENDNSHSQNDDFEFVAFRSHR 60
N N +SS+ S S S YSS++ + ++ ++ N +S S N + S
Sbjct: 130 NSSSNSNSSSSSSSSYSSSSSSYSSSSNSSISSSSSSYISNSSSGSSNSSSSSNSSSSSS 189
Query: 61 RNAADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLF 120
+++ S I +S + IS S + + NS ++ S
Sbjct: 190 YSSSSNSSI------SSSSSSYISNSSSSSSSSSSSSSSSYSSSSNSSSSNSSSSSSSSS 243
Query: 121 IGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSS 180
+ SS SI + S SS+ +S +S NS+ C S+S+ SSS
Sbjct: 244 NSSNSSSSNSSSSNSSNSSISSSSIS----NSSNSSSSSNSSSSNSSSSCSYSSSSNSSS 299
Query: 181 NTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGN 222
+S+S S S S S + + + N ++S N
Sbjct: 300 GSSNSSSNSNS---SSSSSSSYSSSSSSYSSSSNSSISSSSN 338
Score = 37.0 bits (84), Expect = 0.59
Identities = 41/185 (22%), Positives = 71/185 (38%), Gaps = 2/185 (1%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
+ + +SST S+ S S SS++ N ++ NS S + + S N
Sbjct: 1 SNSSSSNSSSTSSSYSSKSNSSSSSSSSNSSNSSSSSSKSNSSSSSYSSNRSSSNSSSSN 60
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEK--PKLNGGGRRNSDAAEILYSLKKLF 120
++ S + + +S + S + N I N NS ++ S
Sbjct: 61 SSSNSSSSSSNSSNSSSSNSSSSNSSNSSISSSSISNSSNSSSSSNSSSSNSSSSCSYSS 120
Query: 121 IGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSS 180
N ++S + S + SY + SS S +S +S I S S+ SSS
Sbjct: 121 SSNSSSGSSNSSSNSNSSSSSSSSYSSSSSSYSSSSNSSISSSSSSYISNSSSGSSNSSS 180
Query: 181 NTSSS 185
+++SS
Sbjct: 181 SSNSS 185
Score = 36.6 bits (83), Expect = 0.77
Identities = 47/225 (20%), Positives = 89/225 (38%), Gaps = 17/225 (7%)
Query: 3 NQQQNEETSSTMDSFLCPSFS--VYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHR 60
+ N SS+ S++ S S SS++ N + + + NS + +++ S
Sbjct: 153 SSSSNSSISSSSSSYISNSSSGSSNSSSSSNSSSSSSYSSSSNSSISSSSSSYISNSSSS 212
Query: 61 RNAADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLF 120
+++ S + NS + + S S + N +S++ S+
Sbjct: 213 SSSSSSSSSSSYSSSSNSSSSNSSSSSSSSSNSSNSSSSNSSSSNSSNS-----SISSSS 267
Query: 121 IGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSP-MNSTIKCKKSNSTGSS 179
I N +SS + S + S C ++ S+S S +S NS+ S S+ SS
Sbjct: 268 ISNSSNSSSSS----NSSSSNSSSSCSYSSSSNSSSGSSNSSSNSNSSSSSSSSYSSSSS 323
Query: 180 SNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGN 224
S +SSS S S+ S + ++ + + +S G+ N
Sbjct: 324 SYSSSSNSSI-----SSSSNISSSNSSISSSSSRYISNSSSGSSN 363
Score = 33.5 bits (75), Expect = 6.5
Identities = 49/241 (20%), Positives = 95/241 (39%), Gaps = 19/241 (7%)
Query: 3 NQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN 62
N N +SS+ S S S S+++ + ++ + + NS S ++ + S +
Sbjct: 60 NSSSNSSSSSSNSSNSSSSNSSSSNSSNSSISSSSISNSSNSSSSSNSSSSNSSSSCSYS 119
Query: 63 AADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
++ S + + NS++ S S + + +S + + S +I
Sbjct: 120 SSSNSSSGSSNSSSNSNSSSSSSSSYS----------SSSSSYSSSSNSSISSSSSSYIS 169
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT 182
N SS +S + SY + +SS+ +S + NS+ S+S+ SSS +
Sbjct: 170 NSSSG--SSNSSSSSNSSSSSSYS--SSSNSSISSSSSSYISNSSSSSSSSSSSSSSSYS 225
Query: 183 SSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVS 242
SSS S S S S N + + NS N ++ I + + ++ S
Sbjct: 226 SSSN----SSSSNSSSSSSSSSNSSNSSSSNSSSSNS-SNSSISSSSISNSSNSSSSSNS 280
Query: 243 A 243
+
Sbjct: 281 S 281
>UniRef100_UPI000036CC67 UPI000036CC67 UniRef100 entry
Length = 1148
Score = 43.1 bits (100), Expect = 0.008
Identities = 57/276 (20%), Positives = 105/276 (37%), Gaps = 46/276 (16%)
Query: 3 NQQQNEETSSTMDSFLCPS-------FSVYSSNNINDVAQQVTNENDNSHSQNDDF---- 51
N ++NEE + T + L P + + ++ +++ SQN D
Sbjct: 140 NMKKNEENTKTKNKPLSPIKLTPTSVLDYFGTGSVQRSNKKMVASKRKELSQNTDESGLQ 199
Query: 52 -EFVAFRSHRRNAADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAA 110
E +A + A++ + DE + L + DE+PK R++++A
Sbjct: 200 DEAIAKQLQLDEDAELERQLHEDEE----------FARTLAMLDEEPKTKKA-RKDTEAG 248
Query: 111 EILYSLKKLFIGNEKEKWT----SSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNS 166
E S++ EK K+ +++V D+ S ++P+ S S K S +S
Sbjct: 249 ETFSSVQANLSKAEKHKYLHKVKTAQVSDERKS--------YSPRKQSKYESSKESQQHS 300
Query: 167 TIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVV 226
K S +S L++++R + + + V +KEN G
Sbjct: 301 KSSADKIGEVSSPKASSK-----LAIMKRKEESSYKEIEPVASKRKENAIKLKGET---- 351
Query: 227 GKKIPATEKKTPATVSAMEVFYGRKKETRVKSYLPY 262
K P K +PA ++ KK T ++Y Y
Sbjct: 352 --KTPKKTKSSPAKKESVSPEDSEKKRTNYQAYRSY 385
>UniRef100_UPI000036CC66 UPI000036CC66 UniRef100 entry
Length = 1147
Score = 43.1 bits (100), Expect = 0.008
Identities = 57/276 (20%), Positives = 105/276 (37%), Gaps = 46/276 (16%)
Query: 3 NQQQNEETSSTMDSFLCPS-------FSVYSSNNINDVAQQVTNENDNSHSQNDDF---- 51
N ++NEE + T + L P + + ++ +++ SQN D
Sbjct: 140 NMKKNEENTKTKNKPLSPIKLTPTSVLDYFGTGSVQRSNKKMVASKRKELSQNTDESGLQ 199
Query: 52 -EFVAFRSHRRNAADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAA 110
E +A + A++ + DE + L + DE+PK R++++A
Sbjct: 200 DEAIAKQLQLDEDAELERQLHEDEE----------FARTLAMLDEEPKTKKA-RKDTEAG 248
Query: 111 EILYSLKKLFIGNEKEKWT----SSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNS 166
E S++ EK K+ +++V D+ S ++P+ S S K S +S
Sbjct: 249 ETFSSVQANLSKAEKHKYLHKVKTAQVSDERKS--------YSPRKQSKYESSKESQQHS 300
Query: 167 TIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVV 226
K S +S L++++R + + + V +KEN G
Sbjct: 301 KSSADKIGEVSSPKASSK-----LAIMKRKEESSYKEIEPVASKRKENAIKLKGET---- 351
Query: 227 GKKIPATEKKTPATVSAMEVFYGRKKETRVKSYLPY 262
K P K +PA ++ KK T ++Y Y
Sbjct: 352 --KTPKKTKSSPAKKESVSPEDSEKKRTNYQAYRSY 385
>UniRef100_UPI0000465A1A UPI0000465A1A UniRef100 entry
Length = 1314
Score = 43.1 bits (100), Expect = 0.008
Identities = 55/231 (23%), Positives = 96/231 (40%), Gaps = 32/231 (13%)
Query: 31 NDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAADVSPIFNRDERRNSDAMDISISLKNL 90
N++ V N +NS + D + ++ SH+ ++A D R N ++S +
Sbjct: 1062 NEIENGVENGIENSMNSLGDPDVISKESHQNDSAI-------DNRTNDRYGNLSRKSRYR 1114
Query: 91 LIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFWTP 150
I ++ +N R + EI +K NEKE T S ++DDL SI E++
Sbjct: 1115 SISNKSYNINEMYYRKENQREIENKMK-----NEKENNTKSNIDDDLKSIMTENFV---- 1165
Query: 151 KSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPA 210
+ + +N IK + G+ N S KF + K+ +++LN +
Sbjct: 1166 ---------EQNIINDEIKINE----GALDNLSKISQKFRYSFLKIKNKFRQNLNASENS 1212
Query: 211 KKENLKLNSGGNGNVVGKKIPATEKKTPA---TVSAMEVFYGRKKETRVKS 258
KK K + N + K TE++ + S E YG+K + + S
Sbjct: 1213 KKGIEKDKAIKNSFLNIKHSILTEEENKSYSEEGSTYEEEYGKKNKEKKNS 1263
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 479,856,116
Number of Sequences: 2790947
Number of extensions: 20913480
Number of successful extensions: 82867
Number of sequences better than 10.0: 229
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 198
Number of HSP's that attempted gapping in prelim test: 77209
Number of HSP's gapped (non-prelim): 1397
length of query: 285
length of database: 848,049,833
effective HSP length: 126
effective length of query: 159
effective length of database: 496,390,511
effective search space: 78926091249
effective search space used: 78926091249
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC124951.5


