

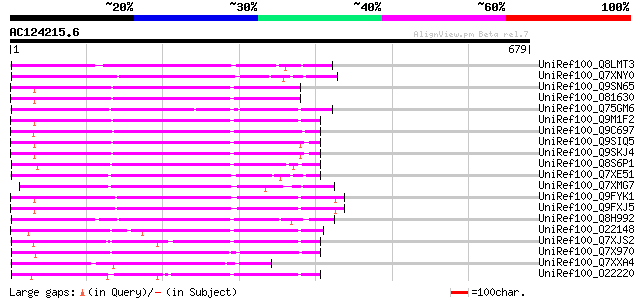
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.6 - phase: 0 /pseudo
(679 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LMT3 Putative retroelement [Oryza sativa] 258 3e-67
UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa] 255 3e-66
UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis tha... 249 2e-64
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 249 2e-64
UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcrip... 245 4e-63
UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis tha... 239 2e-61
UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [A... 234 5e-60
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 233 2e-59
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 232 2e-59
UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa] 228 6e-58
UniRef100_Q7XE51 Putative non-LTR retroelement reverse transcrip... 227 1e-57
UniRef100_Q7XMG7 OSJNBa0028I23.15 protein [Oryza sativa] 224 5e-57
UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana] 223 1e-56
UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana] 223 1e-56
UniRef100_Q8H992 Oryza sativa (japonica cultivar-group) retrotra... 221 6e-56
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 220 9e-56
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 218 4e-55
UniRef100_Q7X970 BZIP-like protein [Oryza sativa] 216 1e-54
UniRef100_Q7XXA4 OSJNBa0019G23.12 protein [Oryza sativa] 213 1e-53
UniRef100_O22220 Putative non-LTR retroelement reverse transcrip... 211 7e-53
>UniRef100_Q8LMT3 Putative retroelement [Oryza sativa]
Length = 764
Score = 258 bits (660), Expect = 3e-67
Identities = 146/425 (34%), Positives = 234/425 (54%), Gaps = 23/425 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q+AFI GRQ LDG+++ +EV+ + +K KK ++FK+DFEKAYD V ++L +V+ K GF
Sbjct: 84 QTAFIPGRQNLDGVVILHEVLHELKKEKKSGIIFKLDFEKAYDKVQWSFLFDVLHKKGFS 143
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
W +W+K +V +NG F +RG+RQGDPLSP LF L A+
Sbjct: 144 DRWIQWVKMATIGGKMAVNINGEVKDFFKTYRGVRQGDPLSPLLFNLVAD---------A 194
Query: 123 VNNLFNGYKVGSHEMV--GVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNF 180
++ + N K +V G+THLQ+ADDTI+ + NI ++ +L ++ +SGLK+N+
Sbjct: 195 LSEMLNNAKQAVPHLVPGGLTHLQYADDTILFMTNTEENIVTVKFLLYCYEAMSGLKINY 254
Query: 181 SKSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLS 240
KS + + A + NC AG +PF YLG+PI N N+I RL+
Sbjct: 255 QKSEIMVIGGDEMETQRVADLFNCQAGKMPFTYLGIPISMNKLTNADLDIPPNKIEKRLA 314
Query: 241 SWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSAD 300
+WK +LS GG+ +L+ S LSS+P+Y + + P G+ + ++SI FF+ G
Sbjct: 315 TWKCGYLSYGGKAILINSCLSSIPLYMMGVYLLPEGVHNKMDSIRARFFW-----EGLEK 369
Query: 301 HRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRV- 359
RK + W ++CR +E GGL ++ N+ALL KW +R+ + ++ + +RN+
Sbjct: 370 KRKYHMIKWEALCRPKEFGGLGFIDTRKMNIALLCKWIYRLESGKEDPC--CVLLRNKYM 427
Query: 360 --EGGHLCSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLRDR 417
GG S S +W+ + + + W S +G+G + FW+DV +GE L+ +
Sbjct: 428 KDGGGFFQSKAEESSQFWKGLHEV--KKWMDLGSSYKVGNGKATNFWSDVWIGETPLKTQ 485
Query: 418 FSRLY 422
+ +Y
Sbjct: 486 YPNIY 490
>UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa]
Length = 1026
Score = 255 bits (652), Expect = 3e-66
Identities = 149/430 (34%), Positives = 231/430 (53%), Gaps = 17/430 (3%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q+AF+K R I++G+++ +EV++ ++K+ +LFKVDFEKAYD V+ ++ ++ GFP
Sbjct: 348 QTAFLKNRFIMEGVVILHEVLNSIHQKKQSGILFKVDFEKAYDKVNWVFIYRMLKAKGFP 407
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
W WI K V +V VN F H+GLRQGDPLSP LF LAAE +L++
Sbjct: 408 DQWCDWIMKVVMGGKVAVKVNDQIGSFFKTHKGLRQGDPLSPLLFNLAAEALTLLVQRAE 467
Query: 123 VNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSK 182
N+L G + + LQ+ADDTI L + + + L+ IL LF++LSGLK+NF+K
Sbjct: 468 ENSLIEGLGTNGDNKIAI--LQYADDTIFLINDKLDHAKNLKYILCLFEQLSGLKINFNK 525
Query: 183 SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSW 242
S + + C GS+P YLG+PI WK N++ +L W
Sbjct: 526 SEVFCFGEAKEKQDLYSNIFTCKVGSLPLKYLGIPIDQKRILNKDWKLAENKMEHKLGCW 585
Query: 243 KSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHR 302
+ + S+GGRL+LL S LSS+P+Y +SF++ P G+ I+ F + G R
Sbjct: 586 QGRLQSIGGRLILLNSTLSSVPMYMISFYRLPKGVQERIDYFRKRFLWQEDQG-----IR 640
Query: 303 KIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR----NR 358
K V+W VC ++ GGL V ++ N A+LGKW WR L + W ++ +
Sbjct: 641 KYHLVNWPLVCSPRDQGGLGVLDLEAMNKAMLGKWIWR-LENEEGWWQEIIYAKYCSDKP 699
Query: 359 VEGGHLCSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLRDRF 418
+ G L +G S +W+ ++ + + F + ++ +G+G LFW D +G L +F
Sbjct: 700 LSGLRLKAG---SSHFWQGVMEVKDD--FFSFCTKIVGNGEKTLFWEDSWLGGKPLAIQF 754
Query: 419 SRLYDLSVLK 428
LY + + K
Sbjct: 755 PSLYGIVITK 764
>UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis thaliana]
Length = 1294
Score = 249 bits (635), Expect = 2e-64
Identities = 146/385 (37%), Positives = 216/385 (55%), Gaps = 11/385 (2%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRKK---DLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR + D +++A+E++ + RK+ + K D KAYD V+ N+LE M
Sbjct: 897 DSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMR 956
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W KWI V + SVLVNG P RG+RQGDPLSP+LF+L A+ + L
Sbjct: 957 LFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHL 1016
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
++ V G ++G + + GVTHLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 1017 IKNRVAEGDIRGIRIG-NGVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQK 1075
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N SKS+ G VHG+ +L + YLGLP + + +I R+
Sbjct: 1076 INMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVK 1135
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R SSW +K+LS G+ ++LKSV S+PVYA+S FK P IVS IE++L NF W
Sbjct: 1136 KRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNF-----WWE 1190
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+A R+I W+ W + S++ GGL R + +FN ALL K WR++ +SL+ R++ R
Sbjct: 1191 KNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKAR 1250
Query: 357 NRVEGGHL-CSGGRNESMWWRNIVA 380
E L R +S W +++A
Sbjct: 1251 YFREDSILDAKRQRYQSYGWTSMLA 1275
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 249 bits (635), Expect = 2e-64
Identities = 146/385 (37%), Positives = 216/385 (55%), Gaps = 11/385 (2%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRKK---DLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR + D +++A+E++ + RK+ + K D KAYD V+ N+LE M
Sbjct: 917 DSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMR 976
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W KWI V + SVLVNG P RG+RQGDPLSP+LF+L A+ + L
Sbjct: 977 LFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHL 1036
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
++ V G ++G + + GVTHLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 1037 IKNRVAEGDIRGIRIG-NGVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQK 1095
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N SKS+ G VHG+ +L + YLGLP + + +I R+
Sbjct: 1096 INMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVK 1155
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R SSW +K+LS G+ ++LKSV S+PVYA+S FK P IVS IE++L NF W
Sbjct: 1156 KRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNF-----WWE 1210
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+A R+I W+ W + S++ GGL R + +FN ALL K WR++ +SL+ R++ R
Sbjct: 1211 KNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKAR 1270
Query: 357 NRVEGGHL-CSGGRNESMWWRNIVA 380
E L R +S W +++A
Sbjct: 1271 YFREDSILDAKRQRYQSYGWTSMLA 1295
>UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1614
Score = 245 bits (625), Expect = 4e-63
Identities = 145/420 (34%), Positives = 220/420 (51%), Gaps = 9/420 (2%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q+AFI GR IL+G ++ +EV+ + ++ + ++ K+DFEKAYD V ++L EVMV+ GFP
Sbjct: 1167 QTAFIPGRFILEGCVIIHEVLHEMNRKNLEGIILKIDFEKAYDKVSWDFLIEVMVRKGFP 1226
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
+ W WIK CV + +NG T F RGLRQGDPLSP LF L ++ ++++
Sbjct: 1227 SKWVNWIKTCVMGGRVCININGERTDFFRTFRGLRQGDPLSPLLFNLISDALAAMLDSAK 1286
Query: 123 VNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSK 182
+ +G V G+THLQ+ADDT++ I A + IL F+E++ LKVN+ K
Sbjct: 1287 REGVLSGL-VPDIFPGGITHLQYADDTVLFVANDDKQIVATKFILYCFEEMAVLKVNYHK 1345
Query: 183 SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSW 242
S + + + A + NC G P YLGLPIG + + + L ++ RL+SW
Sbjct: 1346 SEIFTLGLSDNDTNRVAMMFNCPVGQFPMKYLGLPIGPDKILNLGFDFLGQKLEKRLNSW 1405
Query: 243 KSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHR 302
+LS GR V + + LSS+P YA+ F++ P G+ SI ++ W
Sbjct: 1406 -GNNLSHAGRAVQINTCLSSIPSYAMCFYQLPEGVHQKFGSIRGRYY----WARNRL-KG 1459
Query: 303 KIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRVEGG 362
K V W + ++ GGL + N ALL KW ++ +E DSL +L + +GG
Sbjct: 1460 KYHMVKWEDLAFPKDYGGLGFTETRRMNTALLAKWIMKIESEDDSLCIELLRRKYLQDGG 1519
Query: 363 HLCSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLRDRFSRLY 422
R S +W+ + L+ W +GDGS + FW DV G+ LR F ++
Sbjct: 1520 FFQCKERYASQFWKGL--LNIRRWLSLGSVWQVGDGSHISFWRDVWWGQCPLRTLFPAIF 1577
>UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis thaliana]
Length = 851
Score = 239 bits (610), Expect = 2e-61
Identities = 149/411 (36%), Positives = 223/411 (54%), Gaps = 13/411 (3%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRK---KDLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR I D +++A+E++ + RK K + K D KAYD V+ ++LE M
Sbjct: 156 DSQAAFIPGRIINDNVMIAHEIMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMR 215
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W WI V + SVL+NGSP S RG+RQGDPLSP+LF+L + L
Sbjct: 216 LFGFCDKWIGWIMAAVKSVHYSVLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSHL 275
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
++ + G ++G + +THLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 276 IKVKASSGDIRGVRIG-NGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQK 334
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N KSL G V+GS T+LN YLGLP + + +I+R+
Sbjct: 335 INVQKSLITFGSRVYGSTQTRLKTLLNIPNQGGGGKYLGLPEQFGRKKKEMFNYIIDRVK 394
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R +SW +K LS G+ +LLKSV ++PVYA+S FK P GIVS IES+L NF W
Sbjct: 395 ERTASWSAKFLSPAGKEILLKSVALAMPVYAMSCFKLPQGIVSEIESLLMNF-----WWE 449
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+++ R I WV W + S++ GGL R + +FN ALL K WR++ +SL+ RV+ R
Sbjct: 450 KASNKRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRIIQYPNSLFARVMKAR 509
Query: 357 NRVEGGHLCSGGRN-ESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ + + R+ +S W ++ L + +GDG ++ D
Sbjct: 510 YFKDNSIIDAKTRSQQSYGWSSL--LSGIALLRKGTRYVIGDGKTIRLGID 558
>UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
Length = 1142
Score = 234 bits (598), Expect = 5e-60
Identities = 144/411 (35%), Positives = 214/411 (52%), Gaps = 13/411 (3%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKR---KKDLLLFKVDFEKAYDSVD*NYLEEVMV 57
+ QSAF+ GR I D IL+A E+ R K + K D KAYD V+ N++E ++
Sbjct: 319 ETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKFMAIKTDMSKAYDQVEWNFIEALLR 378
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
KMGF W WI C+ T VL+NG P RGLRQGDPLSP+LF+L E
Sbjct: 379 KMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERGLRQGDPLSPYLFILCTEVLIAN 438
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ NL G KV + V+HL FADD++ C + + IL ++ +SG +
Sbjct: 439 IRKAERQNLITGIKVAT-PSPAVSHLLFADDSLFFCKANKEQCGIILEILKQYESVSGQQ 497
Query: 178 VNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+NFSK S+ G V S A+ +L + YLGLP S+ + + +R+
Sbjct: 498 INFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSFVRDRLQ 557
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
SR++ W +K LS GG+ V++KSV ++LP Y +S F+ P I S + S + F W
Sbjct: 558 SRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPKAITSKLTSAVAKF-----WWS 612
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+ D R + W+ W+ +C S+ GGL R + +FN ALL K WR++T DSL+ +V R
Sbjct: 613 SNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGR 672
Query: 357 NRVEGGHLCS-GGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ L S + S WR++++ S + + +G G+S+ W D
Sbjct: 673 YFRKSNPLDSIKSYSPSYGWRSMISARS--LVYKGLIKRVGSGASISVWND 721
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 233 bits (593), Expect = 2e-59
Identities = 147/414 (35%), Positives = 225/414 (53%), Gaps = 19/414 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRK---KDLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR I D +++A+EV+ + RK K + K D KAYD V+ ++LE M
Sbjct: 691 DSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMR 750
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W WI V + SVL+NGSP + RG+RQGDPLSP+LF+L + L
Sbjct: 751 LFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHL 810
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ + G ++G + +THLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 811 INGRASSGDLRGVRIG-NGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQK 869
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N KS+ G V+GS ++ +L YLGLP + ++ +I+R+
Sbjct: 870 INVQKSMITFGSRVYGSTQSKLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVK 929
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R S+W ++ LS G+ ++LKSV ++PVYA+S FK P GIVS IES+L NF W
Sbjct: 930 KRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNF-----WWE 984
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+++ R I WV W + S++ GGL R + +FN ALL K WR++ +SL+ RV+ R
Sbjct: 985 KASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKAR 1044
Query: 357 NRVEGGHLCSGGR-NESMWWRNI---VALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ L + R +S W ++ +AL +G +GDG ++ D
Sbjct: 1045 YFKDVSILDAKVRKQQSYGWASLLDGIALLKKG-----TRHLIGDGQNIRIGLD 1093
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 232 bits (592), Expect = 2e-59
Identities = 147/414 (35%), Positives = 224/414 (53%), Gaps = 19/414 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRK---KDLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR I D +++A+EV+ + RK K + K D KAYD V+ ++LE M
Sbjct: 917 DSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMR 976
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W WI V + SVL+NGSP + RG+RQGDPLSP+LF+L + L
Sbjct: 977 LFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHL 1036
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ + G ++G + +THLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 1037 INGRASSGDLRGVRIG-NGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQK 1095
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N KS+ G V+GS + +L YLGLP + ++ +I+R+
Sbjct: 1096 INVQKSMITFGSRVYGSTQSRLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVK 1155
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R S+W ++ LS G+ ++LKSV ++PVYA+S FK P GIVS IES+L NF W
Sbjct: 1156 KRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNF-----WWE 1210
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+++ R I WV W + S++ GGL R + +FN ALL K WR++ +SL+ RV+ R
Sbjct: 1211 KASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKAR 1270
Query: 357 NRVEGGHLCSGGR-NESMWWRNI---VALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ L + R +S W ++ +AL +G +GDG ++ D
Sbjct: 1271 YFKDVSILDAKVRKQQSYGWASLLDGIALLKKG-----TRHLIGDGQNIRIGLD 1319
>UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa]
Length = 1509
Score = 228 bits (580), Expect = 6e-58
Identities = 146/413 (35%), Positives = 210/413 (50%), Gaps = 21/413 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLL---LFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ GR I D IL+A E+ R ++ + FK+D KAYD V+ ++L ++M+K+
Sbjct: 738 QSAFVPGRLISDNILIAYEMTHYMRNKRSGQVGYAAFKLDMSKAYDRVEWSFLHDMMLKL 797
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLME 119
GF T W I KCV T T + VNG + FS RGLRQGDPLSP+LFLL AEGF L+
Sbjct: 798 GFHTDWVNLIMKCVSTVTYRIRVNGELSESFSPERGLRQGDPLSPYLFLLCAEGFSALLS 857
Query: 120 ALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVN 179
+G ++ V+HL FADD++ILC + + L+ IL +++E SG +N
Sbjct: 858 KTEEEGRLHGIRI-CQGAPSVSHLLFADDSLILCRANGGEAQQLQTILQIYEECSGQVIN 916
Query: 180 FSKS-LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSR 238
KS + N LN + YLGLP+ SR + L RI R
Sbjct: 917 KDKSAVMFSPNTSSLEKGAVMAALNMQRETTNEKYLGLPVFVGRSRTKIFSYLKERIWQR 976
Query: 239 LSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGS 298
+ WK K LS G+ +L+K+V +P +A+ F+ + I ++ + W
Sbjct: 977 IQGWKEKLLSRAGKEILIKAVAQVIPTFAMGCFELTKDLCDQISKMIAKY-----WWSNQ 1031
Query: 299 ADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNR 358
K+ W+ WN + + +GGL R I FNLA+L K WR++ + DSL RVL +
Sbjct: 1032 EKDNKMHWLSWNKLTLPKNMGGLGFRDIYIFNLAMLAKQGWRLIQDPDSLCSRVLRAKYF 1091
Query: 359 VEGGHLCSGGR---NESMWWRNIVALHSEGW--FQNHVSRSLGDGSSVLFWTD 406
G C + N S WR+I +G QN + +GDGS + W D
Sbjct: 1092 PLGD--CFRPKQTSNVSYTWRSI----QKGLRVLQNGMIWRVGDGSKINIWAD 1138
>UniRef100_Q7XE51 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1652
Score = 227 bits (578), Expect = 1e-57
Identities = 135/409 (33%), Positives = 216/409 (52%), Gaps = 18/409 (4%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q+ F+ GR I++G+++ +E + + K+KK+ ++ K+DFEKAYD VD +L++ + GF
Sbjct: 1256 QTTFLSGRNIMEGVVILHETLHELHKKKKNGVILKLDFEKAYDKVDWKFLQQSLRMKGFS 1315
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
+ W WI V + +V VN F +GLRQGDPLSP LF L + +L++
Sbjct: 1316 SKWCDWIDSIVRGGSVAVKVNDEIGSYFQTRKGLRQGDPLSPILFNLVVDMLAILIQRAK 1375
Query: 123 VNNLFNGYKVGSHEM-VGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFS 181
F G V H + G++ LQ+ADDTI+ D R L+ +L F++LS LK+NF
Sbjct: 1376 DQGRFKG--VVPHLVDNGLSILQYADDTILFMDHDLDEARDLKLVLSTFEKLSSLKINFY 1433
Query: 182 KSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSS 241
KS E + C+ PF YLG+ + W+ + RI +LSS
Sbjct: 1434 KSELFCYGKAKDVEHEYVKLFGCDTEDYPFKYLGIRMHHKRINNKDWQGVEERIQKKLSS 1493
Query: 242 WKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADH 301
WK K LS+GGRLVL+ SVLS+L ++ LSFF+ P GI+ ++ + FF+
Sbjct: 1494 WKGKFLSVGGRLVLINSVLSNLAIFMLSFFEIPKGILKKLDYYRSRFFW-----QCDEHK 1548
Query: 302 RKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVL----AVRN 357
+K W+ +C+ +E GGL ++ ++ N LL KW ++++ E + +W +L +
Sbjct: 1549 KKYRLARWSVLCKPKECGGLGIQNLEIQNKCLLSKWLYKLINE-EGVWQDILRNKYLTKK 1607
Query: 358 RVEGGHLCSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ C G +S +W ++ + F + +GDGS + FW D
Sbjct: 1608 TITQVEKCPG---DSHFWAGLMGVRDV--FFKGGAFKVGDGSQIRFWED 1651
>UniRef100_Q7XMG7 OSJNBa0028I23.15 protein [Oryza sativa]
Length = 593
Score = 224 bits (572), Expect = 5e-57
Identities = 145/418 (34%), Positives = 221/418 (52%), Gaps = 26/418 (6%)
Query: 13 LDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFPTLWRKWIKKC 72
++G ++ +E + + KRKKD ++ K+DFEKAYD VD +L++ + GF W KWI +
Sbjct: 1 MEGAIILHETLHEMHKRKKDGVILKLDFEKAYDKVDWKFLQQTLRMKGFEPRWCKWIDQV 60
Query: 73 VGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYKV 132
V + +V VN F +GLRQGDPLSP LF L A+ VL++ + +G V
Sbjct: 61 VRGGSVAVKVNDEIGNFFQTKKGLRQGDPLSPLLFNLVADMLAVLIQRTSEHGKIHG--V 118
Query: 133 GSHEM-VGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKSLFVGVNVH 191
H + G++ LQ+ADDTI+ + + + L+ +L F+ LSGLK+NF KS +
Sbjct: 119 IPHLVDDGLSILQYADDTILFMEHDLEDAKNLKLVLSAFERLSGLKINFHKSELLCFGKA 178
Query: 192 GSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLSLGG 251
E A + C GS YLGLP+ WK + R RL SWK K LS+GG
Sbjct: 179 IEVEREYALLFGCKTGSYTLKYLGLPMHYRKLSNKDWKEVEERFQKRLGSWKGKLLSVGG 238
Query: 252 RLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADH-RKIIWVDWN 310
RLVL+ SVLSSL +Y LSFF+ P GI+ ++ + FF+ S +H +K W+
Sbjct: 239 RLVLINSVLSSLAMYMLSFFEVPKGIIKKLDYYRSRFFW------QSDEHKKKYRLARWS 292
Query: 311 SVCRSQEVGGLEVRRIKEFNLAL---LGKWCWRVLTERDSLWFR-VLAVRNRVEGGHLCS 366
+C+ +E GGL ++ ++ N L L K W+ L + L + ++ V+ +
Sbjct: 293 VLCKPKECGGLGIQNLEVQNKCLLRGLAKLVWQNLLRKKYLAKKTIMQVQKQ-------- 344
Query: 367 GGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLRDRFSRLYDL 424
R +S +W ++ + + F + S L DG + FW D +G SL + LY+L
Sbjct: 345 --RGDSHFWTGLMGV--KDTFSSFGSFKLQDGLQIRFWEDCWLGNQSLEKIYPSLYNL 398
>UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 223 bits (568), Expect = 1e-56
Identities = 156/451 (34%), Positives = 226/451 (49%), Gaps = 22/451 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRKK---DLLLFKVDFEKAYDSVD*NYLEEVMV 57
D QSAF+ R I D ILVA+E+V + + + + K D KAYD V+ +YL +++
Sbjct: 498 DTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRSLLL 557
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
+GF W WI CV + T SVL+N P L RGLRQGDPLSPFLF+L EG L
Sbjct: 558 SLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHL 617
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ G + + + V HL FADD++ LC S L+ IL ++ +G
Sbjct: 618 LNKAQWEGALEGIQFSENGPM-VHHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQT 676
Query: 178 VNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N +K S+ G V T L YLGLP + S++ L +R+
Sbjct: 677 INLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLK 736
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
+L W ++ LS GG+ VLLKSV ++PV+A+S FK P ++ES + +F++
Sbjct: 737 EKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWW------ 790
Query: 297 GSADH-RKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAV 355
S DH RKI W W +C ++ GGL R I+ FN ALL K WR+L D L R+L
Sbjct: 791 DSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKS 850
Query: 356 RNRVEGGHLCSG-GRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSL 414
R L + + S WR+I L + + +GDG+S+ W D + +
Sbjct: 851 RYFDATDFLDAALSQRPSFGWRSI--LFGRELLSKGLQKRVGDGASLFVWIDPWIDDNGF 908
Query: 415 RD--RFSRLYDL-----SVLKGGVGRYDESV 438
R R + +YD+ ++L G +DE V
Sbjct: 909 RAPWRKNLIYDVTLKVKALLNPRTGFWDEEV 939
>UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 223 bits (568), Expect = 1e-56
Identities = 156/451 (34%), Positives = 226/451 (49%), Gaps = 22/451 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRKK---DLLLFKVDFEKAYDSVD*NYLEEVMV 57
D QSAF+ R I D ILVA+E+V + + + + K D KAYD V+ +YL +++
Sbjct: 501 DTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRSLLL 560
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
+GF W WI CV + T SVL+N P L RGLRQGDPLSPFLF+L EG L
Sbjct: 561 SLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHL 620
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ G + + + V HL FADD++ LC S L+ IL ++ +G
Sbjct: 621 LNKAQWEGALEGIQFSENGPM-VHHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQT 679
Query: 178 VNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N +K S+ G V T L YLGLP + S++ L +R+
Sbjct: 680 INLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLK 739
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
+L W ++ LS GG+ VLLKSV ++PV+A+S FK P ++ES + +F++
Sbjct: 740 EKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWW------ 793
Query: 297 GSADH-RKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAV 355
S DH RKI W W +C ++ GGL R I+ FN ALL K WR+L D L R+L
Sbjct: 794 DSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKS 853
Query: 356 RNRVEGGHLCSG-GRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSL 414
R L + + S WR+I L + + +GDG+S+ W D + +
Sbjct: 854 RYFDATDFLDAALSQRPSFGWRSI--LFGRELLSKGLQKRVGDGASLFVWIDPWIDDNGF 911
Query: 415 RD--RFSRLYDL-----SVLKGGVGRYDESV 438
R R + +YD+ ++L G +DE V
Sbjct: 912 RAPWRKNLIYDVTLKVKALLNPRTGFWDEEV 942
>UniRef100_Q8H992 Oryza sativa (japonica cultivar-group) retrotransposon Karma DNA,
complete sequence [Oryza sativa]
Length = 1197
Score = 221 bits (563), Expect = 6e-56
Identities = 136/426 (31%), Positives = 230/426 (53%), Gaps = 22/426 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q+ F+KGR I + ++ A E++ RK L+ K+DF KA+DSV L +++ GFP
Sbjct: 550 QTGFLKGRCISENLIYATELIQACHARKCKTLIIKLDFAKAFDSVLWTSLFQILAVRGFP 609
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
W WIK + T+ ++VL+NG P + +GLRQGDPLSP+LF+L A+ +++ L+
Sbjct: 610 NNWISWIKSLLQTSKSAVLLNGIPGKWINCKKGLRQGDPLSPYLFILVAD----VLQRLL 665
Query: 123 VNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSK 182
N + + +Q+ADDT+++C ++ ALR+ LL F + +GL++NF+K
Sbjct: 666 EKNFQIRHPIYQDRPCAT--IQYADDTLVICRAEEDDVLALRSTLLQFSKATGLQINFAK 723
Query: 183 SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSW 242
S + +++ S + + +L C S+P YLGLP+ + +P++ +++S L+ W
Sbjct: 724 STMISLHIDRSKESSLSELLQCKLESLPMSYLGLPLSLHKLTNNDLQPIVVKVDSFLTGW 783
Query: 243 KSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHR 302
++ LS RL+L+ +VLSS+PVYA+S FK P ++ +I+ FF W G
Sbjct: 784 EASLLSQAERLILVNAVLSSVPVYAMSAFKLPPKVIEAIDKRRRAFF----WTGDDTCSG 839
Query: 303 KIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRVEGG 362
V W+ VC ++E GGL ++ +K N ALL K + + ++ S W + +G
Sbjct: 840 AKCLVAWDEVCTAKEKGGLGIKSLKTQNEALLLKRLFNLFSDNSS-WTNW--IWKEFDGR 896
Query: 363 HLCSG---GRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLRDRFS 419
L G++ S++ + + L + +GDGS FW D G ++L +F
Sbjct: 897 SLLKSLPLGQHWSVFQKLLPELFKI------TTVHIGDGSRTSFWHDRWTGNMTLAAQFE 950
Query: 420 RLYDLS 425
L+ S
Sbjct: 951 SLFSHS 956
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 220 bits (561), Expect = 9e-56
Identities = 140/418 (33%), Positives = 219/418 (51%), Gaps = 19/418 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVV---DDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMV 57
+ Q+AF+KGR I D IL+A+E++ K ++ + K D KAYD V+ +LE+ M
Sbjct: 535 ETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMR 594
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
+GF W + I +CV + VL+NG+P E RGLRQGDPLSP+LF++ E +
Sbjct: 595 GLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEMLVKM 654
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQE---LS 174
+++ N G KV + ++HL FADD++ C N AL I+ + +E S
Sbjct: 655 LQSAEQKNQITGLKV-ARGAPPISHLLFADDSMFYCK---VNDEALGQIIRIIEEYSLAS 710
Query: 175 GLKVNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLIN 233
G +VN+ K S++ G ++ L +YLGLP S++ L +
Sbjct: 711 GQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKVATLSYLKD 770
Query: 234 RINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGG 293
R+ ++ W+S LS GG+ +LLK+V +LP Y +S FK P I IES++ F
Sbjct: 771 RLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESVMAEF----- 825
Query: 294 WGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVL 353
W + R + W W + R + VGGL + I+ FN+ALLGK WR++TE+DSL +V
Sbjct: 826 WWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKVF 885
Query: 354 AVRNRVEGGHLCSG-GRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VG 410
R + L + G S W++I ++ + + +G+G ++ WTD +G
Sbjct: 886 KSRYFSKSDPLNAPLGSRPSFAWKSI--YEAQVLIKQGIRAVIGNGETINVWTDPWIG 941
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 218 bits (556), Expect = 4e-55
Identities = 145/414 (35%), Positives = 209/414 (50%), Gaps = 23/414 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKR---KKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ R I D ILVA+E++ R K+ + FK D KAYD V+ +LE +M +
Sbjct: 515 QSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTAL 574
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEG-FHVLM 118
GF W WI CV + + SVL+NG P RG+RQGDPLSP LF+L E H+L
Sbjct: 575 GFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILN 634
Query: 119 EALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKV 178
+A + G + + V V HL FADDT+++C + L L + +LSG +
Sbjct: 635 KAEQAGKI-TGIQF-QDKKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMI 692
Query: 179 NFSKS-LFVGVNVH---GSWLAEAATV-LNCNAGSIPFLYLGLPIGGNASRMVFWKPLIN 233
N +KS + G NV W+ + + L G YLGLP + S+ + +
Sbjct: 693 NLNKSAITFGKNVDIQIKDWIKSRSGISLEGGTGK----YLGLPECLSGSKRDLFGFIKE 748
Query: 234 RINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGG 293
++ SRL+ W +K LS GG+ VLLKS+ +LPVY +S FK P + + +++ +F
Sbjct: 749 KLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDF----- 803
Query: 294 WGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVL 353
W RKI W+ W + ++ GG + ++ FN ALL K WRVL E+ SL+ RV
Sbjct: 804 WWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVF 863
Query: 354 AVRNRVEGGHL-CSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
R L + G S WR+I L + +G+G WTD
Sbjct: 864 QSRYFSNSDFLSATRGSRPSYAWRSI--LFGRELLMQGLRTVIGNGQKTFVWTD 915
>UniRef100_Q7X970 BZIP-like protein [Oryza sativa]
Length = 2367
Score = 216 bits (551), Expect = 1e-54
Identities = 137/409 (33%), Positives = 204/409 (49%), Gaps = 13/409 (3%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKD---LLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF++GR I D L+A E +K KK +K+D KAYD VD +LE M K+
Sbjct: 1271 QSAFVQGRMITDNALLAFECFHAMQKNKKANHAACAYKLDLSKAYDRVDWRFLEMAMNKL 1330
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLME 119
GF W WI KCV + V NG+ F+ RGLRQGDPL PFLFL A+G +L++
Sbjct: 1331 GFARRWVNWIMKCVTSVRYMVKFNGTLLQSFAPTRGLRQGDPLLPFLFLFVADGLSLLLK 1390
Query: 120 ALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVN 179
V N +KV G++HL FADDT++ ++ +L + +G +N
Sbjct: 1391 EKVAQNSLTPFKV-CRAAPGISHLLFADDTLLFFKAHQREAEVVKEVLSSYAMGTGQLIN 1449
Query: 180 FSKSLFVGVNVHGSWLAEA-ATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSR 238
+K + ++EA + +L YLG P ++ L +I R
Sbjct: 1450 PAKCSILMGGASTPAVSEAISEILQVERDRFEDRYLGFPTPEGRMHKGRFQSLQAKIWKR 1509
Query: 239 LSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGS 298
+ W HLS GG+ VL+K+V+ ++PVY + FK P ++ + + N F+W G
Sbjct: 1510 VIQWGENHLSTGGKEVLIKAVIQAIPVYVMGIFKLPESVIDDLTKLTKN-FWWDSMNG-- 1566
Query: 299 ADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNR 358
RK W W+S+ + + +GGL R + FN ALL + WR++T DSL RVL +
Sbjct: 1567 --QRKTHWKAWDSLTKPKSLGGLGFRDYRLFNQALLARQAWRLITYPDSLCARVLKAKYF 1624
Query: 359 VEGGHL-CSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
G + S G N S WR+I + + + +G+G+S+ W D
Sbjct: 1625 PHGSLIDTSFGSNSSPAWRSIE--YGLDLLKKGIIWRVGNGNSIRIWRD 1671
>UniRef100_Q7XXA4 OSJNBa0019G23.12 protein [Oryza sativa]
Length = 1140
Score = 213 bits (543), Expect = 1e-53
Identities = 123/344 (35%), Positives = 188/344 (53%), Gaps = 14/344 (4%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q+AF++GR ILDG+ + +E V + ++K + ++FK+DFEKAYD V +L + + GF
Sbjct: 773 QTAFMRGRNILDGVAIIHETVHELHRKKLNGVIFKIDFEKAYDKVKWPFLLQTLRMKGFS 832
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
W WIK + + +V VN F +GL+QGDPLSP LF F V M A +
Sbjct: 833 PKWISWIKSFIVGGSVAVKVNDDVGPFFQTKKGLQQGDPLSPILF-----NFIVDMLATL 887
Query: 123 VNNLFNGYKVGS---HEMVG-VTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKV 178
+N +V H + G ++ LQ+ DD ++ + + ++++LL F++LSGLK+
Sbjct: 888 INRAKTQGQVDGLIPHLIDGGLSILQYVDDIVLFMNHDLEKAQNMKSVLLAFEQLSGLKI 947
Query: 179 NFSKSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSR 238
NF KS + + A + C G+ PF YLG+PI R WK ++ R +
Sbjct: 948 NFHKSELYCFGEALEYRDQYAQLFGCQVGNFPFRYLGIPIHCRKLRNAEWKEVVERFEKK 1007
Query: 239 LSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGS 298
LSSWK K LSLGGRL L+ SVLSSLP+Y +SF PSG++ ++ L + F+W G G
Sbjct: 1008 LSSWKGKLLSLGGRLTLINSVLSSLPMYMMSFLAIPSGVLKKLD-YLRSRFYWQGDG--- 1063
Query: 299 ADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVL 342
+K W+ +CR ++ G L + ++ L W +L
Sbjct: 1064 -HKKKYQLAKWDIICRPKDQGRLGIHDLEVITLVTRWLRTWAIL 1106
>UniRef100_O22220 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1094
Score = 211 bits (536), Expect = 7e-53
Identities = 136/417 (32%), Positives = 218/417 (51%), Gaps = 29/417 (6%)
Query: 3 QSAFIKGRQILDGILVANEVVDDAR---KRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ R + D I++A+E+V + R K KD ++FK D KAYD V+ +L+ +++ +
Sbjct: 266 QSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDMSKAYDRVEWPFLKGILLAL 325
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEG-FHVLM 118
GF + W W+ CV + + SVL+NG P + HRGLRQGDPLSPFLF+L E H+L
Sbjct: 326 GFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLRQGDPLSPFLFVLCTEALIHILN 385
Query: 119 EALVVNNL----FNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELS 174
+A + + FNG V HL FADDT+++C S + L + +S
Sbjct: 386 QAEKIGKISGIQFNG------TGPSVNHLLFADDTLLICKASQLECAEIMHCLSQYGHIS 439
Query: 175 GLKVNFSKS-LFVGVNVH---GSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKP 230
G +N KS + G V+ W+ + + G+ YLGLP S+ V +
Sbjct: 440 GQMINSEKSAITFGAKVNEETKQWIMNRSGI-QTEGGT--GKYLGLPECFQGSKQVLFGF 496
Query: 231 LINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFF 290
+ ++ SRLS W +K LS GG+ +LLKS+ + PVYA++ F+ + + + S++ +F
Sbjct: 497 IKEKLQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLSKTLCTKLTSVMMDF-- 554
Query: 291 WGGWGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWF 350
W D +KI W+ + + +GG + ++ FN ALL K R+ T+ DSL
Sbjct: 555 ---WWNSVQDKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLHTDSDSLLS 611
Query: 351 RVLAVRNRVEGGHL-CSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
++L R + L + G S W++I L+ + + + +G+G + W D
Sbjct: 612 QILKSRYYMNSDFLSATKGTRPSYAWQSI--LYGRELLVSGLKKIIGNGENTYVWMD 666
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.340 0.151 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,071,135,810
Number of Sequences: 2790947
Number of extensions: 43801390
Number of successful extensions: 131784
Number of sequences better than 10.0: 884
Number of HSP's better than 10.0 without gapping: 649
Number of HSP's successfully gapped in prelim test: 235
Number of HSP's that attempted gapping in prelim test: 129761
Number of HSP's gapped (non-prelim): 1217
length of query: 679
length of database: 848,049,833
effective HSP length: 134
effective length of query: 545
effective length of database: 474,062,935
effective search space: 258364299575
effective search space used: 258364299575
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC124215.6


