

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.9 - phase: 0
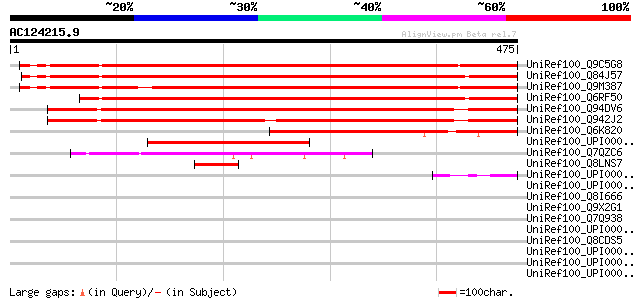
(475 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C5G8 Hypothetical protein At3g54190 [Arabidopsis tha... 730 0.0
UniRef100_Q84J57 Hypothetical protein At2g38630 [Arabidopsis tha... 708 0.0
UniRef100_Q9M387 Hypothetical protein F24B22.150 [Arabidopsis th... 699 0.0
UniRef100_Q6RF50 Hypothetical protein [Arabidopsis thaliana] 678 0.0
UniRef100_Q94DV6 Hypothetical protein P0454H12.18 [Oryza sativa] 627 e-178
UniRef100_Q942J2 B1148D12.19 protein [Oryza sativa] 603 e-171
UniRef100_Q6K820 Hypothetical protein OJ1369_G08.13-1 [Oryza sat... 318 2e-85
UniRef100_UPI00002C2F53 UPI00002C2F53 UniRef100 entry 211 3e-53
UniRef100_Q7QZC6 GLP_43_57301_56045 [Giardia lamblia ATCC 50803] 67 1e-09
UniRef100_Q8LNS7 Hypothetical protein OSJNBa0042E19.8 [Oryza sat... 60 2e-07
UniRef100_UPI000026C8F0 UPI000026C8F0 UniRef100 entry 49 3e-04
UniRef100_UPI000046BDB4 UPI000046BDB4 UniRef100 entry 43 0.023
UniRef100_Q8I666 Hypothetical protein PFB0177c [Plasmodium falci... 40 0.11
UniRef100_Q9X2G1 Beta transducin-related protein [Thermotoga mar... 40 0.19
UniRef100_Q7Q938 ENSANGP00000001275 [Anopheles gambiae str. PEST] 40 0.19
UniRef100_UPI000046D271 UPI000046D271 UniRef100 entry 39 0.43
UniRef100_Q8CDS5 Mus musculus adult male testis cDNA, RIKEN full... 38 0.73
UniRef100_UPI000031AA0C UPI000031AA0C UniRef100 entry 37 0.95
UniRef100_UPI00002D467B UPI00002D467B UniRef100 entry 37 0.95
UniRef100_UPI00002D1DA4 UPI00002D1DA4 UniRef100 entry 37 0.95
>UniRef100_Q9C5G8 Hypothetical protein At3g54190 [Arabidopsis thaliana]
Length = 467
Score = 730 bits (1884), Expect = 0.0
Identities = 368/466 (78%), Positives = 412/466 (87%), Gaps = 12/466 (2%)
Query: 10 SCRRIVATKGESSNNGSKPNIKINNKNIIKKLQTREISPKPHRSFVAATSPHRFQNMRLT 69
S RRIVA K S+P+ +N+ +KKLQ REIS + R+F +T+ RF+NMRL
Sbjct: 14 SGRRIVAKKR------SRPDGFVNS---VKKLQRREISSRKDRAFSISTAQERFRNMRLV 64
Query: 70 HQFDTHDPKHHSSPSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERICFL 129
Q+DTHDPK H + LPFLMKRTKV+EIVAA++IVFALAHSG+CAAFSRE+N+RICFL
Sbjct: 65 EQYDTHDPKGHCLVA--LPFLMKRTKVIEIVAARDIVFALAHSGVCAAFSRESNKRICFL 122
Query: 130 NICPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSES 189
N+ PDEVIRSLFYNKNNDSLITVSVYAS+NFSSLKCRSTRIEYI R +PDAGF LF+SES
Sbjct: 123 NVSPDEVIRSLFYNKNNDSLITVSVYASDNFSSLKCRSTRIEYILRGQPDAGFALFESES 182
Query: 190 LKWPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFN 249
LKWPGFVEFDDVN KVLTYSAQDS+YKVFDLKNYT+LYSISD+NVQEIKISPGIMLLIF
Sbjct: 183 LKWPGFVEFDDVNGKVLTYSAQDSVYKVFDLKNYTMLYSISDKNVQEIKISPGIMLLIFK 242
Query: 250 RASGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRSSE 309
RA+ H+PLKI+SIEDGTVLK+FNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVR++E
Sbjct: 243 RAASHVPLKILSIEDGTVLKSFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRNAE 302
Query: 310 LMEVSRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPDCNTNNI 369
LMEVSR EFMTPSAFIFLYENQLFLTFRNR VSVWNFRGELVTSFEDHLLWHPDCNTNNI
Sbjct: 303 LMEVSRAEFMTPSAFIFLYENQLFLTFRNRNVSVWNFRGELVTSFEDHLLWHPDCNTNNI 362
Query: 370 YITSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVAKINAANCISKVDDCSSTCS 429
YITSDQDLIISYCKA++EDQW+E NAGSIN+SNILTGKC+AKI ++ K DD SS+ +
Sbjct: 363 YITSDQDLIISYCKADTEDQWIEGNAGSINISNILTGKCLAKITPSSGPPK-DDESSSSN 421
Query: 430 CKHTDSSQLRSSVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
C +S Q R++VAEALEDITALFYDE+RNEIYTGNRHG VHVWSN
Sbjct: 422 CMGKNSKQRRNAVAEALEDITALFYDEERNEIYTGNRHGLVHVWSN 467
>UniRef100_Q84J57 Hypothetical protein At2g38630 [Arabidopsis thaliana]
Length = 467
Score = 708 bits (1828), Expect = 0.0
Identities = 355/464 (76%), Positives = 408/464 (87%), Gaps = 14/464 (3%)
Query: 12 RRIVATKGESSNNGSKPNIKINNKNIIKKLQTREISPKPHRSFVAATSPHRFQNMRLTHQ 71
RR++A K S+P+ +N+ +KKLQ REIS + R+F +T+ RF+NMRL Q
Sbjct: 18 RRVIAKKR------SRPDGFVNS---VKKLQRREISSRMDRAFSISTAQERFRNMRLVEQ 68
Query: 72 FDTHDPKHHSSPSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERICFLNI 131
+DTHDPK + S LP L+KR+KV+EIVAA++IVFAL SG+CA+FSRETN+++CFLN+
Sbjct: 69 YDTHDPKGYCLVS--LPNLLKRSKVIEIVAARDIVFALTLSGVCASFSRETNKKVCFLNV 126
Query: 132 CPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSESLK 191
PDEVIRSLFYNKNNDSLITVSVYAS+N+SSLKCRSTRIEYI R + DAGFPLF+SESLK
Sbjct: 127 SPDEVIRSLFYNKNNDSLITVSVYASDNYSSLKCRSTRIEYILRGQADAGFPLFESESLK 186
Query: 192 WPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFNRA 251
WPGFVEFDDVN KVLTYSAQDS+YKVFDLKNY LLYSISD+NVQEIKISPGIMLLIF RA
Sbjct: 187 WPGFVEFDDVNGKVLTYSAQDSVYKVFDLKNYALLYSISDKNVQEIKISPGIMLLIFKRA 246
Query: 252 SGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRSSELM 311
+ H+PLKI+SIEDGT+LK+F+HLLHRNKKVDFIEQFNEKLLVKQENENLQILDVR++EL+
Sbjct: 247 ASHVPLKILSIEDGTLLKSFHHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRNAELI 306
Query: 312 EVSRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPDCNTNNIYI 371
EVSRT+FMTPSAFIFLYENQLFLTFRNR VSVWNFRGELVTSFEDHLLWHPDCNTNNIYI
Sbjct: 307 EVSRTDFMTPSAFIFLYENQLFLTFRNRNVSVWNFRGELVTSFEDHLLWHPDCNTNNIYI 366
Query: 372 TSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVAKINAANCISKVDDCSSTCSCK 431
TSDQDLIISYCKA++EDQW+E NAGSIN+SNILTGKC+AKI A N K +DCSS+
Sbjct: 367 TSDQDLIISYCKADTEDQWIEGNAGSINISNILTGKCLAKIKANNGPPKEEDCSSS---D 423
Query: 432 HTDSSQLRSSVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
+SS+ RS+VAEALEDITALFYDE+RNEIYTGNRHG +HVWSN
Sbjct: 424 LGNSSRRRSAVAEALEDITALFYDEERNEIYTGNRHGLLHVWSN 467
>UniRef100_Q9M387 Hypothetical protein F24B22.150 [Arabidopsis thaliana]
Length = 454
Score = 699 bits (1804), Expect = 0.0
Identities = 359/466 (77%), Positives = 400/466 (85%), Gaps = 25/466 (5%)
Query: 10 SCRRIVATKGESSNNGSKPNIKINNKNIIKKLQTREISPKPHRSFVAATSPHRFQNMRLT 69
S RRIVA K S+P+ +N+ +KKLQ REIS + R+F +T+ RF+NMRL
Sbjct: 14 SGRRIVAKKR------SRPDGFVNS---VKKLQRREISSRKDRAFSISTAQERFRNMRLV 64
Query: 70 HQFDTHDPKHHSSPSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERICFL 129
Q+DTHDPK H + LPFLMKRTKV+EIVAA++IVFALAHSG+CAAFSR
Sbjct: 65 EQYDTHDPKGHCLVA--LPFLMKRTKVIEIVAARDIVFALAHSGVCAAFSR--------- 113
Query: 130 NICPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSES 189
DEVIRSLFYNKNNDSLITVSVYAS+NFSSLKCRSTRIEYI R +PDAGF LF+SES
Sbjct: 114 ----DEVIRSLFYNKNNDSLITVSVYASDNFSSLKCRSTRIEYILRGQPDAGFALFESES 169
Query: 190 LKWPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFN 249
LKWPGFVEFDDVN KVLTYSAQDS+YKVFDLKNYT+LYSISD+NVQEIKISPGIMLLIF
Sbjct: 170 LKWPGFVEFDDVNGKVLTYSAQDSVYKVFDLKNYTMLYSISDKNVQEIKISPGIMLLIFK 229
Query: 250 RASGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRSSE 309
RA+ H+PLKI+SIEDGTVLK+FNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVR++E
Sbjct: 230 RAASHVPLKILSIEDGTVLKSFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRNAE 289
Query: 310 LMEVSRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPDCNTNNI 369
LMEVSR EFMTPSAFIFLYENQLFLTFRNR VSVWNFRGELVTSFEDHLLWHPDCNTNNI
Sbjct: 290 LMEVSRAEFMTPSAFIFLYENQLFLTFRNRNVSVWNFRGELVTSFEDHLLWHPDCNTNNI 349
Query: 370 YITSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVAKINAANCISKVDDCSSTCS 429
YITSDQDLIISYCKA++EDQW+E NAGSIN+SNILTGKC+AKI ++ K DD SS+ +
Sbjct: 350 YITSDQDLIISYCKADTEDQWIEGNAGSINISNILTGKCLAKITPSSGPPK-DDESSSSN 408
Query: 430 CKHTDSSQLRSSVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
C +S Q R++VAEALEDITALFYDE+RNEIYTGNRHG VHVWSN
Sbjct: 409 CMGKNSKQRRNAVAEALEDITALFYDEERNEIYTGNRHGLVHVWSN 454
>UniRef100_Q6RF50 Hypothetical protein [Arabidopsis thaliana]
Length = 405
Score = 678 bits (1750), Expect = 0.0
Identities = 334/410 (81%), Positives = 375/410 (91%), Gaps = 5/410 (1%)
Query: 66 MRLTHQFDTHDPKHHSSPSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNER 125
MRL Q+DTHDPK + S LP L+KR+KV+EIVAA++IVFAL SG+CA+FSRETN++
Sbjct: 1 MRLVEQYDTHDPKGYCLVS--LPNLLKRSKVIEIVAARDIVFALTLSGVCASFSRETNKK 58
Query: 126 ICFLNICPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDAGFPLF 185
+CFLN+ PDEVIRSLFYNKNNDSLITVSVYAS+N+SSLKCRSTRIEYI R + DAGFPLF
Sbjct: 59 VCFLNVSPDEVIRSLFYNKNNDSLITVSVYASDNYSSLKCRSTRIEYILRGQADAGFPLF 118
Query: 186 QSESLKWPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIML 245
+SESLKWPGFVEFDDVN KVLTYSAQDS+YKVFDLKNY LLYSISD+NVQEIKISPGIML
Sbjct: 119 ESESLKWPGFVEFDDVNGKVLTYSAQDSVYKVFDLKNYALLYSISDKNVQEIKISPGIML 178
Query: 246 LIFNRASGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDV 305
LIF RA+ H+PLKI+SIEDGT+LK+F+HLLHRNKKVDFIEQFNEKLLVKQENENLQILDV
Sbjct: 179 LIFKRAASHVPLKILSIEDGTLLKSFHHLLHRNKKVDFIEQFNEKLLVKQENENLQILDV 238
Query: 306 RSSELMEVSRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPDCN 365
R++EL+EVSRT+FMTPSAFIFLYENQLFLTFRNR VSVWNFRGELVTSFEDHLLWHPDCN
Sbjct: 239 RNAELIEVSRTDFMTPSAFIFLYENQLFLTFRNRNVSVWNFRGELVTSFEDHLLWHPDCN 298
Query: 366 TNNIYITSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVAKINAANCISKVDDCS 425
TNNIYITSDQDLIISYCKA++EDQW+E NAGSIN+SNILTGKC+AKI A N K +DCS
Sbjct: 299 TNNIYITSDQDLIISYCKADTEDQWIEGNAGSINISNILTGKCLAKIKANNGPPKEEDCS 358
Query: 426 STCSCKHTDSSQLRSSVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
S+ +SS+ RS+VAEALEDITALFYDE+RNEIYTGNRHG +HVWSN
Sbjct: 359 SS---DLGNSSRRRSAVAEALEDITALFYDEERNEIYTGNRHGLLHVWSN 405
>UniRef100_Q94DV6 Hypothetical protein P0454H12.18 [Oryza sativa]
Length = 454
Score = 627 bits (1617), Expect = e-178
Identities = 315/440 (71%), Positives = 366/440 (82%), Gaps = 16/440 (3%)
Query: 36 NIIKKLQTREISPKPHRSFVAATSPHRFQNMRLTHQFDTHDPKHHSSPSPFLPFLMKRTK 95
N +KLQ REIS P R+F A+T+ RF+N++L +FDTHDPK S LP+LMKR++
Sbjct: 31 NSARKLQRREISALPCRAFSASTTRERFRNIQLQEEFDTHDPKEMGS---LLPYLMKRSE 87
Query: 96 VVEIVAAKNIVFALAHSGLCAAFSRETNERICFLNICPDEVIRSLFYNKNNDSLITVSVY 155
++EIV A +I+FAL+ SG+CAAFSR +N+RICFLN PDEVIRSLFYNKNNDSLITVSVY
Sbjct: 88 IIEIVGASDIIFALSQSGVCAAFSRVSNQRICFLNGRPDEVIRSLFYNKNNDSLITVSVY 147
Query: 156 ASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSESLKWPGFVEFDDVNAKVLTYSAQDSIY 215
SENFS+L+CR+TRIEYI+R KPDAGFPLF++ESLKWPGFVEFDDVN KVLTYSAQDS Y
Sbjct: 148 GSENFSALRCRTTRIEYIRRGKPDAGFPLFETESLKWPGFVEFDDVNGKVLTYSAQDSTY 207
Query: 216 KVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFNRASGHIPLKIISIEDGTVLKAFNHLL 275
KVFDLKNYTLLY+ISD+NVQEIKISPGIMLLI++R G IPL I+SIEDG LK+F HLL
Sbjct: 208 KVFDLKNYTLLYTISDKNVQEIKISPGIMLLIYSRKKGCIPLDILSIEDGKRLKSFKHLL 267
Query: 276 HRNKKVDFIEQFNEKLLVKQENENLQILDVRSSELMEVSRTEFMTPSAFIFLYENQLFLT 335
HRNKKVDFIEQFNEKLL+KQE ENLQILDVR+ + +EVSR+EF+TPSAFIFLYE QLFLT
Sbjct: 268 HRNKKVDFIEQFNEKLLIKQEGENLQILDVRNFQSIEVSRSEFVTPSAFIFLYEMQLFLT 327
Query: 336 FRNRTVSVWNFRGELVTSFEDHLLWHPDCNTNNIYITSDQDLIISYCKAESEDQWMEANA 395
FR+R+VSVWNFRGELVTSFEDH+LWHPDCNTN+IYITS+QDLIISYCKA+ D E NA
Sbjct: 328 FRSRSVSVWNFRGELVTSFEDHMLWHPDCNTNSIYITSNQDLIISYCKADPNDPSSEENA 387
Query: 396 GSINVSNILTGKCVAKINAANCISKVDDCSSTCSCKHTDSSQLRSSVAEALEDITALFYD 455
SIN+S ILTGKC+AKI A N S+ +S+ +EAL DITAL+YD
Sbjct: 388 CSINISEILTGKCLAKIKAGN-------------LNKQRVSKFQSTPSEALGDITALYYD 434
Query: 456 EDRNEIYTGNRHGFVHVWSN 475
E+R EIYTGNR G VHVWSN
Sbjct: 435 EEREEIYTGNRLGLVHVWSN 454
>UniRef100_Q942J2 B1148D12.19 protein [Oryza sativa]
Length = 444
Score = 603 bits (1554), Expect = e-171
Identities = 307/440 (69%), Positives = 356/440 (80%), Gaps = 26/440 (5%)
Query: 36 NIIKKLQTREISPKPHRSFVAATSPHRFQNMRLTHQFDTHDPKHHSSPSPFLPFLMKRTK 95
N +KLQ REIS P R+F A+T+ RF+N++L +FDTHDPK S LP+LMKR++
Sbjct: 31 NSARKLQRREISALPCRAFSASTTRERFRNIQLQEEFDTHDPKEMGS---LLPYLMKRSE 87
Query: 96 VVEIVAAKNIVFALAHSGLCAAFSRETNERICFLNICPDEVIRSLFYNKNNDSLITVSVY 155
++EIV A +I+FAL+ SG+CAAFSR +N+RICFLN PDEVIRSLFYNKNNDSLITVSVY
Sbjct: 88 IIEIVGASDIIFALSQSGVCAAFSRVSNQRICFLNGRPDEVIRSLFYNKNNDSLITVSVY 147
Query: 156 ASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSESLKWPGFVEFDDVNAKVLTYSAQDSIY 215
SENFS+L+CR+TRIEYI+R KPDAGFPLF++ESLKWPGFVEFDDVN KVLTYSAQDS Y
Sbjct: 148 GSENFSALRCRTTRIEYIRRGKPDAGFPLFETESLKWPGFVEFDDVNGKVLTYSAQDSTY 207
Query: 216 KVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFNRASGHIPLKIISIEDGTVLKAFNHLL 275
KVFDLKNYTLLY+ISD+NVQEIKI R G IPL I+SIEDG LK+F HLL
Sbjct: 208 KVFDLKNYTLLYTISDKNVQEIKI----------RKKGCIPLDILSIEDGKRLKSFKHLL 257
Query: 276 HRNKKVDFIEQFNEKLLVKQENENLQILDVRSSELMEVSRTEFMTPSAFIFLYENQLFLT 335
HRNKKVDFIEQFNEKLL+KQE ENLQILDVR+ + +EVSR+EF+TPSAFIFLYE QLFLT
Sbjct: 258 HRNKKVDFIEQFNEKLLIKQEGENLQILDVRNFQSIEVSRSEFVTPSAFIFLYEMQLFLT 317
Query: 336 FRNRTVSVWNFRGELVTSFEDHLLWHPDCNTNNIYITSDQDLIISYCKAESEDQWMEANA 395
FR+R+VSVWNFRGELVTSFEDH+LWHPDCNTN+IYITS+QDLIISYCKA+ D E NA
Sbjct: 318 FRSRSVSVWNFRGELVTSFEDHMLWHPDCNTNSIYITSNQDLIISYCKADPNDPSSEENA 377
Query: 396 GSINVSNILTGKCVAKINAANCISKVDDCSSTCSCKHTDSSQLRSSVAEALEDITALFYD 455
SIN+S ILTGKC+AKI A N S+ +S+ +EAL DITAL+YD
Sbjct: 378 CSINISEILTGKCLAKIKAGN-------------LNKQRVSKFQSTPSEALGDITALYYD 424
Query: 456 EDRNEIYTGNRHGFVHVWSN 475
E+R EIYTGNR G VHVWSN
Sbjct: 425 EEREEIYTGNRLGLVHVWSN 444
>UniRef100_Q6K820 Hypothetical protein OJ1369_G08.13-1 [Oryza sativa]
Length = 234
Score = 318 bits (815), Expect = 2e-85
Identities = 162/240 (67%), Positives = 191/240 (79%), Gaps = 14/240 (5%)
Query: 244 MLLIFNRASGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQIL 303
ML+I+ +++ H+PLKI+SIEDGT LK F LLHR++KVDFIEQFNEKLLVKQ+ ENLQI+
Sbjct: 1 MLVIYQKSANHVPLKILSIEDGTPLKTFTQLLHRSRKVDFIEQFNEKLLVKQDKENLQII 60
Query: 304 DVRSSELMEVSRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPD 363
DVR+S L+EV++TEFMTPSAFIFLYEN LFLTF NRTV+ WNFRGELVTSF+DH LWH +
Sbjct: 61 DVRNSNLIEVNKTEFMTPSAFIFLYENNLFLTFCNRTVAAWNFRGELVTSFDDHELWHSN 120
Query: 364 CNTNNIYITSDQDLIISYCKAESE------DQWMEANAGSINVSNILTGKCVAKINAANC 417
CNTNNIYIT+DQDLIISYCKA E + + GSIN+SNI TGKCVAK
Sbjct: 121 CNTNNIYITADQDLIISYCKASKEVRDSGGCEGIAPPTGSINMSNIFTGKCVAK------ 174
Query: 418 ISKVDDCSSTCSCKHTDSSQ--LRSSVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
IS +D + K DSS+ +RS+V++ALEDITALFYDEDRNEIYTGN G VHVWSN
Sbjct: 175 ISPLDPTLTIAPRKRGDSSRSTIRSTVSDALEDITALFYDEDRNEIYTGNSKGLVHVWSN 234
>UniRef100_UPI00002C2F53 UPI00002C2F53 UniRef100 entry
Length = 167
Score = 211 bits (538), Expect = 3e-53
Identities = 97/152 (63%), Positives = 127/152 (82%)
Query: 130 NICPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSES 189
N+ PDEVIRSLF+NK ++SL+TVSVY ++NFSSL+CR+T +EYIKR + D GF +F+SES
Sbjct: 1 NLTPDEVIRSLFFNKASNSLVTVSVYRADNFSSLRCRNTPLEYIKRGQTDRGFAIFESES 60
Query: 190 LKWPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFN 249
LKWPGFVEFDDVN KVLTYS ++ Y+V+D+ N+ LY+I D +V E+KISPG+MLLI+
Sbjct: 61 LKWPGFVEFDDVNNKVLTYSVENKAYRVWDMTNFEPLYTIPDDDVTEVKISPGVMLLIYG 120
Query: 250 RASGHIPLKIISIEDGTVLKAFNHLLHRNKKV 281
R G++PLK++S+EDG+ LK FNHLL R KKV
Sbjct: 121 RQGGYVPLKLLSMEDGSTLKKFNHLLERTKKV 152
>UniRef100_Q7QZC6 GLP_43_57301_56045 [Giardia lamblia ATCC 50803]
Length = 418
Score = 66.6 bits (161), Expect = 1e-09
Identities = 68/310 (21%), Positives = 136/310 (42%), Gaps = 30/310 (9%)
Query: 58 TSPHRFQNMRLTHQFDTHDPKHHSSPSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAA 117
T+P F ++ L H F + + + L R+ V EIVA ++ V AL ++G +
Sbjct: 15 TAPRLFTSIGLKHIF--YADCFEAPADSTVGLLPARSGVKEIVAGESTVAALTYAGTTSV 72
Query: 118 FSRETNERICFLNICPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAK 177
FS E +C +N + ++ ++Y++ + S I+ SV S++ S L + + A
Sbjct: 73 FSAE-GRYLCTINPGDEFAVKCIWYSRRSRSYISASVALSDDCSCLHIHAASEQECLDAI 131
Query: 178 PDAGFPLFQSESLKWPGFVEFDDVNAKVLTY--------------SAQDSIYKVFDLKNY 223
+ PG+VE+DDV+ +L Y S ++VF
Sbjct: 132 ERPLMCTLNIGRIVRPGYVEYDDVSDVILAYEPAGEKPTGSGRAVSKVQGCFRVFCYTGT 191
Query: 224 TL--LYSISDRNVQEIKISPGIMLLIFNRASGHIPLKIISIEDGTVLKAFNHL------- 274
TL + +S NV++I++S ++ + +K+++++ + + N L
Sbjct: 192 TLDCIVKLSAENVKDIRLSGEHLIATMLGPKSILYIKVLTLQTSQEVTSANELEKRASLK 251
Query: 275 --LHRNKKVDFIEQFNEKLLVKQENENLQILDVRSSELME--VSRTEFMTPSAFIFLYEN 330
L + + F+E L ++QE L +++ S E+ +S T P++ + + +
Sbjct: 252 LRLPHSSPIQFMEVTGSYLYIQQEGAPLLAINLGSREIYRYLMSTTVKDLPTSQLAVTTS 311
Query: 331 QLFLTFRNRT 340
+L F + T
Sbjct: 312 GDYLAFSSGT 321
>UniRef100_Q8LNS7 Hypothetical protein OSJNBa0042E19.8 [Oryza sativa]
Length = 417
Score = 59.7 bits (143), Expect = 2e-07
Identities = 28/41 (68%), Positives = 33/41 (80%)
Query: 174 KRAKPDAGFPLFQSESLKWPGFVEFDDVNAKVLTYSAQDSI 214
+RAK +A P F+SESL WPG +EFDDVN KV TYSAQDS+
Sbjct: 110 RRAKLNACSPHFESESLNWPGLMEFDDVNRKVPTYSAQDSV 150
>UniRef100_UPI000026C8F0 UPI000026C8F0 UniRef100 entry
Length = 52
Score = 48.9 bits (115), Expect = 3e-04
Identities = 29/79 (36%), Positives = 42/79 (52%), Gaps = 27/79 (34%)
Query: 397 SINVSNILTGKCVAKINAANCISKVDDCSSTCSCKHTDSSQLRSSVAEALEDITALFYDE 456
SI+VS+IL+G+C+AKI+ K D + L TAL+Y+E
Sbjct: 1 SIHVSHILSGRCLAKID----------------WKSADPNNL-----------TALYYNE 33
Query: 457 DRNEIYTGNRHGFVHVWSN 475
+R +I TGN G +HVW+N
Sbjct: 34 ERGDIITGNTQGVLHVWTN 52
>UniRef100_UPI000046BDB4 UPI000046BDB4 UniRef100 entry
Length = 164
Score = 42.7 bits (99), Expect = 0.023
Identities = 28/96 (29%), Positives = 50/96 (51%), Gaps = 3/96 (3%)
Query: 85 PFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERICFLNICPDEVIRSLFYNK 144
PF +RTK+ EI+ AK+++ L SG+ A+ + +C +N V+ ++ YN
Sbjct: 46 PFNQLNNERTKIKEIILAKDVIVVLLISGISRAYDIISGLFLCEINPNTFSVVHTIVYNS 105
Query: 145 NNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDA 180
N++LI YAS + L+C+ +K K ++
Sbjct: 106 YNNTLIV--AYASFP-AHLQCKIINYNDLKVGKTNS 138
>UniRef100_Q8I666 Hypothetical protein PFB0177c [Plasmodium falciparum]
Length = 147
Score = 40.4 bits (93), Expect = 0.11
Identities = 22/65 (33%), Positives = 37/65 (56%), Gaps = 2/65 (3%)
Query: 93 RTKVVEIVAAKNIVFALAHSGLCAAFSRETNERICFLNICPDEVIRSLFYNKNNDSLITV 152
RTK+ EI+ A +++ L SG+ A++ E +C +N V+ ++ YN N++LI
Sbjct: 54 RTKIKEIILANDVLVVLLISGISRAYNMINGEFLCEINPNNFSVVHTIVYNSYNNTLII- 112
Query: 153 SVYAS 157
YAS
Sbjct: 113 -AYAS 116
>UniRef100_Q9X2G1 Beta transducin-related protein [Thermotoga maritima]
Length = 580
Score = 39.7 bits (91), Expect = 0.19
Identities = 41/197 (20%), Positives = 92/197 (45%), Gaps = 21/197 (10%)
Query: 194 GFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQ-EIKISP-GIMLLIFNR- 250
G + + V+ +A ++ Y V L+ +S+++ +K P GI ++F+
Sbjct: 137 GEISSQKLGPSVVQIAAHENRYVVGLANGLALIRDLSNQSFSIPLKAHPEGIKKIVFSEN 196
Query: 251 -----ASGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDV 305
G +K+ + +G ++ ++H++ N V F+E N+ L+ ++ +L++
Sbjct: 197 GQLIATCGGNTVKVWNASNGNLVLQYDHVITVND-VAFLE--NDSLIFVADDYKAVVLNI 253
Query: 306 RSSELMEV--SRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPD 363
+ +E+++ + F+ F+ + + ++TV VWN EL S H L
Sbjct: 254 QKNEVVKSIDAHNNFVL---FVSSDDGSIATYGMDKTVKVWNANLELQYSLYGHQL---- 306
Query: 364 CNTNNIYITSDQDLIIS 380
+ N + ++SD+ I+S
Sbjct: 307 -SVNTVALSSDRKFIVS 322
>UniRef100_Q7Q938 ENSANGP00000001275 [Anopheles gambiae str. PEST]
Length = 1703
Score = 39.7 bits (91), Expect = 0.19
Identities = 64/298 (21%), Positives = 125/298 (41%), Gaps = 42/298 (14%)
Query: 203 AKVLTYSAQDSIYKVFDLKNYTLLYSIS-DRNVQEIKISPGIMLLIFNRASGHIPLKIIS 261
++VL S+ D+ ++ L+N+TLL I + + ++IS L+ + A + L+ S
Sbjct: 1021 SEVLVSSSHDATVCLWSLENFTLLNCIQMSQPILNMQISCDSTFLLAHCADNGLYLR--S 1078
Query: 262 IEDGTVLKAFNHLLHRNK-KVDFIEQFNEKLLVKQENENLQILDVRSSELMEVSRTEFMT 320
+ GT L A H++K + I +++ +V E+ I D+ S ++
Sbjct: 1079 LTTGTELHALKG--HKSKVRALCIAMDSQRAIVGGEDTRALIFDMHSGRVIRSLPPNPGA 1136
Query: 321 PSAFIFLYENQLFLTFRNRTVSVWNFRGE--LVTSFEDHLLWHPDCNTNNIYITSDQDLI 378
+A + ++ +T ++ ++FR E +V + + N + + S +
Sbjct: 1137 VTAVYVMEKDDYLITAGGNKITFYSFRNEDSIVNFYPKKKKTLKKFHRNRLALQSTSSPV 1196
Query: 379 ISYCKAESEDQWMEANAGSINVSNILTGKCVA--KINAANCISKVDDCSSTCSC-KHTDS 435
I C S D M A A S +CV ++N+ S ++ S+T +C +
Sbjct: 1197 I--CFDVSRDSTMAAVASS---------RCVQIWQLNSPELTSILEGHSATVTCVSFAPN 1245
Query: 436 SQLRSSVAE---------ALEDITALF-----------YDEDRNEIYTGNRHGFVHVW 473
+L +S +E L +TA F + D I + +R G ++VW
Sbjct: 1246 CELVASGSEDKTVNVWSLGLGTVTATFKGHCTSISSVTFMMDARRIISADRDGLLYVW 1303
>UniRef100_UPI000046D271 UPI000046D271 UniRef100 entry
Length = 80
Score = 38.5 bits (88), Expect = 0.43
Identities = 22/76 (28%), Positives = 41/76 (53%), Gaps = 1/76 (1%)
Query: 220 LKNYTLLYSISDRNVQEIKISPGIMLLIFNRASGHIPLKIISIEDGTVLKAFNHLLHRNK 279
+K Y ++ I++ QEI++S G++ + + IPL + IE+G L + +
Sbjct: 1 MKTYEKVFEIAEE-FQEIRVSDGLVAMFKQPINSTIPLALFDIENGERLVDTTISILPRR 59
Query: 280 KVDFIEQFNEKLLVKQ 295
++ F+E KLLVK+
Sbjct: 60 EMQFLELLVSKLLVKE 75
>UniRef100_Q8CDS5 Mus musculus adult male testis cDNA, RIKEN full-length enriched
library, clone:4930432E11 product:hypothetical Trp-Asp
repeat (WD- repeat) structure containing protein, full
insert sequence [Mus musculus]
Length = 1154
Score = 37.7 bits (86), Expect = 0.73
Identities = 21/56 (37%), Positives = 33/56 (58%), Gaps = 4/56 (7%)
Query: 331 QLFLT-FRNRTVSVWNFRGELVTSFEDHLLWHPDCNTN---NIYITSDQDLIISYC 382
+LF+T + +V VW+FRG+L+T FE L + C N ++ +T DQ + I C
Sbjct: 518 KLFVTGATDGSVRVWDFRGKLITHFESELHFSSPCFANSRGDLLLTFDQSIYIVSC 573
>UniRef100_UPI000031AA0C UPI000031AA0C UniRef100 entry
Length = 268
Score = 37.4 bits (85), Expect = 0.95
Identities = 36/150 (24%), Positives = 64/150 (42%), Gaps = 13/150 (8%)
Query: 212 DSIYKVFDLKNYTLL---YSISDRNVQEIKI----SPGIMLLIFNRASGHIPLKIISIED 264
DSIY D+K Y L YS + + I SP + + +A G + ++
Sbjct: 47 DSIYSERDIKKYKLKDIKYSKAPSKLDVDLIIDFSSPKSSISLAKKAEG-FRVPFVTGTT 105
Query: 265 GTVLKAFNHLLHRNKKVDFIEQFNEKL----LVKQENENLQILDVRSSELMEVSRTEFM- 319
G + N L +KK+ ++ +N L L+K EN+ E+ E + +
Sbjct: 106 GFNSRQINELKKISKKIPVLQSYNMSLGINLLIKIIKENIDYFSATDLEITETHHKDKID 165
Query: 320 TPSAFIFLYENQLFLTFRNRTVSVWNFRGE 349
+PS L + + + + +T + N+RGE
Sbjct: 166 SPSGTALLLADSISSSLQTKTEKIVNYRGE 195
>UniRef100_UPI00002D467B UPI00002D467B UniRef100 entry
Length = 268
Score = 37.4 bits (85), Expect = 0.95
Identities = 36/150 (24%), Positives = 64/150 (42%), Gaps = 13/150 (8%)
Query: 212 DSIYKVFDLKNYTLL---YSISDRNVQEIKI----SPGIMLLIFNRASGHIPLKIISIED 264
DSIY D+K Y L YS + + I SP + + +A G + ++
Sbjct: 47 DSIYSERDIKKYKLKDIKYSKAPSKLDVDLIIDFSSPKSSISLAKKAEG-FRVPFVTGTT 105
Query: 265 GTVLKAFNHLLHRNKKVDFIEQFNEKL----LVKQENENLQILDVRSSELMEVSRTEFM- 319
G + N L +KK+ ++ +N L L+K EN+ E+ E + +
Sbjct: 106 GFNSRQINELKKISKKIPVLQSYNMSLGINLLIKIIKENIDYFSATDLEITETHHKDKID 165
Query: 320 TPSAFIFLYENQLFLTFRNRTVSVWNFRGE 349
+PS L + + + + +T + N+RGE
Sbjct: 166 SPSGTALLLADSISSSLQTKTEKIVNYRGE 195
>UniRef100_UPI00002D1DA4 UPI00002D1DA4 UniRef100 entry
Length = 489
Score = 37.4 bits (85), Expect = 0.95
Identities = 36/157 (22%), Positives = 65/157 (40%), Gaps = 21/157 (13%)
Query: 304 DVRSSELMEVSRTEFMTPSAFIFLYENQLF-------------LTFRNRTVSVWNFRGEL 350
DV EL + R+ + PS +F Y+N+ + L FR + ++ F+ +L
Sbjct: 303 DVEEIELEYIDRSPEIDPSDILFTYDNKKWTVKDFNDLLSSHPLVFRKKKMNEAEFQTQL 362
Query: 351 VTSFEDHLLWHPDCNTNNIYITSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVA 410
+ D L D NI D D + + DQW +A A + N+ T K V+
Sbjct: 363 KFAIADLLR---DMEITNICYNEDLD--DDWRVIANVDQWYDAYASKRYIENMDTIKVVS 417
Query: 411 KINAANCISKVDDCSSTCSCKHTDSSQLRSSVAEALE 447
+ + ++D + ++ S ++ EA+E
Sbjct: 418 E---KELLDRLDPVVDSLQAVYSQSIKINIEAFEAIE 451
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 783,868,825
Number of Sequences: 2790947
Number of extensions: 32280356
Number of successful extensions: 111393
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 111348
Number of HSP's gapped (non-prelim): 64
length of query: 475
length of database: 848,049,833
effective HSP length: 131
effective length of query: 344
effective length of database: 482,435,776
effective search space: 165957906944
effective search space used: 165957906944
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC124215.9


