

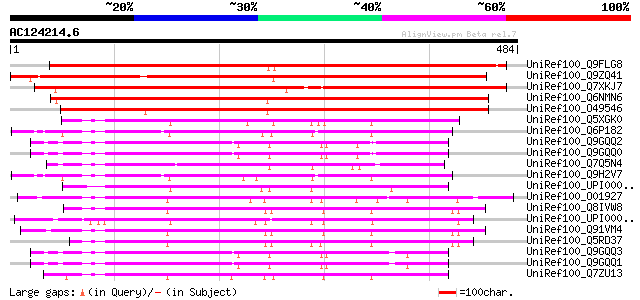
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124214.6 + phase: 0
(484 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FLG8 Similarity to unknown protein [Arabidopsis thal... 678 0.0
UniRef100_Q9ZQ41 Hypothetical protein At2g22730 [Arabidopsis tha... 622 e-177
UniRef100_Q7XKJ7 OSJNBa0038O10.10 protein [Oryza sativa] 589 e-167
UniRef100_Q6NMN6 At5g65687 [Arabidopsis thaliana] 576 e-163
UniRef100_O49546 Predicted protein [Arabidopsis thaliana] 558 e-157
UniRef100_Q5XGK0 Hypothetical protein [Xenopus laevis] 160 8e-38
UniRef100_Q6P182 SPINL protein [Homo sapiens] 149 1e-34
UniRef100_Q9GQQ2 Spinster type II [Drosophila melanogaster] 149 2e-34
UniRef100_Q9GQQ0 Spinster type IV [Drosophila melanogaster] 149 2e-34
UniRef100_Q7Q5N4 ENSANGP00000013028 [Anopheles gambiae str. PEST] 149 2e-34
UniRef100_Q9H2V7 Spinster [Homo sapiens] 149 2e-34
UniRef100_UPI0000318BDA UPI0000318BDA UniRef100 entry 147 7e-34
UniRef100_O01927 Hypothetical protein C13C4.5 [Caenorhabditis el... 144 6e-33
UniRef100_Q8IVW8 Similar to spinster-like protein [Homo sapiens] 142 2e-32
UniRef100_UPI00003374A0 UPI00003374A0 UniRef100 entry 142 2e-32
UniRef100_Q91VM4 BC011467 protein [Mus musculus] 140 1e-31
UniRef100_Q5RD37 Hypothetical protein DKFZp459J1933 [Pongo pygma... 139 2e-31
UniRef100_Q9GQQ3 Spinster type I [Drosophila melanogaster] 138 3e-31
UniRef100_Q9GQQ1 Spinster type III [Drosophila melanogaster] 138 3e-31
UniRef100_Q7ZU13 Spinster-like [Brachydanio rerio] 137 9e-31
>UniRef100_Q9FLG8 Similarity to unknown protein [Arabidopsis thaliana]
Length = 484
Score = 678 bits (1750), Expect = 0.0
Identities = 325/458 (70%), Positives = 385/458 (83%), Gaps = 23/458 (5%)
Query: 39 IPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTCTD-GICKSGTGIQGDFNL 97
I SWFTPK+LL +FCV+N++NY+DRGAIASNG+NG RG+CT G C SG+GIQGDFNL
Sbjct: 27 ISEPSWFTPKKLLFVFCVVNLINYIDRGAIASNGINGSRGSCTSSGTCSSGSGIQGDFNL 86
Query: 98 TNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSIT 157
+NF+DGVLSSAFMVGLL+ASPIFASL+KSVNPFRLIGVGLS+WT+A + CGLSF+FWSIT
Sbjct: 87 SNFEDGVLSSAFMVGLLVASPIFASLAKSVNPFRLIGVGLSIWTLAVIGCGLSFDFWSIT 146
Query: 158 VCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG 217
+CRM VGVGEASF+SLAAPFIDDNAP QK+ WL++FYMCIP GYA GYVYGGVVGS
Sbjct: 147 ICRMFVGVGEASFVSLAAPFIDDNAPHDQKSAWLAVFYMCIPTGYAFGYVYGGVVGSVLP 206
Query: 218 WRYAFWVEAVLMLPFAILGFVMKPLQLK------------DQLSLF---------LKDMK 256
WR AFW EA+LMLPFA+LGFV+KPL LK D L++ +KD+K
Sbjct: 207 WRAAFWGEAILMLPFAVLGFVIKPLHLKGFAPDDTGKPRTDNLNVLPVGYGFSAVMKDLK 266
Query: 257 ELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADMIFGGITIVCGILGTLA 316
LL DKV+V N+LGYIAYNFV+GAYSYWGPKAGY+IY M NADMIFGG+T+VCGI+GTL+
Sbjct: 267 LLLVDKVYVTNILGYIAYNFVLGAYSYWGPKAGYNIYKMENADMIFGGVTVVCGIVGTLS 326
Query: 317 GGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIGELLVFATQGPV 376
GG++LDYM T+SNAFK+LS++T +G FCF AF FKSMY FLALFA+GELLVFATQGPV
Sbjct: 327 GGVILDYMDATISNAFKVLSVSTFIGAIFCFAAFCFKSMYAFLALFAVGELLVFATQGPV 386
Query: 377 NFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTALILTTIFFPA 436
NF+ LHCVKPSLRPL+MAMSTV+IHIFGDVPS+PLVGV+QD++NNWR T+L+LT + FPA
Sbjct: 387 NFIVLHCVKPSLRPLAMAMSTVSIHIFGDVPSSPLVGVLQDYVNNWRVTSLVLTFVLFPA 446
Query: 437 AAIWFIGIFLNSKDKFNEESEHQVSRVEGTTTAPLLEE 474
AAIW IGIFLNS D++NE+SE E +T APLL+E
Sbjct: 447 AAIWSIGIFLNSVDRYNEDSEPDAVTRE-STAAPLLQE 483
>UniRef100_Q9ZQ41 Hypothetical protein At2g22730 [Arabidopsis thaliana]
Length = 507
Score = 622 bits (1604), Expect = e-177
Identities = 318/496 (64%), Positives = 367/496 (73%), Gaps = 47/496 (9%)
Query: 1 MAQKSEDEPKPATTTSSS--STPNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVIN 58
M K ED P T T+S ST + P + E + ++ ++S +P LL IFC+IN
Sbjct: 1 MVTKEEDCLPPVTETTSRCYSTSSSTPLA-ELETVRSLEIVESSSSLSPVWLLVIFCIIN 59
Query: 59 MLNYLDRGAIASNGVNGHRGTCTD-GICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIAS 117
+LNY+DRGAIASNGVNG +C D G C TGIQG FNL+NF+DGVLSS+FMVGLLIAS
Sbjct: 60 LLNYMDRGAIASNGVNGSTRSCNDKGKCTLATGIQGHFNLSNFEDGVLSSSFMVGLLIAS 119
Query: 118 PIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPF 177
PIFASL+K RLIGVGL+VWT+A L CG SF FW I +CRM VGVGEASFISLAAPF
Sbjct: 120 PIFASLAK-----RLIGVGLTVWTIAVLGCGSSFAFWFIVLCRMFVGVGEASFISLAAPF 174
Query: 178 IDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFGWRYAFWVEAVLMLPFAILGF 237
IDDNAP QK WL +FYMCIP G A+GYVYGG VG HF WRYAFW EAVLM PFA+LGF
Sbjct: 175 IDDNAPQEQKAAWLGLFYMCIPSGVALGYVYGGYVGKHFSWRYAFWGEAVLMAPFAVLGF 234
Query: 238 VMKPLQLKDQLSL--------------------------------------FLKDMKELL 259
+MKPLQLK +L F KDMK L
Sbjct: 235 LMKPLQLKGSETLKNNNRLQVDNEIEHDQFEVSIETSKSSYANAVFKSFTGFAKDMKVLY 294
Query: 260 SDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADMIFGGITIVCGILGTLAGGL 319
+KVFVVNVLGY++YNFVIGAYSYWGPKAGY+IY M NADMIFG +TI+CGI+GTL+GG
Sbjct: 295 KEKVFVVNVLGYVSYNFVIGAYSYWGPKAGYNIYKMKNADMIFGAVTIICGIVGTLSGGF 354
Query: 320 VLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIGELLVFATQGPVNFV 379
+LD +T T+ NAFKLLS T +G FCF AF KS+YGF+ALFA+GELLVFATQ PVN+V
Sbjct: 355 ILDRVTATIPNAFKLLSGATFLGAVFCFTAFTLKSLYGFIALFALGELLVFATQAPVNYV 414
Query: 380 CLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTALILTTIFFPAAAI 439
CLHCVKPSLRPLSMA+STVAIHIFGDVPS+PLVG+VQDHIN+WR T LILT+I F AAAI
Sbjct: 415 CLHCVKPSLRPLSMAISTVAIHIFGDVPSSPLVGIVQDHINSWRKTTLILTSILFLAAAI 474
Query: 440 WFIGIFLNSKDKFNEE 455
WFIGIF+NS D+FN+E
Sbjct: 475 WFIGIFINSVDRFNQE 490
>UniRef100_Q7XKJ7 OSJNBa0038O10.10 protein [Oryza sativa]
Length = 472
Score = 589 bits (1518), Expect = e-167
Identities = 302/465 (64%), Positives = 358/465 (76%), Gaps = 18/465 (3%)
Query: 24 VPSSVEPNMLPKSTMIPAT---SWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTC 80
V VE L ST A SWFTPKRLL +FC+INMLNY+DRGAIASNGVNG R +C
Sbjct: 5 VARDVEAGGLDASTSGAADEKPSWFTPKRLLVMFCLINMLNYVDRGAIASNGVNGSRQSC 64
Query: 81 TDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVW 140
T G C SG+GIQGDFNL NF+DGVLSSAFMVGLLIASPIFASL+K NPFRLIGVGL VW
Sbjct: 65 TGGTCTSGSGIQGDFNLNNFEDGVLSSAFMVGLLIASPIFASLAKIHNPFRLIGVGLLVW 124
Query: 141 TVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPG 200
T+AT CG SF+FWSIT+CRMLVGVGEASFISLAAPFIDDNAPA+QKT WL++FYMCIP
Sbjct: 125 TIATAGCGCSFDFWSITICRMLVGVGEASFISLAAPFIDDNAPAAQKTAWLAMFYMCIPT 184
Query: 201 GYAIGYVYGGVVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKD-QLSLFLKDMKELL 259
G A+GYVYGG+VG+ WR AFW E++LMLPF ILGFV+KPL+LK S+ K+ E+L
Sbjct: 185 GIAVGYVYGGLVGNSLHWRAAFWGESILMLPFVILGFVIKPLELKGFNHSVKTKEYGEML 244
Query: 260 SDK----------VFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADMIFGGITIVC 309
+ + + V + + + F I ++ G K I + D++FGGITIVC
Sbjct: 245 NPERQDETKQGASIGVDGLAETLPHKFSISSF---GKKVLTEIKHFMK-DIMFGGITIVC 300
Query: 310 GILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIGELLV 369
GI GTL+GG +LD + +T+SNAFKLLS T +G FCFGAF FKS+YGF+ F++GELLV
Sbjct: 301 GIFGTLSGGFILDKIDSTISNAFKLLSGATFLGAIFCFGAFCFKSLYGFIPFFSVGELLV 360
Query: 370 FATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTALIL 429
FATQ PVN+VCLHCVKPSLRPLSMAMSTVAIHIFGDVPS+PLVG++QD I+NWR+TAL L
Sbjct: 361 FATQAPVNYVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSSPLVGLLQDKIHNWRSTALTL 420
Query: 430 TTIFFPAAAIWFIGIFLNSKDKFNEESEHQVSRVEGTTTAPLLEE 474
T+I F AA WFIGIF+ S D+FNE+SEH V VE + PLL+E
Sbjct: 421 TSILFIAAIFWFIGIFVRSVDRFNEQSEHDVPAVERSNLRPLLDE 465
>UniRef100_Q6NMN6 At5g65687 [Arabidopsis thaliana]
Length = 492
Score = 576 bits (1484), Expect = e-163
Identities = 280/457 (61%), Positives = 339/457 (73%), Gaps = 39/457 (8%)
Query: 40 PATS--WFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTC-TDGICKSGTGIQGDFN 96
PAT + TP R + I C+IN++NY+DRG IASNGVNG C G+C +GTGIQG+FN
Sbjct: 17 PATKKRFLTPGRFVTILCIINLINYVDRGVIASNGVNGSSKVCDAKGVCSAGTGIQGEFN 76
Query: 97 LTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSI 156
LTNF+DG+LSSAFMVGLL+ASPIFA LSK NPF+LIGVGL+VWT+A + CG S+NFW I
Sbjct: 77 LTNFEDGLLSSAFMVGLLVASPIFAGLSKRFNPFKLIGVGLTVWTIAVIGCGFSYNFWMI 136
Query: 157 TVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHF 216
V RM VGVGEASFISLAAP+IDD+AP ++K WL +FYMCIP G A+GYV+GG +G+H
Sbjct: 137 AVFRMFVGVGEASFISLAAPYIDDSAPVARKNFWLGLFYMCIPAGVALGYVFGGYIGNHL 196
Query: 217 GWRYAFWVEAVLMLPFAILGFVMKPLQL-------------------------------- 244
GWR+AF++EA+ M F IL F +KP Q
Sbjct: 197 GWRWAFYIEAIAMAVFVILSFCIKPPQQLKGFADKDSKKPSTSIETVAPTDAEASQIKTK 256
Query: 245 ----KDQLSLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADM 300
K+ + LF KD+K L S+KVF+VNVLGYI YNFVIGAYSYWGPKAG+ IY M NADM
Sbjct: 257 TPKSKNLVVLFGKDLKALFSEKVFIVNVLGYITYNFVIGAYSYWGPKAGFGIYKMKNADM 316
Query: 301 IFGGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLA 360
IFGG+TI+CGI+GTL G VLD + TLSN FKLL+ +TL+G AFCF AF K+MY F+A
Sbjct: 317 IFGGLTIICGIIGTLGGSYVLDRINATLSNTFKLLAASTLLGAAFCFTAFLMKNMYAFIA 376
Query: 361 LFAIGELLVFATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHIN 420
LFA+GE+L+FA Q PVNFVCLHCV+P+LRPLSMA STV IHI GDVPS+PL G +QDH+
Sbjct: 377 LFAVGEILIFAPQAPVNFVCLHCVRPNLRPLSMASSTVLIHILGDVPSSPLYGKMQDHLK 436
Query: 421 NWRTTALILTTIFFPAAAIWFIGIFLNSKDKFNEESE 457
NWR + LI+T+I F AA IW IGIF+NS D+ NE SE
Sbjct: 437 NWRKSTLIITSILFLAAIIWGIGIFMNSVDRSNEVSE 473
>UniRef100_O49546 Predicted protein [Arabidopsis thaliana]
Length = 746
Score = 558 bits (1439), Expect = e-157
Identities = 274/463 (59%), Positives = 333/463 (71%), Gaps = 54/463 (11%)
Query: 49 RLLAIFCVINMLNYLDRGAIASNGVNGHRGTC-TDGICKSGTGIQGDFNLTNFQDGVLSS 107
+ + I C+IN++NY+DRG IASNGVNG C G+C +GTGIQG+FNLTNF+DG+LSS
Sbjct: 265 KFVTILCIINLINYVDRGVIASNGVNGSSKVCDAKGVCSAGTGIQGEFNLTNFEDGLLSS 324
Query: 108 AFMVGLLIASPIFASLSKSVN-----------------PFRLIGVGLSVWTVATLCCGLS 150
AFMVGLL+ASPIFA LSK N PF+LIGVGL+VWT+A + CG S
Sbjct: 325 AFMVGLLVASPIFAGLSKRFNFYQQFHIFVFLFFGVFNPFKLIGVGLTVWTIAVIGCGFS 384
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
+NFW I V RM VGVGEASFISLAAP+IDD+AP ++K WL +FYMCIP G A+GYV+GG
Sbjct: 385 YNFWMIAVFRMFVGVGEASFISLAAPYIDDSAPVARKNFWLGLFYMCIPAGVALGYVFGG 444
Query: 211 VVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQL-------------------------- 244
+G+H GWR+AF++EA+ M F IL F +KP Q
Sbjct: 445 YIGNHLGWRWAFYIEAIAMAVFVILSFCIKPPQQLKGFADKDSKKPSTSIETVAPTDAEA 504
Query: 245 ----------KDQLSLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYN 294
K+ + LF KD+K L S+KVF+VNVLGYI YNFVIGAYSYWGPKAG+ IY
Sbjct: 505 SQIKTKTPKSKNLVVLFGKDLKALFSEKVFIVNVLGYITYNFVIGAYSYWGPKAGFGIYK 564
Query: 295 MTNADMIFGGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKS 354
M NADMIFGG+TI+CGI+GTL G VLD + TLSN FKLL+ +TL+G AFCF AF K+
Sbjct: 565 MKNADMIFGGLTIICGIIGTLGGSYVLDRINATLSNTFKLLAASTLLGAAFCFTAFLMKN 624
Query: 355 MYGFLALFAIGELLVFATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGV 414
MY F+ALFA+GE+L+FA Q PVNFVCLHCV+P+LRPLSMA STV IHI GDVPS+PL G
Sbjct: 625 MYAFIALFAVGEILIFAPQAPVNFVCLHCVRPNLRPLSMASSTVLIHILGDVPSSPLYGK 684
Query: 415 VQDHINNWRTTALILTTIFFPAAAIWFIGIFLNSKDKFNEESE 457
+QDH+ NWR + LI+T+I F AA IW IGIF+NS D+ NE SE
Sbjct: 685 MQDHLKNWRKSTLIITSILFLAAIIWGIGIFMNSVDRSNEVSE 727
>UniRef100_Q5XGK0 Hypothetical protein [Xenopus laevis]
Length = 526
Score = 160 bits (405), Expect = 8e-38
Identities = 126/409 (30%), Positives = 187/409 (44%), Gaps = 45/409 (11%)
Query: 50 LLAIFCVINMLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAF 109
++ I IN+LNY+DR +A G D I+ FN+++ G++ + F
Sbjct: 61 IVIILFYINLLNYMDRFTVA--------GVLPD--------IKKAFNISDSNSGLVQTVF 104
Query: 110 MVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFN--FWSITVCRMLVGVGE 167
+ + +P+F L N ++ VG+S W++ TL N FW + R LVGVGE
Sbjct: 105 ICSYMFLAPVFGYLGDRYNRKLIMCVGISFWSLVTLLSSFVSNQYFWLFLITRGLVGVGE 164
Query: 168 ASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAFWVE- 225
AS+ ++A I D A Q+T LS FY P G +GY+ G + S G W +A V
Sbjct: 165 ASYSTIAPTIIADLFLADQRTRMLSFFYFATPVGCGLGYIVGSEMTSAAGDWHWALRVTP 224
Query: 226 -----AVLMLPFAILGFVMKPLQLKDQLSL----FLKDMKELLSDKVFVVNVLGYIAYNF 276
AVL+L F L+ K L + DMK LL + F+++ G+ F
Sbjct: 225 GLGLLAVLLLIFVAEEPPRGALERKTDRPLTNTSWSSDMKALLKNPSFILSTFGFTTVAF 284
Query: 277 VIGAYSYWGP----KAGYSIY-------NMTNAD--MIFGGITIVCGILGTLAGGLVLDY 323
V GA + WGP ++ IY + N D MIFGGIT + GILG L G +
Sbjct: 285 VTGALALWGPTYLMRSRMVIYKSKPCEGGICNYDDSMIFGGITCITGILGVLTGVEISKR 344
Query: 324 MTNTLSNAFKLLSLTTLVGGA---FCFGAFAFKSMYGFLALFAIGELLVFATQGPVNFVC 380
T A L+ ++ A F AFA S+ IGE L+ V +
Sbjct: 345 YRKTNPRADPLVCAVGMISSAPFLFLSLAFADTSLVATYVFIFIGETLLSLNWALVADIL 404
Query: 381 LHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTALIL 429
L+ V P+ R + A+ V H+ GD S L+GV+ D I + + ++
Sbjct: 405 LYVVIPTRRSTAEALQIVVSHLLGDAGSPYLIGVISDQIQKGKPASFLI 453
>UniRef100_Q6P182 SPINL protein [Homo sapiens]
Length = 528
Score = 149 bits (377), Expect = 1e-34
Identities = 134/460 (29%), Positives = 211/460 (45%), Gaps = 62/460 (13%)
Query: 2 AQKSEDEPKPATTTSSSSTPNPVPSSVEPNMLPKSTMIPATSWFTPKR---LLAIFCVIN 58
A +D P P T ST NP S EP + P + + +P R ++A+ C IN
Sbjct: 13 ADDPDDGPVPGTPGLPGSTGNP--KSEEPEV-PDQEGLQRITGLSPGRSALIVAVLCYIN 69
Query: 59 MLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASP 118
+LNY+DR +A G D I+ FN+ + G++ + F+ ++ +P
Sbjct: 70 LLNYMDRFTVA--------GVLPD--------IEQFFNIGDSSSGLIQTVFISSYMVLAP 113
Query: 119 IFASLSKSVNPFRLIGVGLSVWTVATLCCGLSF----NFWSITVCRMLVGVGEASFISLA 174
+F L N L+ G++ W++ TL G SF +FW + + R LVGVGEAS+ ++A
Sbjct: 114 VFGYLGDRYNRKYLMCGGIAFWSLVTL--GSSFIPGEHFWLLLLTRGLVGVGEASYSTIA 171
Query: 175 APFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAFWVEAVL-MLPF 232
I D A Q++ LSIFY IP G +GY+ G V G W +A V L ++P
Sbjct: 172 PTLIADLFVADQRSRMLSIFYFAIPVGSGLGYIAGSKVKDMAGDWHWALRVTPGLGVVPV 231
Query: 233 AILGFVMK-----PLQLKDQL-----SLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYS 282
+L V++ ++ L + + D++ L + FV++ LG+ A FV G+ +
Sbjct: 232 LLLFLVVREPPRGAVERHSDLPPLNPTSWWADLRALARNPSFVLSSLGFTAVAFVTGSLA 291
Query: 283 YWGPK----------------AGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMTN 326
W P G S ++ +IFG IT + G+LG G + + +
Sbjct: 292 LWAPAFLLRSRVVLGETPPCLPGDSC--SSSDSLIFGLITCLTGVLGVGLGVEISRRLRH 349
Query: 327 TLSNAFKLLSLTTLVGGA---FCFGAFAFKSMYGFLALFAIGELLVFATQGPVNFVCLHC 383
+ A L+ T L+G A F A A S+ IGE L+ V + L+
Sbjct: 350 SNPRADPLVCATGLLGSAPFLFLSLACARGSIVATYIFIFIGETLLSMNWAIVADILLYV 409
Query: 384 VKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHI-NNW 422
V P+ R + A V H+ GD S L+G++ D + NW
Sbjct: 410 VIPTRRSTAEAFQIVLSHLLGDAGSPYLIGLISDRLRRNW 449
>UniRef100_Q9GQQ2 Spinster type II [Drosophila melanogaster]
Length = 630
Score = 149 bits (376), Expect = 2e-34
Identities = 116/423 (27%), Positives = 195/423 (45%), Gaps = 51/423 (12%)
Query: 21 PNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTC 80
P PS+V P+ L + + + WFT + + C +N++NY+DR IA G
Sbjct: 94 PGIPPSAVVPSRL---SSVGRSQWFT----VTVLCFVNLINYMDRFTIA--------GVL 138
Query: 81 TDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVW 140
TD ++ DF++ N G+L + F++ ++ +PIF L + ++ VG+ +W
Sbjct: 139 TD--------VRNDFDIGNDSAGLLQTVFVISYMVCAPIFGYLGDRYSRPWIMAVGVGLW 190
Query: 141 TVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPG 200
+ TL F R LVG+GEAS+ ++A I D ++ L++FY IP
Sbjct: 191 STTTLLGSFMKQFGWFIAFRALVGIGEASYSTIAPTIISDLFVHDMRSKMLALFYFAIPV 250
Query: 201 GYAIGYVYGGVVGSHFG--WRYAFWVEAVLMLPFAILGFVMK-PLQLKD------QLSLF 251
G +GY+ G +H WR+A V +L + L ++K P++ + + +
Sbjct: 251 GSGLGYIVGSKT-AHLANDWRWALRVTPILGIVAVFLILLIKDPVRGHSEGSHNLEATTY 309
Query: 252 LKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMT--NADMI-------F 302
+D+K L+ ++ F+++ G+ FV GA ++WGP Y M N +++ F
Sbjct: 310 KQDIKALVRNRSFMLSTAGFTCVAFVAGALAWWGPSFIYLGMKMQPGNENIVQDDVAFNF 369
Query: 303 GGITIVCGILGTLAGGLVLDYMTNTLSN------AFKLLSLTTLVGGAFCFGAFAFKSMY 356
G IT++ G+LG G + Y+ AF LL L+ GA C +
Sbjct: 370 GVITMLAGLLGVPLGSFLSQYLVKRYPTADPVICAFGLLVSAPLLTGA-CL--LVNSNSV 426
Query: 357 GFLALFAIGELLVFATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQ 416
G AL G+L + V + L+ V P+ R + A + H GD S LVG +
Sbjct: 427 GTYALIFFGQLALNLNWAIVADILLYVVVPTRRSTAEAFQILISHALGDAGSPYLVGAIS 486
Query: 417 DHI 419
+ I
Sbjct: 487 EAI 489
>UniRef100_Q9GQQ0 Spinster type IV [Drosophila melanogaster]
Length = 605
Score = 149 bits (376), Expect = 2e-34
Identities = 116/423 (27%), Positives = 195/423 (45%), Gaps = 51/423 (12%)
Query: 21 PNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTC 80
P PS+V P+ L + + + WFT + + C +N++NY+DR IA G
Sbjct: 94 PGIPPSAVVPSRL---SSVGRSQWFT----VTVLCFVNLINYMDRFTIA--------GVL 138
Query: 81 TDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVW 140
TD ++ DF++ N G+L + F++ ++ +PIF L + ++ VG+ +W
Sbjct: 139 TD--------VRNDFDIGNDSAGLLQTVFVISYMVCAPIFGYLGDRYSRPWIMAVGVGLW 190
Query: 141 TVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPG 200
+ TL F R LVG+GEAS+ ++A I D ++ L++FY IP
Sbjct: 191 STTTLLGSFMKQFGWFIAFRALVGIGEASYSTIAPTIISDLFVHDMRSKMLALFYFAIPV 250
Query: 201 GYAIGYVYGGVVGSHFG--WRYAFWVEAVLMLPFAILGFVMK-PLQLKD------QLSLF 251
G +GY+ G +H WR+A V +L + L ++K P++ + + +
Sbjct: 251 GSGLGYIVGSKT-AHLANDWRWALRVTPILGIVAVFLILLIKDPVRGHSEGSHNLEATTY 309
Query: 252 LKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMT--NADMI-------F 302
+D+K L+ ++ F+++ G+ FV GA ++WGP Y M N +++ F
Sbjct: 310 KQDIKALVRNRSFMLSTAGFTCVAFVAGALAWWGPSFIYLGMKMQPGNENIVQDDVAFNF 369
Query: 303 GGITIVCGILGTLAGGLVLDYMTNTLSN------AFKLLSLTTLVGGAFCFGAFAFKSMY 356
G IT++ G+LG G + Y+ AF LL L+ GA C +
Sbjct: 370 GVITMLAGLLGVPLGSFLSQYLVKRYPTADPVICAFGLLVSAPLLTGA-CL--LVNSNSV 426
Query: 357 GFLALFAIGELLVFATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQ 416
G AL G+L + V + L+ V P+ R + A + H GD S LVG +
Sbjct: 427 GTYALIFFGQLALNLNWAIVADILLYVVVPTRRSTAEAFQILISHALGDAGSPYLVGAIS 486
Query: 417 DHI 419
+ I
Sbjct: 487 EAI 489
>UniRef100_Q7Q5N4 ENSANGP00000013028 [Anopheles gambiae str. PEST]
Length = 449
Score = 149 bits (375), Expect = 2e-34
Identities = 114/405 (28%), Positives = 190/405 (46%), Gaps = 50/405 (12%)
Query: 36 STMIPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDF 95
++ I A +WFT + + C +N++NY+DR IA G T+ IQ F
Sbjct: 10 TSSIGANAWFT----VGVLCFVNLINYMDRFTIA--------GVLTE--------IQDHF 49
Query: 96 NLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNF-W 154
+ + + G+L +AF++ ++ +P+F L + ++ +G+S+W+ TL +F W
Sbjct: 50 KIGDDEGGLLQTAFVLSYMVFAPLFGYLGDRYSRKWIMVLGVSLWSTTTLLGSYMDHFGW 109
Query: 155 SITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGS 214
IT R LVG+GEAS+ ++A I D ++ L++FY IP G +GY+ G + S
Sbjct: 110 FITF-RALVGIGEASYSTIAPTIISDLFVGEMRSRMLALFYFAIPVGSGLGYIVGAKMAS 168
Query: 215 HF-GWRYAFWVEAVLMLPFAILGFVMKPLQLKD-------QLSLFLKDMKELLSDKVFVV 266
W ++ V VL +L +++ Q Q + + +D+K ++ ++ F++
Sbjct: 169 IMNSWVWSLRVTPVLGAIAVVLIVMLRDPQRGQSEGTHHMQTTSYKEDVKAIMRNRSFML 228
Query: 267 NVLGYIAYNFVIGAYSYWGPK---------AGYSIYNMTNADMIFGGITIVCGILGTLAG 317
+ G+ FV GA ++WGPK G + IFG IT+ GI+G G
Sbjct: 229 STAGFTCVAFVTGALAWWGPKFIHLGLISQPGNEHVTLNEVSFIFGAITMTTGIIGVPLG 288
Query: 318 GLVLDYMT--NTLSNAF-----KLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIGELLVF 370
+ + N ++ + LLS LVG F A ++Y AL GEL +
Sbjct: 289 SYLSQRLNAKNVKADPYICATGLLLSAPLLVGAMFSVRA----NIYATYALIFFGELALN 344
Query: 371 ATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVV 415
V + L+ V P+ R + A + H FGD S VGVV
Sbjct: 345 LNWAIVADILLYVVVPTRRSTAEAFQILISHAFGDAGSPYFVGVV 389
>UniRef100_Q9H2V7 Spinster [Homo sapiens]
Length = 528
Score = 149 bits (375), Expect = 2e-34
Identities = 136/460 (29%), Positives = 209/460 (44%), Gaps = 62/460 (13%)
Query: 2 AQKSEDEPKPATTTSSSSTPNPVPSSVEPNMLPKSTMIPATSWFTPKR---LLAIFCVIN 58
A +D P P T ST NP S EP + P + + +P R ++A+ C IN
Sbjct: 13 ADDPDDGPVPGTPGLPGSTGNP--KSEEPEV-PDQEGLQRITGLSPGRSALIVAVLCYIN 69
Query: 59 MLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASP 118
+LNY+DR +A G D I+ FN+ + G++ + F+ ++ +P
Sbjct: 70 LLNYMDRFTVA--------GVLPD--------IEQFFNIGDSSSGLIQTVFISSYMVLAP 113
Query: 119 IFASLSKSVNPFRLIGVGLSVWTVATLCCGLSF----NFWSITVCRMLVGVGEASFISLA 174
+F L N L+ G++ W++ TL G SF +FW + + R LVGVGEAS+ ++A
Sbjct: 114 VFGYLGDRYNRKYLMCGGIAFWSLVTL--GSSFIPGEHFWLLLLTRGLVGVGEASYSTIA 171
Query: 175 APFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYA------FWVEAV 227
I D A Q++ LSIFY IP G +GY+ G V G W +A V AV
Sbjct: 172 PTLIADLFVADQRSRMLSIFYFAIPVGSGLGYIAGSKVKDMAGDWHWALRVTPGLGVVAV 231
Query: 228 LMLPFAI----LGFVMKPLQLKD-QLSLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYS 282
L+L + G V + L + + D++ L + FV++ LG+ A FV G+ +
Sbjct: 232 LLLFLVVREPPRGAVERHSDLPPLNPTSWWADLRALARNPSFVLSSLGFTAVAFVTGSLA 291
Query: 283 YWGPK----------------AGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMTN 326
W P G S ++ +IFG IT + G+LG G + + +
Sbjct: 292 LWAPAFLLRSRVVLGETPPCLPGDSC--SSSDSLIFGLITCLTGVLGVGLGVEISRRLRH 349
Query: 327 TLSNAFKLLSLTTLVGGA---FCFGAFAFKSMYGFLALFAIGELLVFATQGPVNFVCLHC 383
+ A L+ T L+G A F A A S+ IGE L+ V + L+
Sbjct: 350 SNPRADPLVCATGLLGSAPFLFLSLACARGSIVATYIFIFIGETLLSMNWAIVADILLYV 409
Query: 384 VKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHI-NNW 422
V P+ R + A V H+ GD S L+G++ D + NW
Sbjct: 410 VIPTRRSTAEAFQIVLSHLLGDAGSPYLIGLISDRLRRNW 449
>UniRef100_UPI0000318BDA UPI0000318BDA UniRef100 entry
Length = 445
Score = 147 bits (371), Expect = 7e-34
Identities = 112/398 (28%), Positives = 186/398 (46%), Gaps = 45/398 (11%)
Query: 51 LAIFCVINMLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFM 110
+A+ C +N++NY++R IA N IQ F +++ +L + F+
Sbjct: 10 IAVLCYVNLVNYIERYTIAGVLPN----------------IQEYFLISDATAALLQTVFI 53
Query: 111 VGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFN--FWSITVCRMLVGVGEA 168
L+ +P+F L N L+ VGL +WT + CC FW + + R LVG+GEA
Sbjct: 54 CSFLLLAPLFGYLGDRYNRKYLMIVGLIMWTFTSFCCSFVTESYFWVLVLLRALVGIGEA 113
Query: 169 SFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAFWVEAV 227
S+ ++A I D ++++V + +FY+ IP G +G++ G V S G W +AF + +
Sbjct: 114 SYTTIAPTIIGDLFSGARRSVMICVFYILIPVGSGLGFIIGAGVASQTGDWHWAFRINPI 173
Query: 228 L-MLPFAILGFVM--KPLQLKD-------QLSLFLKDMKELLSDKVFVVNVLGYIAYNFV 277
++ A+L F+ P + Q S +L+D+K LL K +V + LG A F
Sbjct: 174 FGVVGVALLVFLCPNPPRGAAETGGEGVRQQSSYLEDIKYLLKIKSYVWSTLGITASTFN 233
Query: 278 IGAYSYWGP------------KAGYSIYNMTNADMI-FGGITIVCGILGTLAGGLVLDYM 324
+GA ++W P G + + + D FG +T+V GILG G L+
Sbjct: 234 LGALAFWMPTFLSRARLLQGLNQGCTNGSCQSTDSYGFGVVTMVTGILGGCVGTLLSRSF 293
Query: 325 TNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFA---IGELLVFATQGPVNFVCL 381
+ + + L+ L+G CF F + A + +G LV + + L
Sbjct: 294 RDRVPHVDPLICAVGLLGSVPCFIISMFTATASIAASYVFAFLGLCLVALNWAVMADILL 353
Query: 382 HCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHI 419
+ V P+ R + A+ + IH+ GD S +VG V D I
Sbjct: 354 YIVIPNRRSTAEALQVMFIHLLGDCGSPYIVGAVSDAI 391
>UniRef100_O01927 Hypothetical protein C13C4.5 [Caenorhabditis elegans]
Length = 531
Score = 144 bits (363), Expect = 6e-33
Identities = 136/530 (25%), Positives = 227/530 (42%), Gaps = 81/530 (15%)
Query: 8 EPKPATTTSSSSTPNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVINMLNYLDRGA 67
EP P TT + S + + S N + P ++AI +IN+LNY+DR
Sbjct: 20 EPPPPYTTPTDSPEDKIRS----NSTATTASQPEFQGCWTIVVVAILFIINLLNYMDRYT 75
Query: 68 IASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSV 127
IA G D +Q +N+++ G++ + FMV +I SPI L
Sbjct: 76 IA--------GVLND--------VQTYYNISDAWAGLIQTTFMVFFIIFSPICGFLGDRY 119
Query: 128 NPFRLIGVGLSVWTVATLCCGL--SFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPAS 185
N + VG+++W A S FW + R +VG+GEAS+ ++ I D
Sbjct: 120 NRKWIFVVGIAIWVSAVFASTFIPSNQFWLFLLFRGIVGIGEASYAIISPTVIADMFTGV 179
Query: 186 QKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAFWVEAVL--MLPFAILGFVMKP- 241
++ L +FY IP G +G+V G V S G W++ V VL + I+ FV +P
Sbjct: 180 LRSRMLMVFYFAIPFGCGLGFVVGSAVASWTGHWQWGVRVTGVLGIVCLLLIIVFVREPE 239
Query: 242 ----------LQLKDQLSLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGP----K 287
+ + + +L DMK+LLS+ +V + LGY A F++G ++W P
Sbjct: 240 RGKAEREKGEIAASTEATSYLDDMKDLLSNATYVTSSLGYTATVFMVGTLAWWAPITIQY 299
Query: 288 AGYSIYNMT-------NADMIFGGITIVCGILGTLAGGLVLDYMTNTLS-----NAFKLL 335
A + N T N +++FG +T V G+LG G LV + + + +
Sbjct: 300 ADSARRNGTITEDQKANINLVFGALTCVGGVLGVAIGTLVSNMWSRGVGPFKHIQTVRAD 359
Query: 336 SLTTLVGGAFCFGA--FAFKSMYGFLALFAIGELLVFATQGPVNF-----VCLHCVKPSL 388
+L +G A C A +++ + FA G L + N+ + L V P
Sbjct: 360 ALVCAIGAAICIPTLILAIQNIESNMN-FAWGMLFICIVASSFNWATNVDLLLSVVVPQR 418
Query: 389 RPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTA-----------------LILTT 431
R + + + H+FGD ++G++ D I TA L+L+
Sbjct: 419 RSSASSWQILISHMFGDASGPYILGLISDAIRGNEDTAQAHYKSLVTSFWLCVGTLVLSV 478
Query: 432 IFFPAAAIWFIGIFLNSKDKFNEESEHQVSRVEGTTTAPLLEEKTAEPKS 481
I F +AI + K +FNE Q ++ ++ +E++ E ++
Sbjct: 479 ILFGISAITVV----KDKARFNEIMLAQANKDNTSSGTLPIEDRNTEDET 524
>UniRef100_Q8IVW8 Similar to spinster-like protein [Homo sapiens]
Length = 619
Score = 142 bits (359), Expect = 2e-32
Identities = 122/450 (27%), Positives = 194/450 (43%), Gaps = 63/450 (14%)
Query: 52 AIFCVINMLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMV 111
AI + N+LNYLDR +A G D IQ F + + G+L S F+
Sbjct: 176 AILSLGNVLNYLDRYTVA--------GVLLD--------IQQHFGVKDRGAGLLQSVFIC 219
Query: 112 GLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGL--SFNFWSITVCRMLVGVGEAS 169
++A+PIF L N ++ G+ W+ T FW + + R LVG+GEAS
Sbjct: 220 SFMVAAPIFGYLGDRFNRKVILSCGIFFWSAVTFSSSFIPQQYFWLLVLSRGLVGIGEAS 279
Query: 170 FISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAFWVEAVL 228
+ ++A I D + +T+ LS+FY IP G +GY+ G V G W +A V VL
Sbjct: 280 YSTIAPTIIGDLFTKNTRTLMLSVFYFAIPLGSGLGYITGSSVKQAAGDWHWALRVSPVL 339
Query: 229 MLPFAILGFVMKPL-------QLKDQL---SLFLKDMKELLSDKVFVVNVLGYIAYNFVI 278
+ L ++ P QL DQL + +L+DMK L+ ++ +V + L A +F
Sbjct: 340 GMITGTLILILVPATKRGHADQLGDQLKARTSWLRDMKALIRNRSYVFSSLATSAVSFAT 399
Query: 279 GAYSYWGPKAGYSIYNMTNA-------------DMIFGGITIVCGILGTLAGGLVLDYMT 325
GA W P + + +IFG IT G LG + G +
Sbjct: 400 GALGMWIPLYLHRAQVVQKTAETCNSPPCGAKDSLIFGAITCFTGFLGVVTGAGATRWCR 459
Query: 326 NTLSNAFKLLSLTTLVGGA--FCFGAFAFK-SMYGFLALFAIGELLVFATQGPVNFVCLH 382
A L+ ++G A C A K S+ G +GE L+F+ + ++
Sbjct: 460 LKTQRADPLVCAVGMLGSAIFICLIFVAAKSSIVGAYICIFVGETLLFSNWAITADILMY 519
Query: 383 CVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINN-------WRTTAL-------- 427
V P+ R ++A+ + H+ GD S L+G + D I W +L
Sbjct: 520 VVIPTRRATAVALQSFTSHLLGDAGSPYLIGFISDLIRQSTKDSPLWEFLSLGYALMLCP 579
Query: 428 ---ILTTIFFPAAAIWFIGIFLNSKDKFNE 454
+L +FF A A++F+ ++ + N+
Sbjct: 580 FVVVLGGMFFLATALFFVSDRARAEQQVNQ 609
>UniRef100_UPI00003374A0 UPI00003374A0 UniRef100 entry
Length = 527
Score = 142 bits (358), Expect = 2e-32
Identities = 134/505 (26%), Positives = 215/505 (42%), Gaps = 72/505 (14%)
Query: 5 SEDEPKPATTTSSSST-PNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVINMLNYL 63
S DE + + + S PNP +P ++ ++ TP I N+LNY+
Sbjct: 6 SSDEVRTLSGSMSPGLKPNPDLHPCKPGQKFRAALLRCR---TPTAAAGILSFGNVLNYM 62
Query: 64 DRGAIASNGVNG--HRGTCTDG--------------ICKSGT--GIQGDFNLTNFQDGVL 105
DR +A + + G H G C SG IQ + +T+ G+L
Sbjct: 63 DRYTVA-DALAGKVHAGECDHAGIVMATKVGAATLVAASSGVLLDIQRHYGVTDSGIGLL 121
Query: 106 SSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFN--FWSITVCRMLV 163
+ F+ ++A+PIF L N ++ G+ W+V TL +W + R LV
Sbjct: 122 QTVFICSFMVAAPIFGYLGDRFNRKVILSCGIFFWSVITLSSSFIGEEYYWLFVLSRGLV 181
Query: 164 GVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAF 222
G+GE+S+ S++ I D + +T LSIFY+ IP G +GY+ G G W +A
Sbjct: 182 GIGESSYSSISPTIIGDLFTNNSRTTMLSIFYLAIPLGSGLGYILGSSAKVAAGDWHWAL 241
Query: 223 WVEAVLMLPFA--ILGFVMKPL---------QLKDQLSLFLKDMKELLSDKVFVVNVLGY 271
V VL + IL FV +P ++K + S ++ DMK L ++ +V + L
Sbjct: 242 RVSPVLGITTGTLILVFVPEPKRGSADQVRGRIKSRTS-WVCDMKALAKNRSYVFSSLAS 300
Query: 272 IAYNFVIGAYSYWGP---KAGYSIYNMTNA----------DMIFGGITIVCGILGTLAGG 318
A +F GA+ P + N A +IFG IT V G+LG + G
Sbjct: 301 AAVSFATGAFGMLIPLYLTRAQMVQNPAEACTKEICSSTESLIFGAITCVTGLLGVVIGA 360
Query: 319 LVLDYMTNTLSNAFKLLSLTTLVGGAF---CFGAFAFKSMYGFLALFAIGELLVFATQGP 375
A L+ +++G A A KS+ G IGE L+F
Sbjct: 361 ATTRLFRQKTERADPLVCAVSMLGSAIFICLIFVVAKKSIVGAYVCIFIGETLLFVNWAI 420
Query: 376 VNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHI-NNWRTTAL------- 427
+ + V P+ R ++A + H+ GD S L+G++ D + N++T+AL
Sbjct: 421 TADILMFVVIPTRRATAVAFQSFTSHLLGDAGSPYLIGLISDALKENYKTSALWQFLSLG 480
Query: 428 ----------ILTTIFFPAAAIWFI 442
+L +FF A A++F+
Sbjct: 481 YALMLCPFIIVLGGMFFLATALFFL 505
>UniRef100_Q91VM4 BC011467 protein [Mus musculus]
Length = 590
Score = 140 bits (352), Expect = 1e-31
Identities = 131/492 (26%), Positives = 208/492 (41%), Gaps = 68/492 (13%)
Query: 11 PATTTSSSSTPNPVPSSVEPNM-LPKSTMIPATSWFTPKRLLAIFCVINMLNYLDRGAIA 69
P+ S STP ++ P+ PK PA+ AI + N+LNYLDR +A
Sbjct: 109 PSGLPSIPSTPGCAAAAKGPSAPQPK----PASLGRGRGAAAAILSLGNVLNYLDRYTVA 164
Query: 70 SNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNP 129
G D IQ F + + G+L S F+ ++A+PIF L N
Sbjct: 165 --------GVLLD--------IQQHFGVKDRGAGLLQSVFICSFMVAAPIFGYLGDRFNR 208
Query: 130 FRLIGVGLSVWTVATLCCGL--SFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQK 187
++ G+ W+ T FW + + R LVG+GEAS+ ++A I D + +
Sbjct: 209 KVILSCGIFFWSAVTFSSSFIPQQYFWLLVLSRGLVGIGEASYSTIAPTIIGDLFTKNTR 268
Query: 188 TVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAFWVEAVLMLPFAILGFVMKPL---- 242
T+ LS+FY IP G +GY+ G V G W +A V VL + L ++ P
Sbjct: 269 TLMLSVFYFAIPLGSGLGYITGSSVKQAAGDWHWALRVSPVLGMITGTLILILVPATKRG 328
Query: 243 ---QLKDQL---SLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMT 296
QL QL + +L+DMK L+ ++ +V + L A +F GA W P + +
Sbjct: 329 HADQLGGQLKARTSWLRDMKALIRNRSYVFSSLATSAVSFATGALGMWIPLYLHRAQVVQ 388
Query: 297 NA-------------DMIFGGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGG 343
+IFG IT G LG + G + A L+ ++G
Sbjct: 389 KTAETCNSPPCGAKDSLIFGAITCFTGFLGVVTGAGATRWCRLRTQRADPLVCAVGMLGS 448
Query: 344 A--FCFGAFAFK-SMYGFLALFAIGELLVFATQGPVNFVCLHCVKPSLRPLSMAMSTVAI 400
A C A K S+ G +GE L+F+ + ++ V P+ R ++A+ +
Sbjct: 449 AIFICLIFVAAKTSIVGAYICIFVGETLLFSNWAITADILMYVVIPTRRATAVALQSFTS 508
Query: 401 HIFGDVPSAPLVGVVQDHINN-------WRTTAL-----------ILTTIFFPAAAIWFI 442
H+ GD S L+G + D I W +L +L +FF A A++F+
Sbjct: 509 HLLGDAGSPYLIGFISDLIRQSTKDSPLWEFLSLGYALMLCPFVVVLGGMFFLATALFFL 568
Query: 443 GIFLNSKDKFNE 454
++ + N+
Sbjct: 569 SDRAKAEQQVNQ 580
>UniRef100_Q5RD37 Hypothetical protein DKFZp459J1933 [Pongo pygmaeus]
Length = 652
Score = 139 bits (349), Expect = 2e-31
Identities = 120/432 (27%), Positives = 185/432 (42%), Gaps = 63/432 (14%)
Query: 58 NMLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIAS 117
N+LNYLDR +A G D IQ F + + G+L S F+ ++A+
Sbjct: 3 NVLNYLDRYTVA--------GVLLD--------IQQHFGVKDRGAGLLQSVFICSFMVAA 46
Query: 118 PIFASLSKSVNPFRLIGVGLSVWTVATLCCGL--SFNFWSITVCRMLVGVGEASFISLAA 175
PIF L N ++ G+ W+ T FW + + R LVG+GEAS+ ++A
Sbjct: 47 PIFGYLGDRFNRKVILSCGIFFWSAVTFSSSFIPQQYFWLLVLSRGLVGIGEASYSTIAP 106
Query: 176 PFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG-WRYAFWVEAVLMLPFAI 234
I D + +T+ LS+FY IP G +GY+ G V G W A V VL +
Sbjct: 107 TIIGDLFTKNTRTLMLSVFYFAIPLGSGLGYITGSSVKQAAGDWHRALRVSPVLGMITGT 166
Query: 235 LGFVMKPL-------QLKDQL---SLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYW 284
L ++ P QL DQL + +L+DMK L+ ++ +V + L A +F GA W
Sbjct: 167 LILILVPATKRGHADQLGDQLKARTSWLRDMKALIRNRSYVFSSLATSAVSFATGALGMW 226
Query: 285 GP---------KAGYSIYNM----TNADMIFGGITIVCGILGTLAGGLVLDYMTNTLSNA 331
P + YN +IFG IT G LG + G + A
Sbjct: 227 IPLYLHRAQVVQKTAETYNSPPCGAKDSLIFGAITCFTGFLGVVTGAGATRWCRLKTQRA 286
Query: 332 FKLLSLTTLVGGA--FCFGAFAFK-SMYGFLALFAIGELLVFATQGPVNFVCLHCVKPSL 388
L+ ++G C A K S+ G +GE L+F+ + ++ V P+
Sbjct: 287 DPLVCAVGMLGSTIFICLIFVAAKSSIVGAYICIFVGETLLFSNWAITADILMYVVIPTR 346
Query: 389 RPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINN-------WRTTAL-----------ILT 430
R ++A+ + H+ GD S L+G + D I W +L +L
Sbjct: 347 RATAVALQSFTSHLLGDAGSPYLIGFISDLIRQSTKDSPLWEFLSLGYALMLCPFVVVLG 406
Query: 431 TIFFPAAAIWFI 442
+FF A A++F+
Sbjct: 407 GMFFLATALFFL 418
>UniRef100_Q9GQQ3 Spinster type I [Drosophila melanogaster]
Length = 630
Score = 138 bits (348), Expect = 3e-31
Identities = 108/425 (25%), Positives = 188/425 (43%), Gaps = 55/425 (12%)
Query: 21 PNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTC 80
P PS+V P+ L + + + WFT + + C +N++NY+DR IA G
Sbjct: 94 PGIPPSAVVPSRL---SSVGRSQWFT----VTVLCFVNLINYMDRFTIA--------GVL 138
Query: 81 TDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVW 140
TD ++ DF++ N G+L + F++ ++ +PIF L + ++ VG+ +W
Sbjct: 139 TD--------VRNDFDIGNDSAGLLQTVFVISYMVCAPIFGYLGDRYSRPWIMAVGVGLW 190
Query: 141 TVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPG 200
+ TL F R LVG+GEAS+ ++A I D ++ L++FY IP
Sbjct: 191 STTTLLGSFMKQFGWFIAFRALVGIGEASYSTIAPTIISDLFVHDMRSKMLALFYFAIPV 250
Query: 201 GYAIGYVYGGVVGSHFG--WRYAFWVEAVLMLPFAILGFVMK-PLQLKD------QLSLF 251
G +GY+ G +H WR+A V +L + L ++K P++ + + +
Sbjct: 251 GSGLGYIVGSKT-AHLANDWRWALRVTPILGIVAVFLILLIKDPVRGHSEGSHNLEATTY 309
Query: 252 LKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMT--NADMI-------F 302
+D+K L+ ++ F+++ G+ FV GA ++WGP Y M N +++ F
Sbjct: 310 KQDIKALVRNRSFMLSTAGFTCVAFVAGALAWWGPSFIYLGMKMQPGNENIVQDDISYKF 369
Query: 303 GGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTL-VGGAFCFGAFAFKSMYGFLAL 361
G + ++ G++G G + + N + L + F A L
Sbjct: 370 GLVAMLAGLIGVPLGSFLAQRLRGRYENCDPYICAVGLFISAPMVFAALVVPQTSESLCF 429
Query: 362 FAIGELLVFATQGPVNF-------VCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGV 414
F VF Q +N + L+ V P+ R + A + H GD S LVG
Sbjct: 430 F-----FVFVAQVALNLCWSIVADILLYVVVPTRRSTAEAFQILISHALGDAGSPYLVGA 484
Query: 415 VQDHI 419
+ + I
Sbjct: 485 ISEAI 489
>UniRef100_Q9GQQ1 Spinster type III [Drosophila melanogaster]
Length = 605
Score = 138 bits (348), Expect = 3e-31
Identities = 108/425 (25%), Positives = 188/425 (43%), Gaps = 55/425 (12%)
Query: 21 PNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTC 80
P PS+V P+ L + + + WFT + + C +N++NY+DR IA G
Sbjct: 94 PGIPPSAVVPSRL---SSVGRSQWFT----VTVLCFVNLINYMDRFTIA--------GVL 138
Query: 81 TDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVW 140
TD ++ DF++ N G+L + F++ ++ +PIF L + ++ VG+ +W
Sbjct: 139 TD--------VRNDFDIGNDSAGLLQTVFVISYMVCAPIFGYLGDRYSRPWIMAVGVGLW 190
Query: 141 TVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPG 200
+ TL F R LVG+GEAS+ ++A I D ++ L++FY IP
Sbjct: 191 STTTLLGSFMKQFGWFIAFRALVGIGEASYSTIAPTIISDLFVHDMRSKMLALFYFAIPV 250
Query: 201 GYAIGYVYGGVVGSHFG--WRYAFWVEAVLMLPFAILGFVMK-PLQLKD------QLSLF 251
G +GY+ G +H WR+A V +L + L ++K P++ + + +
Sbjct: 251 GSGLGYIVGSKT-AHLANDWRWALRVTPILGIVAVFLILLIKDPVRGHSEGSHNLEATTY 309
Query: 252 LKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMT--NADMI-------F 302
+D+K L+ ++ F+++ G+ FV GA ++WGP Y M N +++ F
Sbjct: 310 KQDIKALVRNRSFMLSTAGFTCVAFVAGALAWWGPSFIYLGMKMQPGNENIVQDDISYKF 369
Query: 303 GGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTL-VGGAFCFGAFAFKSMYGFLAL 361
G + ++ G++G G + + N + L + F A L
Sbjct: 370 GLVAMLAGLIGVPLGSFLAQRLRGRYENCDPYICAVGLFISAPMVFAALVVPQTSESLCF 429
Query: 362 FAIGELLVFATQGPVNF-------VCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGV 414
F VF Q +N + L+ V P+ R + A + H GD S LVG
Sbjct: 430 F-----FVFVAQVALNLCWSIVADILLYVVVPTRRSTAEAFQILISHALGDAGSPYLVGA 484
Query: 415 VQDHI 419
+ + I
Sbjct: 485 ISEAI 489
>UniRef100_Q7ZU13 Spinster-like [Brachydanio rerio]
Length = 506
Score = 137 bits (344), Expect = 9e-31
Identities = 115/419 (27%), Positives = 180/419 (42%), Gaps = 48/419 (11%)
Query: 33 LPKSTMIPATSWFTPKRLLA---IFCVINMLNYLDRGAIASNGVNGHRGTCTDGICKSGT 89
LP+ + S T +R + + C IN+LNY+DR +A G D
Sbjct: 30 LPEDEEEESPSGVTDRRAIMTVIVLCYINLLNYMDRFTVA--------GVLPD------- 74
Query: 90 GIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGL 149
I+ F + + G+L + F+ + +P+F L N ++ VG+ W+V TL
Sbjct: 75 -IEHFFGIGDGTSGLLQTVFICSYMFLAPLFGYLGDRYNRKLIMCVGIFFWSVVTLASSF 133
Query: 150 --SFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYV 207
+FW++ + R LVGVGEAS+ ++A I D ++T LSIFY IP G +GY+
Sbjct: 134 IGKDHFWALLLTRGLVGVGEASYSTIAPTIIADLFVKEKRTNMLSIFYFAIPVGSGMGYI 193
Query: 208 YGG---VVGSHFGWRYAFWVEAVLMLPFAILGFVMKP----LQLKDQLSL----FLKDMK 256
G V + W L+ F ++ V +P ++ + +L +L DMK
Sbjct: 194 VGSKVDTVAKDWHWALRVTPGLGLLAVFLLMLVVQEPKRGAIEAHPEHTLHRTSWLADMK 253
Query: 257 ELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNA-------------DMIFG 303
L + F+++ G+ A FV G+ + W P + T +IFG
Sbjct: 254 ALCRNPSFILSTFGFTAVAFVTGSLALWAPAFLFRAGVFTGVKQPCFKAPCDDSDSLIFG 313
Query: 304 GITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGGA---FCFGAFAFKSMYGFLA 360
IT+V GILG +G + A L+ L+ A + FA S
Sbjct: 314 AITVVTGILGVASGVQASKLLRTRTPRADPLVCAAGLLLAAPFLYLSIMFAQASTVATYV 373
Query: 361 LFAIGELLVFATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHI 419
+GE + V + L+ V P+ R + A V H+ GD S L+GVV D I
Sbjct: 374 FIFLGETFLSMNWAIVADILLYVVIPTRRSTAEAFQIVLSHLLGDAISPYLIGVVSDSI 432
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.139 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 828,670,531
Number of Sequences: 2790947
Number of extensions: 36352480
Number of successful extensions: 165841
Number of sequences better than 10.0: 3593
Number of HSP's better than 10.0 without gapping: 1392
Number of HSP's successfully gapped in prelim test: 2206
Number of HSP's that attempted gapping in prelim test: 161830
Number of HSP's gapped (non-prelim): 4847
length of query: 484
length of database: 848,049,833
effective HSP length: 131
effective length of query: 353
effective length of database: 482,435,776
effective search space: 170299828928
effective search space used: 170299828928
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 77 (34.3 bits)
Medicago: description of AC124214.6


