

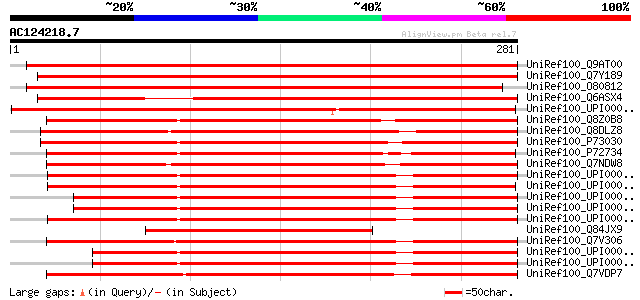
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124218.7 + phase: 0 /partial
(281 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9AT00 At1g65410/T8F5_19 [Arabidopsis thaliana] 465 e-130
UniRef100_Q7Y189 Putative ABC transporter, 5'-partial [Oryza sat... 459 e-128
UniRef100_O80812 T8F5.19 protein [Arabidopsis thaliana] 446 e-124
UniRef100_Q6ASX4 ABC transporter ATP-binding protein, putative [... 409 e-113
UniRef100_UPI00003396EA UPI00003396EA UniRef100 entry 345 8e-94
UniRef100_Q8Z0B8 ATP-binding protein of ABC transporter [Anabaen... 284 2e-75
UniRef100_Q8DLZ8 ABC transporter ATP-binding protein [Synechococ... 275 1e-72
UniRef100_P73030 ABC transporter [Synechocystis sp.] 267 2e-70
UniRef100_P72734 ABC transporter [Synechocystis sp.] 263 5e-69
UniRef100_Q7NDW8 Glr4114 protein [Gloeobacter violaceus] 259 4e-68
UniRef100_UPI0000337A0B UPI0000337A0B UniRef100 entry 229 6e-59
UniRef100_UPI00002D9610 UPI00002D9610 UniRef100 entry 229 8e-59
UniRef100_UPI00002DDDDD UPI00002DDDDD UniRef100 entry 219 8e-56
UniRef100_UPI000030F3D3 UPI000030F3D3 UniRef100 entry 217 2e-55
UniRef100_UPI00002DC4B9 UPI00002DC4B9 UniRef100 entry 217 2e-55
UniRef100_Q84JX9 Hypothetical protein At1g65410 [Arabidopsis tha... 216 4e-55
UniRef100_Q7V306 Possible ABC transporter, ATP binding component... 215 1e-54
UniRef100_UPI00002A4963 UPI00002A4963 UniRef100 entry 209 6e-53
UniRef100_UPI00002F0731 UPI00002F0731 UniRef100 entry 208 1e-52
UniRef100_Q7VDP7 ABC-type transport system involved in resistanc... 201 2e-50
>UniRef100_Q9AT00 At1g65410/T8F5_19 [Arabidopsis thaliana]
Length = 345
Score = 465 bits (1196), Expect = e-130
Identities = 234/272 (86%), Positives = 256/272 (94%)
Query: 10 KARDHEDDSDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAG 69
K R E+DSDVLIECRDVYKSFGEK IL GVSFKIRHGEAVG+IGPSGTGKSTILKI+AG
Sbjct: 73 KERGLENDSDVLIECRDVYKSFGEKHILKGVSFKIRHGEAVGVIGPSGTGKSTILKIMAG 132
Query: 70 LLAPDKGEVYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMP 129
LLAPDKGEVYIRG+KR GLISD+EISGLRIGLVFQSAALFDSL+VRENVGFLLYE S M
Sbjct: 133 LLAPDKGEVYIRGKKRAGLISDEEISGLRIGLVFQSAALFDSLSVRENVGFLLYERSKMS 192
Query: 130 EEEISELVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDE 189
E +ISELV +TLAAVGLKGVENRLPSELSGGMKKRVALARS+IFDTTK+ IEPEVLLYDE
Sbjct: 193 ENQISELVTQTLAAVGLKGVENRLPSELSGGMKKRVALARSLIFDTTKEVIEPEVLLYDE 252
Query: 190 PTAGLDPIASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKG 249
PTAGLDPIASTVVEDLIRSVH+ DA+GKPG I+SY+VVTHQHSTI+RA+DRLLFL++G
Sbjct: 253 PTAGLDPIASTVVEDLIRSVHMTDEDAVGKPGKIASYLVVTHQHSTIQRAVDRLLFLYEG 312
Query: 250 KLVWEGMTHEFTTSTNPIVQQFASGSLDGPIK 281
K+VW+GMTHEFTTSTNPIVQQFA+GSLDGPI+
Sbjct: 313 KIVWQGMTHEFTTSTNPIVQQFATGSLDGPIR 344
>UniRef100_Q7Y189 Putative ABC transporter, 5'-partial [Oryza sativa]
Length = 281
Score = 459 bits (1180), Expect = e-128
Identities = 225/266 (84%), Positives = 254/266 (94%)
Query: 16 DDSDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDK 75
DD D+LIECRDV+KSFG KK+LNG+SFKIRHGEAVGIIGPSGTGKST+LK++AGLLAPDK
Sbjct: 14 DDHDILIECRDVHKSFGNKKVLNGISFKIRHGEAVGIIGPSGTGKSTVLKVMAGLLAPDK 73
Query: 76 GEVYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISE 135
G+V I GRKR GL+SD++ISG+RIGLVFQSAALFDSLTVRENVGFLLYE+SS+PEE I+
Sbjct: 74 GDVIICGRKRHGLVSDEDISGVRIGLVFQSAALFDSLTVRENVGFLLYENSSLPEERIAT 133
Query: 136 LVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLD 195
LV ETLAAVGLKGVE+R+PSELSGGMKKRVALARSII+D TK++IEPEV+LYDEPTAGLD
Sbjct: 134 LVTETLAAVGLKGVEDRMPSELSGGMKKRVALARSIIYDDTKETIEPEVILYDEPTAGLD 193
Query: 196 PIASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEG 255
PIASTVVEDLIRSVH+ G+DALGKPG I+SYVVVTHQHSTIKRA+DRLLFLH+GK+VWEG
Sbjct: 194 PIASTVVEDLIRSVHVTGKDALGKPGKIASYVVVTHQHSTIKRAVDRLLFLHEGKVVWEG 253
Query: 256 MTHEFTTSTNPIVQQFASGSLDGPIK 281
MT EFTTSTNPIV+QFASGSLDGPI+
Sbjct: 254 MTQEFTTSTNPIVKQFASGSLDGPIR 279
>UniRef100_O80812 T8F5.19 protein [Arabidopsis thaliana]
Length = 364
Score = 446 bits (1148), Expect = e-124
Identities = 226/264 (85%), Positives = 246/264 (92%)
Query: 10 KARDHEDDSDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAG 69
K R E+DSDVLIECRDVYKSFGEK IL GVSFKIRHGEAVG+IGPSGTGKSTILKI+AG
Sbjct: 73 KERGLENDSDVLIECRDVYKSFGEKHILKGVSFKIRHGEAVGVIGPSGTGKSTILKIMAG 132
Query: 70 LLAPDKGEVYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMP 129
LLAPDKGEVYIRG+KR GLISD+EISGLRIGLVFQSAALFDSL+VRENVGFLLYE S M
Sbjct: 133 LLAPDKGEVYIRGKKRAGLISDEEISGLRIGLVFQSAALFDSLSVRENVGFLLYERSKMS 192
Query: 130 EEEISELVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDE 189
E +ISELV +TLAAVGLKGVENRLPSELSGGMKKRVALARS+IFDTTK+ IEPEVLLYDE
Sbjct: 193 ENQISELVTQTLAAVGLKGVENRLPSELSGGMKKRVALARSLIFDTTKEVIEPEVLLYDE 252
Query: 190 PTAGLDPIASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKG 249
PTAGLDPIASTVVEDLIRSVH+ DA+GKPG I+SY+VVTHQHSTI+RA+DRLLFL++G
Sbjct: 253 PTAGLDPIASTVVEDLIRSVHMTDEDAVGKPGKIASYLVVTHQHSTIQRAVDRLLFLYEG 312
Query: 250 KLVWEGMTHEFTTSTNPIVQQFAS 273
K+VW+GMTHEFTTSTNPIVQQ S
Sbjct: 313 KIVWQGMTHEFTTSTNPIVQQVPS 336
>UniRef100_Q6ASX4 ABC transporter ATP-binding protein, putative [Oryza sativa]
Length = 279
Score = 409 bits (1050), Expect = e-113
Identities = 207/266 (77%), Positives = 231/266 (86%), Gaps = 26/266 (9%)
Query: 16 DDSDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDK 75
DD D+LIECRDV+KSFG KK+LNG+SFKIRHGEAVGIIGPSGTGKST+LK++AGLLAPDK
Sbjct: 38 DDHDILIECRDVHKSFGNKKVLNGISFKIRHGEAVGIIGPSGTGKSTVLKVMAGLLAPDK 97
Query: 76 GEVYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISE 135
VFQSAALFDSLTVRENVGFLLYE+SS+PEE I+
Sbjct: 98 --------------------------VFQSAALFDSLTVRENVGFLLYENSSLPEERIAT 131
Query: 136 LVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLD 195
LV ETLAAVGLKGVE+R+PSELSGGMKKRVALARSII+D TK++IEPEV+LYDEPTAGLD
Sbjct: 132 LVTETLAAVGLKGVEDRMPSELSGGMKKRVALARSIIYDDTKETIEPEVILYDEPTAGLD 191
Query: 196 PIASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEG 255
PIASTVVEDLIRSVH+ G+DALGKPG I+SYVVVTHQHSTIKRA+DRLLFLH+GK+VWEG
Sbjct: 192 PIASTVVEDLIRSVHVTGKDALGKPGKIASYVVVTHQHSTIKRAVDRLLFLHEGKVVWEG 251
Query: 256 MTHEFTTSTNPIVQQFASGSLDGPIK 281
MT EFTTSTNPIV+QFASGSLDGPI+
Sbjct: 252 MTQEFTTSTNPIVKQFASGSLDGPIR 277
>UniRef100_UPI00003396EA UPI00003396EA UniRef100 entry
Length = 317
Score = 345 bits (885), Expect = 8e-94
Identities = 172/282 (60%), Positives = 226/282 (79%), Gaps = 4/282 (1%)
Query: 2 SSKSEQLNKARDHEDDSDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKS 61
SS + + + +D+ +VLIE + V K+FG K +L+G SFKIR GEAVGIIGPSGTGKS
Sbjct: 35 SSTANGAEASTNEDDEGEVLIELKGVVKTFGTKVVLDGASFKIRRGEAVGIIGPSGTGKS 94
Query: 62 TILKIIAGLLAPDKGEVYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFL 121
T+L+I+AGLL PD+GEVYIRG +R GL SD+ L +G+VFQSAALFDSLTV ENVGF
Sbjct: 95 TVLRIMAGLLVPDEGEVYIRGERRKGLASDEAHPKLHVGMVFQSAALFDSLTVGENVGFK 154
Query: 122 LYEHSSMPEEEISELVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTK---D 178
LYEHS++ + ++ + V+ +LAAVGL+G E++ P++LSGGMKKR ALAR+II + + D
Sbjct: 155 LYEHSTLSKRKVEKAVRNSLAAVGLQGSEDKYPAQLSGGMKKRAALARAIIKEEREHEDD 214
Query: 179 SIEPEVLLYDEPTAGLDPIASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKR 238
+IE EV++YDEPTAGLDP+ASTVVEDL+RS+H K ++ + G +SSY+VVTHQHSTI+R
Sbjct: 215 AIE-EVVMYDEPTAGLDPVASTVVEDLMRSLHCKPNESCEEHGGVSSYIVVTHQHSTIRR 273
Query: 239 AIDRLLFLHKGKLVWEGMTHEFTTSTNPIVQQFASGSLDGPI 280
++DRL+FLH+GK+ WEG EF T+T PIV+QFA+GSL GPI
Sbjct: 274 SVDRLVFLHQGKVAWEGTVEEFDTTTEPIVRQFATGSLKGPI 315
>UniRef100_Q8Z0B8 ATP-binding protein of ABC transporter [Anabaena sp.]
Length = 260
Score = 284 bits (727), Expect = 2e-75
Identities = 147/261 (56%), Positives = 190/261 (72%), Gaps = 8/261 (3%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
LI+ + V KSFG ++L+ V I GEA+GIIGPSGTGKSTIL+IIAGL+ PD GE+Y+
Sbjct: 5 LIQLKGVSKSFGNNQVLDNVDLTIYRGEALGIIGPSGTGKSTILRIIAGLITPDAGEIYV 64
Query: 81 RGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKET 140
+G KR+GLI D + + IG+VFQ AALFDSLTV ENVGFLLY+HS + I ELV E
Sbjct: 65 QGVKREGLIEDGQ-DPVGIGMVFQQAALFDSLTVEENVGFLLYQHSQLERSRIRELVAEK 123
Query: 141 LAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAST 200
L VGL+G+ + PSELSGGM+KRV+ AR+I+ + + PEVLLYDEPTAGLDPIAST
Sbjct: 124 LEMVGLRGIGHLYPSELSGGMRKRVSFARAIMSNPDNPAEGPEVLLYDEPTAGLDPIAST 183
Query: 201 VVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEF 260
V+EDLI RD G S+Y +VTHQ STI+R DRL+FL++G++ WEG +
Sbjct: 184 VIEDLI-------RDLQCTHGVCSTYAIVTHQDSTIRRTSDRLVFLYQGQVPWEGTVSDI 236
Query: 261 TTSTNPIVQQFASGSLDGPIK 281
++ +P++ QF SGS+ GPI+
Sbjct: 237 DSTDDPLINQFVSGSVQGPIQ 257
>UniRef100_Q8DLZ8 ABC transporter ATP-binding protein [Synechococcus elongatus]
Length = 268
Score = 275 bits (702), Expect = 1e-72
Identities = 143/264 (54%), Positives = 189/264 (71%), Gaps = 10/264 (3%)
Query: 18 SDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGE 77
++ L+E R V K+FG K +L+ V I +A+ I+GPSGTGKSTIL+IIAGLLAPD+GE
Sbjct: 5 AEPLVELRGVCKAFGNKPVLSDVDLVIYPQDALVILGPSGTGKSTILRIIAGLLAPDRGE 64
Query: 78 VYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELV 137
VY+ G +R GL D + LR+G+VFQ +ALFDSLTV ENVGF LY+H+ +PE I E+V
Sbjct: 65 VYVAGERRQGL-RQDGLCRLRMGMVFQQSALFDSLTVAENVGFYLYQHTRLPEARIREIV 123
Query: 138 KETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPI 197
E LA VGL G+E+ P++LSGGM+KRV+ AR+I+ + +P +LLYDEPTAGLDPI
Sbjct: 124 SEKLAMVGLSGMEDLYPAQLSGGMRKRVSFARAIVDNPEDPDDDPYLLLYDEPTAGLDPI 183
Query: 198 ASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMT 257
ASTV+EDLIR + K A YVVVTHQ STI R DRL+ L++G++ WEG
Sbjct: 184 ASTVIEDLIRELQEKTGHA---------YVVVTHQKSTIDRTGDRLILLYQGRICWEGTQ 234
Query: 258 HEFTTSTNPIVQQFASGSLDGPIK 281
E T+ NP ++QF SG ++GP++
Sbjct: 235 REIQTTDNPYLRQFLSGDVNGPMR 258
>UniRef100_P73030 ABC transporter [Synechocystis sp.]
Length = 260
Score = 267 bits (683), Expect = 2e-70
Identities = 137/264 (51%), Positives = 186/264 (69%), Gaps = 8/264 (3%)
Query: 18 SDVLIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGE 77
++ LI+ R + KSFG+++IL+ V + GEA+ +IGPSGTGKSTIL+IIAGLL PD GE
Sbjct: 2 TEPLIQLRGISKSFGDRQILDSVDLDLYPGEALVVIGPSGTGKSTILRIIAGLLPPDSGE 61
Query: 78 VYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELV 137
V I+ + R GL+ DD +RI LVFQ +ALFDSLTV ENVGF L EHSS+ E I E V
Sbjct: 62 VLIKTQPRRGLVEDDN-ERIRIALVFQQSALFDSLTVAENVGFFLLEHSSLSREVIREQV 120
Query: 138 KETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPI 197
+ LA VGL G+E+ P++LSGGM+KRV+ AR+I+ + + PEV+LYDEPTAGLDPI
Sbjct: 121 ADKLALVGLTGIEDYYPAQLSGGMRKRVSFARAIMSNPHQPEDNPEVILYDEPTAGLDPI 180
Query: 198 ASTVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMT 257
ASTV+EDL+R + PG +Y++V+HQ STI+R DR++ L+ GK+ W+G
Sbjct: 181 ASTVIEDLVRRLQ-------KSPGGCGTYIMVSHQESTIRRTADRVVLLYDGKIQWQGPV 233
Query: 258 HEFTTSTNPIVQQFASGSLDGPIK 281
+ NP+V+QF ++GPI+
Sbjct: 234 SAIDQTDNPLVRQFFEAKVEGPIR 257
>UniRef100_P72734 ABC transporter [Synechocystis sp.]
Length = 260
Score = 263 bits (671), Expect = 5e-69
Identities = 139/260 (53%), Positives = 181/260 (69%), Gaps = 8/260 (3%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
+IE R V +SFG K IL+ V KI GEAVG+IGPSGTGKSTIL+I+AGLL PD GEV +
Sbjct: 8 IIEFRGVSQSFGRKVILDDVDLKIYPGEAVGVIGPSGTGKSTILRIVAGLLTPDSGEVIV 67
Query: 81 RGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKET 140
G +R I + E L +GLVFQ +ALFDSLTV ENVGF LY S + EI +V+E
Sbjct: 68 HGHRRQRSIEEGE-KALGVGLVFQQSALFDSLTVAENVGFTLYRDSDLRPREIRAIVEEN 126
Query: 141 LAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAST 200
L VGL G+ +R P+ELSGGM+KRV+LAR+I+ + + +LLYDEPTAGLDP+AST
Sbjct: 127 LELVGLPGIGDRFPAELSGGMRKRVSLARAIVINPEQHQQYKNILLYDEPTAGLDPVAST 186
Query: 201 VVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEF 260
+E LIR H+ +D + Y++VTHQ STI DR++FL+ GK+ W+G T +
Sbjct: 187 RIESLIR--HLLSQDHV-----CCCYLIVTHQFSTIDNTTDRIIFLYDGKIQWDGSTADA 239
Query: 261 TTSTNPIVQQFASGSLDGPI 280
S +P+++QF SGS+DGPI
Sbjct: 240 YKSEHPLLKQFFSGSIDGPI 259
>UniRef100_Q7NDW8 Glr4114 protein [Gloeobacter violaceus]
Length = 262
Score = 259 bits (663), Expect = 4e-68
Identities = 138/262 (52%), Positives = 183/262 (69%), Gaps = 11/262 (4%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
+IE R V KSFG ++IL+G+ K+ +GEA+ I+GPSGTGKSTIL+++ GLL PD GEV +
Sbjct: 11 IIEFRGVSKSFGTQRILDGLDLKVHYGEALVIVGPSGTGKSTILRLMCGLLEPDSGEVLL 70
Query: 81 RGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKET 140
G + +++G+VFQ AALFDSLTV ENVGFLLYEHS +P +I ELV+E
Sbjct: 71 GGLPLQA--EQLQRKPIQVGMVFQQAALFDSLTVEENVGFLLYEHSKLPRTKIGELVREK 128
Query: 141 LAAVGLKGVENR-LPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAS 199
L VGL V R +P+ELSGG +KRV+ AR+I+ D T E ++LLYDEPTAGLDPIAS
Sbjct: 129 LEMVGLDPVLARQMPAELSGGQRKRVSFARAIMEDPTVPDDETQLLLYDEPTAGLDPIAS 188
Query: 200 TVVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHE 259
TV+E+LIRS L + G+ SYVV+THQ +TI+ DR+L +++GK+ WEG E
Sbjct: 189 TVIENLIRS--------LKQRGSCDSYVVITHQETTIRSTADRILLIYQGKVRWEGQAPE 240
Query: 260 FTTSTNPIVQQFASGSLDGPIK 281
+P V+QF G +DGPI+
Sbjct: 241 IDACQDPYVRQFFDGKIDGPIQ 262
>UniRef100_UPI0000337A0B UPI0000337A0B UniRef100 entry
Length = 261
Score = 229 bits (584), Expect = 6e-59
Identities = 124/260 (47%), Positives = 176/260 (67%), Gaps = 10/260 (3%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
+E +++ S+GE +LN ++F++ GE + I+GPSG+GKSTILKI+AGL+ P KGE+ I
Sbjct: 8 VETKNLSISWGEVNVLNQINFELNDGEKLAIVGPSGSGKSTILKILAGLILPTKGELRIF 67
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKETL 141
G K+ L D + + LVFQ+ AL SLT+ ENVGFLL + ++ ++ I E+V+E L
Sbjct: 68 GEKQTYLRIDQN-NPPDVRLVFQNPALLGSLTIEENVGFLLKRNKNLSKKSIHEIVRECL 126
Query: 142 AAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTV 201
A VGL +EN+LP+ELSGGM+KRV+ AR++I D T ++ +LL+DEPTAGLDPIAS+
Sbjct: 127 AEVGLFNIENKLPNELSGGMQKRVSFARALITDQTLNAKSKPLLLFDEPTAGLDPIASSR 186
Query: 202 VEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEFT 261
+EDLI + K S +VV+H STI+R D++L L+ GK W G EF
Sbjct: 187 IEDLINKTNHKAS---------GSSIVVSHVLSTIERTSDKVLMLYGGKFRWAGSIDEFK 237
Query: 262 TSTNPIVQQFASGSLDGPIK 281
ST+P V QF +G LDGP++
Sbjct: 238 KSTDPYVFQFRNGKLDGPMQ 257
>UniRef100_UPI00002D9610 UPI00002D9610 UniRef100 entry
Length = 256
Score = 229 bits (583), Expect = 8e-59
Identities = 125/259 (48%), Positives = 174/259 (66%), Gaps = 10/259 (3%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
+E +++ +GE +LN ++F++ HGE + I+GPSG+GKSTILKI+AGL+ P KGE+ I
Sbjct: 8 VETKNLSIGWGEVNVLNQINFELNHGEKLAIVGPSGSGKSTILKILAGLILPTKGELRIC 67
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKETL 141
G K+ L D + + LVFQ+ AL SLT+ ENVGFLL + ++ ++ I E+V+E L
Sbjct: 68 GEKQTYLRLDQN-NPPDVRLVFQNPALLGSLTIEENVGFLLKRNKNLSKKLIHEIVRECL 126
Query: 142 AAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTV 201
A VGL VEN+LP+ELSGGM+KRV+ AR++I D T +S +LL+DEPTAGLDPIAS+
Sbjct: 127 AEVGLFNVENKLPNELSGGMQKRVSFARALITDQTLNSKSKPLLLFDEPTAGLDPIASSR 186
Query: 202 VEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEFT 261
+EDLI + K S +VV+H STI+R D++L L+ GK W G EF
Sbjct: 187 IEDLINKTNDKAS---------GSSIVVSHVLSTIERTSDKVLMLYGGKFRWAGSIDEFK 237
Query: 262 TSTNPIVQQFASGSLDGPI 280
S +P V QF +G LDGP+
Sbjct: 238 KSKDPYVFQFRNGKLDGPM 256
>UniRef100_UPI00002DDDDD UPI00002DDDDD UniRef100 entry
Length = 241
Score = 219 bits (557), Expect = 8e-56
Identities = 120/246 (48%), Positives = 166/246 (66%), Gaps = 10/246 (4%)
Query: 36 ILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIRGRKRDGLISDDEIS 95
+LN ++F++ GE + I+GPSG+GKSTILKI+AGL+ P KGE+ I G K+ L D +
Sbjct: 2 VLNQINFELNDGEKLAIVGPSGSGKSTILKILAGLILPTKGELRIFGEKQTYLRLDQN-N 60
Query: 96 GLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKETLAAVGLKGVENRLPS 155
+ LVFQ+ AL SLT+ ENVGFLL + ++ ++ I E+V+E LA VGL VEN+LP+
Sbjct: 61 PPDVRLVFQNPALLGSLTIEENVGFLLKRNKNLSKKLIHEIVRECLAEVGLFNVENKLPN 120
Query: 156 ELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTVVEDLIRSVHIKGRD 215
ELSGGM+KRV+ AR++I D T ++ +LL+DEPTAGLDPIAS+ +EDLI + K
Sbjct: 121 ELSGGMQKRVSFARALITDQTLNAKSKPLLLFDEPTAGLDPIASSRIEDLINKTNDKAN- 179
Query: 216 ALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEFTTSTNPIVQQFASGS 275
S +VV+H STI+R D++L L+ GK W G EF S +P V QF +G
Sbjct: 180 --------GSSIVVSHVLSTIERTSDKVLMLYGGKFRWAGAIDEFKKSKDPYVFQFRNGK 231
Query: 276 LDGPIK 281
LDGP++
Sbjct: 232 LDGPMQ 237
>UniRef100_UPI000030F3D3 UPI000030F3D3 UniRef100 entry
Length = 240
Score = 217 bits (553), Expect = 2e-55
Identities = 120/246 (48%), Positives = 165/246 (66%), Gaps = 10/246 (4%)
Query: 36 ILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIRGRKRDGLISDDEIS 95
+LN ++F++ GE + I+GPSG+GKSTILKI+AGL+ P KGE+ I G K+ L D +
Sbjct: 1 VLNQINFELNDGEKLAIVGPSGSGKSTILKILAGLILPTKGELRIFGEKQTYLRLDQN-N 59
Query: 96 GLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKETLAAVGLKGVENRLPS 155
+ LVFQ+ AL SLT+ ENVGFLL + ++ ++ I E+V E LA VGL VEN+LP+
Sbjct: 60 PPDVRLVFQNPALLGSLTIEENVGFLLKRNKNLSKKLIHEIVHECLAEVGLFNVENKLPN 119
Query: 156 ELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTVVEDLIRSVHIKGRD 215
ELSGGM+KRV+ AR++I D T ++ +LL+DEPTAGLDPIAS+ +EDLI + K
Sbjct: 120 ELSGGMQKRVSFARALISDQTLNTKSKPLLLFDEPTAGLDPIASSRIEDLINKTNDKAS- 178
Query: 216 ALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEFTTSTNPIVQQFASGS 275
S +VV+H STI+R D++L L+ GK W G EF S +P V QF +G
Sbjct: 179 --------GSSIVVSHVLSTIERTSDKVLMLYGGKFRWAGSIDEFKKSKDPYVFQFRNGK 230
Query: 276 LDGPIK 281
LDGP++
Sbjct: 231 LDGPMQ 236
>UniRef100_UPI00002DC4B9 UPI00002DC4B9 UniRef100 entry
Length = 261
Score = 217 bits (553), Expect = 2e-55
Identities = 123/260 (47%), Positives = 171/260 (65%), Gaps = 10/260 (3%)
Query: 22 IECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIR 81
+E +++ S+G +LN ++ ++ GE + I+GPSG+GKSTILKIIAGL+ P KGE+ I
Sbjct: 8 VEAKNLSISWGGVNVLNQLNLELNDGEKLAIVGPSGSGKSTILKIIAGLILPTKGELRIF 67
Query: 82 GRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKETL 141
G K+ L D + + LVFQ+ AL SLT+ ENVGFLL + ++ ++ I E+V E L
Sbjct: 68 GEKQIYLRLDQN-NPPDVRLVFQNPALLGSLTIAENVGFLLTRNKNLSKKLIHEIVCECL 126
Query: 142 AAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTV 201
A VGL VEN+LP+ELSGGM+KRV+ AR++I D T ++ +LL+DEPTAGLDPIAS+
Sbjct: 127 AEVGLFNVENKLPNELSGGMQKRVSFARALITDHTLNAKTKPLLLFDEPTAGLDPIASSR 186
Query: 202 VEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEFT 261
+EDLI + K S +VV+H STI+R D++L L+ GK W G EF
Sbjct: 187 IEDLINKTNNKAN---------GSSIVVSHVLSTIERTSDKVLMLYGGKFRWAGSIDEFK 237
Query: 262 TSTNPIVQQFASGSLDGPIK 281
S +P V QF G LDGP++
Sbjct: 238 KSKDPYVFQFRHGKLDGPMQ 257
>UniRef100_Q84JX9 Hypothetical protein At1g65410 [Arabidopsis thaliana]
Length = 126
Score = 216 bits (551), Expect = 4e-55
Identities = 112/126 (88%), Positives = 119/126 (93%)
Query: 76 GEVYIRGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISE 135
GEVYIRG+KR GLISD+EISGLRIGLVFQSAALFDSL+VRENVGFLLYE S M E +ISE
Sbjct: 1 GEVYIRGKKRAGLISDEEISGLRIGLVFQSAALFDSLSVRENVGFLLYERSKMSENQISE 60
Query: 136 LVKETLAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLD 195
LV +TLAAVGLKGVENRLPSELSGGMKKRVALARS+IFDTTK+ IEPEVLLYDEPTAGLD
Sbjct: 61 LVTQTLAAVGLKGVENRLPSELSGGMKKRVALARSLIFDTTKEVIEPEVLLYDEPTAGLD 120
Query: 196 PIASTV 201
PIASTV
Sbjct: 121 PIASTV 126
>UniRef100_Q7V306 Possible ABC transporter, ATP binding component [Prochlorococcus
marinus subsp. pastoris]
Length = 272
Score = 215 bits (547), Expect = 1e-54
Identities = 119/261 (45%), Positives = 173/261 (65%), Gaps = 10/261 (3%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
++E ++ S+GE +LN ++ K+ GE + I+GPSG+GKSTILKI+AGLL P GE+ I
Sbjct: 18 VVETNNLSISWGELNVLNKINLKLYEGEKLAIVGPSGSGKSTILKILAGLLLPTAGELSI 77
Query: 81 RGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKET 140
G+K+ L D + + + LVFQ+ AL SLT+ ENVGFLL ++ ++ ++ I E+V+E
Sbjct: 78 FGQKQTYLRLD-QTNPPDVRLVFQNPALLGSLTIEENVGFLLQKNKNLSKKFIHEIVREC 136
Query: 141 LAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAST 200
L VGL VE +LP+ELSGGM+KRV+ AR++I D D +LL+DEPTAGLDPIAS+
Sbjct: 137 LEEVGLFNVEKKLPNELSGGMQKRVSFARALITDQDLDKNSHPLLLFDEPTAGLDPIASS 196
Query: 201 VVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEF 260
+EDLI + + K S +VV+H STI+R ++++ L+ GK W G EF
Sbjct: 197 RIEDLINNTNNKAN---------GSSIVVSHVLSTIERTSEKVVMLYGGKFRWAGSIDEF 247
Query: 261 TTSTNPIVQQFASGSLDGPIK 281
S +P V QF +G+L GP++
Sbjct: 248 KESNDPYVFQFRNGNLAGPMQ 268
>UniRef100_UPI00002A4963 UPI00002A4963 UniRef100 entry
Length = 231
Score = 209 bits (532), Expect = 6e-53
Identities = 117/235 (49%), Positives = 157/235 (66%), Gaps = 10/235 (4%)
Query: 47 GEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIRGRKRDGLISDDEISGLRIGLVFQSA 106
GE + I+GPSG+GKSTILKI+AGL+ P KGE+ I G K+ L D + + LVFQ+
Sbjct: 3 GEKLAIVGPSGSGKSTILKILAGLILPTKGELRIFGEKQTYLRLDQN-NPPDVRLVFQNP 61
Query: 107 ALFDSLTVRENVGFLLYEHSSMPEEEISELVKETLAAVGLKGVENRLPSELSGGMKKRVA 166
AL SLT+ ENVGFLL + ++ ++ I E+V E LA VGL VEN+LP+ELSGGM+KRV+
Sbjct: 62 ALLGSLTIEENVGFLLKRNKNLSKKSIHEIVGECLAEVGLFNVENKLPNELSGGMQKRVS 121
Query: 167 LARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTVVEDLIRSVHIKGRDALGKPGNISSY 226
AR++I D T ++ +LL+DEPTAGLDPIAS+ +EDLI + K S
Sbjct: 122 FARALITDQTLNAKSKPLLLFDEPTAGLDPIASSRIEDLINKTNHKAS---------GSS 172
Query: 227 VVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEFTTSTNPIVQQFASGSLDGPIK 281
+VV+H STI+R D++L L+ GK W G EF S +P V QF +G LDGP++
Sbjct: 173 IVVSHVLSTIERTSDKVLMLYGGKFRWAGSIDEFKKSKDPYVFQFRNGKLDGPMQ 227
>UniRef100_UPI00002F0731 UPI00002F0731 UniRef100 entry
Length = 229
Score = 208 bits (529), Expect = 1e-52
Identities = 117/235 (49%), Positives = 157/235 (66%), Gaps = 10/235 (4%)
Query: 47 GEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYIRGRKRDGLISDDEISGLRIGLVFQSA 106
GE + I+GPSG GKSTILKI+AGL+ P KGE+ I G K+ L D + + LVFQ+
Sbjct: 1 GEKLAIVGPSGPGKSTILKILAGLILPTKGELRIFGEKQTYLRLDQN-NPPDVRLVFQNP 59
Query: 107 ALFDSLTVRENVGFLLYEHSSMPEEEISELVKETLAAVGLKGVENRLPSELSGGMKKRVA 166
AL SLT+ ENVGFLL + ++ ++ I+E+V E LA VGL VE +LP+ELSGGM+KRV+
Sbjct: 60 ALLGSLTIEENVGFLLKRNKNLSKKLINEIVCECLAEVGLFNVEKKLPNELSGGMQKRVS 119
Query: 167 LARSIIFDTTKDSIEPEVLLYDEPTAGLDPIASTVVEDLIRSVHIKGRDALGKPGNISSY 226
AR++I D T ++ +LL+DEPTAGLDPIAS+ +EDLI + K R S
Sbjct: 120 FARALITDQTLNANSKPLLLFDEPTAGLDPIASSRIEDLINKTNDKAR---------GSS 170
Query: 227 VVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEFTTSTNPIVQQFASGSLDGPIK 281
+VV+H STI+R D++L L+ GK W G EF S +P V QF +G LDGP++
Sbjct: 171 IVVSHVLSTIERTSDKVLMLYGGKFRWAGSIEEFKKSKDPYVFQFRNGKLDGPMQ 225
>UniRef100_Q7VDP7 ABC-type transport system involved in resistance to organic
solvents ATPase component [Prochlorococcus marinus]
Length = 270
Score = 201 bits (511), Expect = 2e-50
Identities = 107/261 (40%), Positives = 165/261 (62%), Gaps = 10/261 (3%)
Query: 21 LIECRDVYKSFGEKKILNGVSFKIRHGEAVGIIGPSGTGKSTILKIIAGLLAPDKGEVYI 80
++E + + + +LN VS K++ GE + IIGPSG GKST+L+++AGLL P G++ +
Sbjct: 16 VVEMDSLSMKWADNSVLNKVSLKMKPGERIAIIGPSGCGKSTVLRLLAGLLLPSSGDLKL 75
Query: 81 RGRKRDGLISDDEISGLRIGLVFQSAALFDSLTVRENVGFLLYEHSSMPEEEISELVKET 140
G ++ L D + LVFQ+ AL SLTV ENVGFLL ++SS+ ++ E+V
Sbjct: 76 FGLMQNYLRLDQTFPP-DVRLVFQNPALIGSLTVEENVGFLLRKNSSITDKRAREIVISC 134
Query: 141 LAAVGLKGVENRLPSELSGGMKKRVALARSIIFDTTKDSIEPEVLLYDEPTAGLDPIAST 200
L VGL + ++ P +LSGGM+KRV+ AR++I D K+S +LL+DEPTAGLDP+A T
Sbjct: 135 LEEVGLYNIADKFPGQLSGGMQKRVSFARALINDPNKESDSMPLLLFDEPTAGLDPVACT 194
Query: 201 VVEDLIRSVHIKGRDALGKPGNISSYVVVTHQHSTIKRAIDRLLFLHKGKLVWEGMTHEF 260
+EDLI + +VV+H STI+R+ +R++ L+ G+ W+G +F
Sbjct: 195 RIEDLIVKTTTLA---------MGCSIVVSHVMSTIERSAERVVMLYGGEFQWDGSIDDF 245
Query: 261 TTSTNPIVQQFASGSLDGPIK 281
+ NP ++QF +GSL+GP++
Sbjct: 246 KNTQNPFIKQFRTGSLEGPMQ 266
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.137 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 459,811,422
Number of Sequences: 2790947
Number of extensions: 19177323
Number of successful extensions: 132371
Number of sequences better than 10.0: 26586
Number of HSP's better than 10.0 without gapping: 22928
Number of HSP's successfully gapped in prelim test: 3658
Number of HSP's that attempted gapping in prelim test: 63355
Number of HSP's gapped (non-prelim): 31868
length of query: 281
length of database: 848,049,833
effective HSP length: 126
effective length of query: 155
effective length of database: 496,390,511
effective search space: 76940529205
effective search space used: 76940529205
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 74 (33.1 bits)
Medicago: description of AC124218.7


