

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124214.2 + phase: 0
(402 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
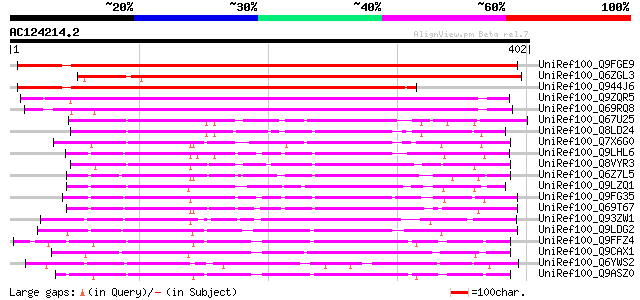
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FGE9 Gb|AAD15463.1 [Arabidopsis thaliana] 584 e-165
UniRef100_Q6ZGL3 Leaf senescence related protein-like [Oryza sat... 468 e-130
UniRef100_Q944J6 AT5g64470/T12B11_6 [Arabidopsis thaliana] 431 e-119
UniRef100_Q9ZQR5 Hypothetical protein At2g14530 [Arabidopsis tha... 317 3e-85
UniRef100_Q69RQ8 Leaf senescence related protein-like [Oryza sat... 311 2e-83
UniRef100_Q67U25 Lustrin A-like [Oryza sativa] 175 2e-42
UniRef100_Q8LD24 Hypothetical protein [Arabidopsis thaliana] 173 7e-42
UniRef100_Q7X6G0 OSJNBa0043L24.14 protein [Oryza sativa] 168 2e-40
UniRef100_Q9LHL6 Gb|AAF30301.1 [Arabidopsis thaliana] 166 1e-39
UniRef100_Q8VYR3 Hypothetical protein At1g60790 [Arabidopsis tha... 163 9e-39
UniRef100_Q6Z7L5 Hypothetical protein OJ1448_G06.4 [Oryza sativa] 160 5e-38
UniRef100_Q9LZQ1 Hypothetical protein T12C14_90 [Arabidopsis tha... 160 6e-38
UniRef100_Q9FG35 Emb|CAB82953.1 [Arabidopsis thaliana] 160 8e-38
UniRef100_Q69T67 Lustrin A-like [Oryza sativa] 159 1e-37
UniRef100_Q93ZW1 Hypothetical protein At5g20680 [Arabidopsis tha... 155 2e-36
UniRef100_Q9LDG2 Hypothetical protein F24F17.6 [Arabidopsis thal... 153 7e-36
UniRef100_Q9FFZ4 Gb|AAF00670.1 [Arabidopsis thaliana] 153 7e-36
UniRef100_Q9CAX1 Hypothetical protein F24K9.24 [Arabidopsis thal... 152 1e-35
UniRef100_Q6YWS2 Leaf senescence protein-like [Oryza sativa] 151 4e-35
UniRef100_Q9ASZ0 AT5g06230/MBL20_11 [Arabidopsis thaliana] 146 9e-34
>UniRef100_Q9FGE9 Gb|AAD15463.1 [Arabidopsis thaliana]
Length = 407
Score = 584 bits (1505), Expect = e-165
Identities = 263/388 (67%), Positives = 316/388 (80%), Gaps = 7/388 (1%)
Query: 7 SHLFPFLIILTLASLYYFSSFISLNTLSSSSSTSPSLTISTCNLFKGTWVFDPNHT-PLY 65
S L P +++L+L L ++S + S+ +S P C+LF G WVF+P PLY
Sbjct: 20 SSLLPRILLLSLLLLLFYSLILRRPITSNIASPPP------CDLFSGRWVFNPETPKPLY 73
Query: 66 DDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSL 125
D+TCPFHRNAWNC+RN+R N+ +INSW+W P GC ++RIDP RFLG MRNKNVG VGDSL
Sbjct: 74 DETCPFHRNAWNCLRNKRDNMDVINSWRWEPNGCGLSRIDPTRFLGMMRNKNVGFVGDSL 133
Query: 126 NENLLVSFLCTLRVADGGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLSKYKWQPKQSES 185
NEN LVSFLC LRVAD A KWKKK AWRGAYFPKFNVTVAYHRAVLL+KY+WQ + S
Sbjct: 134 NENFLVSFLCILRVADPSAIKWKKKKAWRGAYFPKFNVTVAYHRAVLLAKYQWQARSSAE 193
Query: 186 GMQDGSEGIHRVDVDVPADEWAKIAGFYDVLLFNTGHWWNHDKFPKEKPLVFYKAGQPIV 245
QDG +G +RVDVDVPA+EW + FYDVL+FN+GHWW +DKFPKE PLVFY+ G+PI
Sbjct: 194 ANQDGVKGTYRVDVDVPANEWINVTSFYDVLIFNSGHWWGYDKFPKETPLVFYRKGKPIN 253
Query: 246 PPLEMLDGLKVVLGNMITYIEKEFPRNTLKFWRLQSPRHFYGGDWNQNGSCLFNKPLEEN 305
PPL++L G ++VL NM++YI++E P TLKFWRLQSPRHFYGGDWNQNGSCL +KPLEEN
Sbjct: 254 PPLDILPGFELVLQNMVSYIQREVPAKTLKFWRLQSPRHFYGGDWNQNGSCLLDKPLEEN 313
Query: 306 ELDLWFEPRNNGVNKEARQMNFVIEKVLQGTNIHVVDFTHLSEFRADAHPAIWLGRKDAV 365
+LDLWF+PRNNGVNKEAR++N +I+ LQ T I ++D THLSEFRADAHPAIWLG++DAV
Sbjct: 314 QLDLWFDPRNNGVNKEARKINQIIKNELQTTKIKLLDLTHLSEFRADAHPAIWLGKQDAV 373
Query: 366 AIWGQDCMHWCLPGVPDTWVDILSQLII 393
AIWGQDCMHWCLPGVPDTWVDIL++LI+
Sbjct: 374 AIWGQDCMHWCLPGVPDTWVDILAELIL 401
>UniRef100_Q6ZGL3 Leaf senescence related protein-like [Oryza sativa]
Length = 403
Score = 468 bits (1204), Expect = e-130
Identities = 214/350 (61%), Positives = 268/350 (76%), Gaps = 9/350 (2%)
Query: 53 GTWV--FDPNHTPLYDDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCD---MNRIDPF 107
G WV +P PLY +CPFHRNAWNC RN R ++ ++ W P C + RID
Sbjct: 52 GRWVPTREPLPPPLYTSSCPFHRNAWNCPRNSRPPVAALS---WAPARCGGGAVPRIDAA 108
Query: 108 RFLGSMRNKNVGIVGDSLNENLLVSFLCTLRVADGGARKWKKKGAWRGAYFPKFNVTVAY 167
FL R + +G+VGDSL+ENL+V+ LC LR ADGGA KWK++GAWRG YFP+ +V VAY
Sbjct: 109 EFLAVARGRRIGLVGDSLSENLVVALLCALRSADGGASKWKRRGAWRGGYFPRDDVIVAY 168
Query: 168 HRAVLLSKYKWQPKQSESGM-QDGSEGIHRVDVDVPADEWAKIAGFYDVLLFNTGHWWNH 226
HRAVLL+KY WQP ++ + +DG +G +RVDVD+PADEW + FYDVL+FNTGHWW
Sbjct: 169 HRAVLLAKYTWQPVENSKELHKDGIKGTYRVDVDIPADEWVNVTRFYDVLIFNTGHWWGL 228
Query: 227 DKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEFPRNTLKFWRLQSPRHFY 286
DKFPKE PLVFY+ G+PI PPL + DGLKVVL +M +YIE+E P TLK WR QSPRHFY
Sbjct: 229 DKFPKETPLVFYRGGKPIEPPLGIYDGLKVVLKSMASYIEREVPSKTLKLWRSQSPRHFY 288
Query: 287 GGDWNQNGSCLFNKPLEENELDLWFEPRNNGVNKEARQMNFVIEKVLQGTNIHVVDFTHL 346
GG+W+ NGSC+ ++ L+E+ELDLWF+PR GVNKEAR +N I++ L GT+I +++ T++
Sbjct: 289 GGEWDHNGSCVSDRLLQEHELDLWFDPRFGGVNKEARLVNSAIQEALIGTDIQLLNLTYM 348
Query: 347 SEFRADAHPAIWLGRKDAVAIWGQDCMHWCLPGVPDTWVDILSQLIIDGF 396
SEFRADAHPAIWLG+KDAVA+WGQDCMHWCLPGVPDTWVDIL+ I+ F
Sbjct: 349 SEFRADAHPAIWLGKKDAVAVWGQDCMHWCLPGVPDTWVDILAARILHHF 398
>UniRef100_Q944J6 AT5g64470/T12B11_6 [Arabidopsis thaliana]
Length = 325
Score = 431 bits (1107), Expect = e-119
Identities = 197/310 (63%), Positives = 240/310 (76%), Gaps = 8/310 (2%)
Query: 7 SHLFPFLIILTLASLYYFSSFISLNTLSSSSSTSPSLTISTCNLFKGTWVFDPNHT-PLY 65
S L P +++L+L L ++S + S+ +S P C+LF G WVF+P PLY
Sbjct: 20 SSLLPRILLLSLLLLLFYSLILRRPITSNIASPPP------CDLFSGRWVFNPETPKPLY 73
Query: 66 DDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSL 125
D+TCPFHRNAWNC+RN+R N+ +INSW+W P GC ++RIDP RFLG MRNKNVG VGDSL
Sbjct: 74 DETCPFHRNAWNCLRNKRDNMDVINSWRWEPNGCGLSRIDPTRFLGMMRNKNVGFVGDSL 133
Query: 126 NENLLVSFLCTLRVADGGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLSKYKWQPKQSES 185
NEN LVSFLC LRVAD A KWKKK AWRGAYFPKFNVTVAYHRAVLL+KY+WQ + S
Sbjct: 134 NENFLVSFLCILRVADPSAIKWKKKKAWRGAYFPKFNVTVAYHRAVLLAKYQWQARSSAE 193
Query: 186 GMQDGSEGIHRVDVDVPADEWAKIAGFYDVLLFNTGHWWNHDKFPKEKPLVFYKAGQPIV 245
QD +G +RVDVDVPA+EW + FYDVL+FN+GHWW +DKFPKE PLVFY+ G+PI
Sbjct: 194 ANQDEVKGTYRVDVDVPANEWINVTSFYDVLIFNSGHWWGYDKFPKETPLVFYRKGKPIN 253
Query: 246 PPLEMLDGLKVVLGNMITYIEKEFPRNTLKFWRLQSPRHFYGGDWNQNGSCLFNKPLEEN 305
PPL++L G ++VL NM++YI++E P TLKFWRLQSPRHFYGGDWNQNGSCL +KPLEEN
Sbjct: 254 PPLDILPGFELVLQNMVSYIQREVPAKTLKFWRLQSPRHFYGGDWNQNGSCLLDKPLEEN 313
Query: 306 ELDLWFEPRN 315
+ + W E R+
Sbjct: 314 Q-EQWSEQRS 322
>UniRef100_Q9ZQR5 Hypothetical protein At2g14530 [Arabidopsis thaliana]
Length = 412
Score = 317 bits (813), Expect = 3e-85
Identities = 163/398 (40%), Positives = 234/398 (57%), Gaps = 26/398 (6%)
Query: 9 LFPFLIILTLASLYYFSSFISLNTLSSSSSTSPSLTI-------------STCNLFKGTW 55
LFP L +L S++ S +S SS T S T+ +TC+ +G+W
Sbjct: 11 LFPLLSLLCFISIFLLLS-LSKRASLSSPKTHRSATVFPPKPDGSLSPLSATCDFSEGSW 69
Query: 56 VFDPN-HTPLYDDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRIDPFRFLGSMR 114
++DPN + Y +C WNCIRN + N I++W+W P+ CD+ DP +FL S R
Sbjct: 70 IYDPNPRSTRYTSSCKEIFKGWNCIRNNKTNGFEISNWRWKPKHCDLPSFDPLKFLQSHR 129
Query: 115 NKNVGIVGDSLNENLLVSFLCTLRVADGGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLS 174
N N+G VGDSLN N+ VS C L+ G +KW+ GA RG F ++N+T+AYHR LL+
Sbjct: 130 NTNIGFVGDSLNRNMFVSLFCMLKSVTGELKKWRPAGADRGFTFSQYNLTIAYHRTNLLA 189
Query: 175 KY-KWQPKQSESGMQD-GSEGIHRVDVDVPADEWAKIAGFYDVLLFNTGH-WWNHDKF-P 230
+Y +W ++ G + +RVDVD+P WAK + F+D+L+ NTGH WW KF P
Sbjct: 190 RYGRWSANAKGGELESLGFKEGYRVDVDIPDSSWAKASSFHDILILNTGHWWWAPSKFDP 249
Query: 231 KEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEFPRNTLKFWRLQSPRHFYGGDW 290
+ P++F++ G+PI+PP+ GL VL NM+ ++EK + F+R QSPRHF GGDW
Sbjct: 250 VKSPMLFFEGGRPILPPIPPATGLDRVLNNMVNFVEKTKRPGGIIFFRTQSPRHFEGGDW 309
Query: 291 NQNGSCLFNKPLEENELDLWFEPRNNGVNKEARQMNFVIEKVLQGTN-IHVVDFTHLSEF 349
+Q G+C +PL +++ +F NNG N E R +N + L+ + HV+D T +SE+
Sbjct: 310 DQGGTCQRLQPLLPGKVEEFFSVGNNGTNVEVRLVNQHLYNSLKSRSAFHVLDITRMSEY 369
Query: 350 RADAHPAIWLGRKDAVAIWGQDCMHWCLPGVPDTWVDI 387
RADAHPA G+ DCMHWCLPG+ DTW D+
Sbjct: 370 RADAHPAAAGGKNH------DDCMHWCLPGLTDTWNDL 401
>UniRef100_Q69RQ8 Leaf senescence related protein-like [Oryza sativa]
Length = 413
Score = 311 bits (798), Expect = 2e-83
Identities = 167/396 (42%), Positives = 232/396 (58%), Gaps = 31/396 (7%)
Query: 12 FLIILTLASLYYFSSFISLNTLSSSSSTSPSLTIS---------TCNLFKGTWVFDPNHT 62
FL++L L L+ SS + S SP L S C+ G WV D +
Sbjct: 23 FLLLLLLLLLHLTSS-------PARSPNSPLLRGSGDGEPRQRGPCDYASGEWVPDDDDP 75
Query: 63 PL-----YDDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRIDPFRFLGSMRNKN 117
+ YD TC WNC+ N ++N + W+W PRGC++ R+DP RFL RN +
Sbjct: 76 AVSGGLRYDQTCREIFKGWNCLANGKRNGRELLRWRWRPRGCELPRLDPLRFLECHRNTS 135
Query: 118 VGIVGDSLNENLLVSFLCTLRVADGGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLSKY- 176
+G VGDSLN N+ VS +C LR A G RKW+ GA RG F ++N+T+AYHR LL +Y
Sbjct: 136 IGFVGDSLNRNMFVSLVCMLRGASGEVRKWRPAGADRGFTFLRYNLTLAYHRTNLLVRYG 195
Query: 177 KWQPKQSESGMQD-GSEGIHRVDVDVPADEWAKIAGFYDVLLFNTGH-WWNHDKF-PKEK 233
+W + ++ G + +RVDVD+P WA+ F+DVL+FNTGH WW KF P +
Sbjct: 196 RWSASPNGGPLESLGYKQGYRVDVDIPDQTWAEAPSFHDVLIFNTGHWWWAPSKFNPVQS 255
Query: 234 PLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEFPRNTLKFWRLQSPRHFYGGDWNQN 293
P++F++ G P++PPL GL + L +MI ++ K N +K +R QSPRHF GGDWN+
Sbjct: 256 PMLFFEKGIPVIPPLLPPAGLDLALKHMIIFVNKAMRPNGVKLFRTQSPRHFEGGDWNEG 315
Query: 294 GSCLFNKPLEENELDLWFEPRNNGVNKEARQMNFVIEKVLQGTNIHVVDFTHLSEFRADA 353
GSC +KPL E++ F NNG N EAR +N + + L+ + +V++ T +SEFRADA
Sbjct: 316 GSCQRDKPLSAEEVEELFSLDNNGTNVEARLVNQHLVRALEKSTFNVLNITGMSEFRADA 375
Query: 354 HPAIWLGRKDAVAIWGQDCMHWCLPGVPDTWVDILS 389
HP+ G+K DCMHWCLPG DTW D+L+
Sbjct: 376 HPSTTGGKKH------DDCMHWCLPGPTDTWNDLLA 405
>UniRef100_Q67U25 Lustrin A-like [Oryza sativa]
Length = 463
Score = 175 bits (443), Expect = 2e-42
Identities = 119/376 (31%), Positives = 187/376 (49%), Gaps = 46/376 (12%)
Query: 46 STCNLFKGTWVFDPNHTPLY-DDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRI 104
+TC+L+ G W D PLY TCP+ A+ C N R + + W+W PR C + R
Sbjct: 112 TTCDLYDGEWARDEAARPLYAPGTCPYVDEAYACASNGRPDAAYTR-WRWAPRRCRLPRF 170
Query: 105 DPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLRVA-DGGARKWKKKG----AWRGAY-- 157
+ FL ++R K + +VGDS+N N S LC LR A R ++ G RG +
Sbjct: 171 NATDFLATLRGKRLMLVGDSMNRNQFESLLCILREAIPDKTRMFETHGYRISKGRGYFVF 230
Query: 158 -FPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVD-VDVPADEWAKIAGFYDV 215
F ++ TV + R+ L + + Q S I ++D +D A W K+ DV
Sbjct: 231 KFVDYDCTVEFVRSHFLVR-----EGVRYNRQKNSNPILQIDRIDKSASRWRKV----DV 281
Query: 216 LLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEF-PRNTL 274
L+FNTGHWW H K + + +YK G + P + + + L +I+K P ++
Sbjct: 282 LVFNTGHWWTHGK--TARGINYYKEGDTLYPQFDSTEAYRRALKTWARWIDKNMDPAKSV 339
Query: 275 KFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNNG--VNKEARQMNFVIEKV 332
F+R S HF GG+W+ GSC + EP G + R+M ++E+V
Sbjct: 340 VFYRGYSTAHFRGGEWDSGGSCSGER-----------EPTLRGAVIGSYPRKMR-IVEEV 387
Query: 333 LQGTN--IHVVDFTHLSEFRADAHPAIW----LGRKDAVAIWGQDCMHWCLPGVPDTWVD 386
+ + +++ T L+ FR D HP+++ G+K V+ QDC HWCLPGVPD W +
Sbjct: 388 IGRMRFPVRLLNVTKLTNFRRDGHPSVYGKAAAGKK--VSRRKQDCSHWCLPGVPDAWNE 445
Query: 387 IL-SQLIIDGFGRTSK 401
++ + L+++ R+ K
Sbjct: 446 LIYASLVLEPKPRSWK 461
>UniRef100_Q8LD24 Hypothetical protein [Arabidopsis thaliana]
Length = 485
Score = 173 bits (439), Expect = 7e-42
Identities = 116/354 (32%), Positives = 182/354 (50%), Gaps = 40/354 (11%)
Query: 48 CNLFKGTWVFDPNHTPLYDD-TCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRIDP 106
C+L++G+WV D + PLY +CP+ +A++C RN R++ +N W+W P GCD+ R +
Sbjct: 141 CDLYRGSWVKDDDEYPLYQPGSCPYVDDAFDCQRNGRRDSDYLN-WRWKPDGCDLPRFNA 199
Query: 107 FRFLGSMRNKNVGIVGDSLNENLLVSFLCTLRVA-DGGARKWKKKG----AWRGAY---F 158
FL +R K++ +VGDS+N N S LC LR +R ++ G RG + F
Sbjct: 200 TDFLVKLRGKSLMLVGDSMNRNQFESMLCVLREGLSDKSRMYEVHGHNITKGRGYFVFKF 259
Query: 159 PKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGFYDVLLF 218
+N TV + R+ L + + + G + + I R+D +W + D+L+F
Sbjct: 260 EDYNCTVEFVRSHFL--VREGVRANAQGNTNPTLSIDRIDKS--HAKWKRA----DILVF 311
Query: 219 NTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEF-PRNTLKFW 277
NTGHWW H K + K +YK G I P + + + L +I++ P+ L F+
Sbjct: 312 NTGHWWVHGKTARGKN--YYKEGDYIYPKFDATEAYRRSLKTWAKWIDQNVNPKKQLVFY 369
Query: 278 RLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNNG--VNKEARQMNFVIEKVLQ- 334
R S HF GG+W+ GSC E+ EP G ++ +M V E + +
Sbjct: 370 RGYSSAHFRGGEWDSGGSC-------NGEV----EPVKKGSIIDSYPLKMKIVQEAIKEM 418
Query: 335 GTNIHVVDFTHLSEFRADAHPAIWLGRKDA----VAIWGQDCMHWCLPGVPDTW 384
+ +++ T L+ FR D HP+I+ G+ + V+ QDC HWCLPGVPD W
Sbjct: 419 QVPVILLNVTKLTNFRKDGHPSIY-GKTNTDGKKVSTRRQDCSHWCLPGVPDVW 471
>UniRef100_Q7X6G0 OSJNBa0043L24.14 protein [Oryza sativa]
Length = 711
Score = 168 bits (426), Expect = 2e-40
Identities = 116/379 (30%), Positives = 186/379 (48%), Gaps = 49/379 (12%)
Query: 35 SSSSTSPSLTISTCNLFKGTWVFDPNHT----PLYDD-TCPFHRNAWNCIRNQRQNLSLI 89
+S S ++ + C++F G WV D + P Y +CP + +NC +N R + +
Sbjct: 343 TSGVQSGLVSFAKCDVFSGRWVRDDDEGGGAYPFYPPGSCPHIDDDFNCHKNGRADTGFL 402
Query: 90 NSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLR--VAD------ 141
W+W P GCD+ R++P FL +R + + VGDSLN N+ S +C LR V D
Sbjct: 403 R-WRWQPHGCDIPRLNPIDFLERLRGQRIIFVGDSLNRNMWESLVCILRHGVRDKRRMYE 461
Query: 142 -GGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVD 200
G ++K +G + F +N +V + R++ L K M + ++G VD
Sbjct: 462 ASGRNQFKTRG-YYSFRFRDYNCSVDFIRSIFLVK----------EMINETKGGAVVDAK 510
Query: 201 VPADEWAKIAGFY---DVLLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVV 257
+ DE + Y D+++FNTGHWW H K + L +Y+ G + P LE++D K
Sbjct: 511 LRLDELDETTPAYRTADIVVFNTGHWWTHWK--TSRGLNYYQEGNYVHPSLEVMDAYKRA 568
Query: 258 LGNMITYIEKEF-PRNTLKFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNN 316
L +++K T +R S HF GG WN G C E + F N
Sbjct: 569 LTTWARWVDKNIDSTRTQVVFRGYSLTHFRGGQWNSGGRC-------HRETEPIF---NR 618
Query: 317 GVNKEARQMNFVIEKVL--QGTNIHVVDFTHLSEFRADAHPAIWLGRKDA-----VAIWG 369
E + ++E+VL T + ++ + ++++R DAHP+++ R + A+
Sbjct: 619 THLAEYPEKMRILEQVLGRMRTPVIYLNISAMTDYRKDAHPSVYRVRYETEEERMAAVAK 678
Query: 370 QDCMHWCLPGVPDTWVDIL 388
QDC HWCLPGVPD+W ++L
Sbjct: 679 QDCSHWCLPGVPDSWNELL 697
>UniRef100_Q9LHL6 Gb|AAF30301.1 [Arabidopsis thaliana]
Length = 556
Score = 166 bits (419), Expect = 1e-39
Identities = 111/362 (30%), Positives = 182/362 (49%), Gaps = 41/362 (11%)
Query: 44 TISTCNLFKGTWVFDPNHTPLYDD-TCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMN 102
++ +C F+G WV D ++ PLY +C +NCI N R ++ KW P+ C +
Sbjct: 192 SLKSCEFFEGDWVKDDSY-PLYKPGSCNLIDEQFNCISNGRPDVDF-QKLKWKPKQCSLP 249
Query: 103 RIDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLR--VADGGA---RKWKKKGAWRGAY 157
R++ + L +R + + VGDSLN N+ S +C L+ V D + + W Y
Sbjct: 250 RLNGGKLLEMIRGRRLVFVGDSLNRNMWESLVCILKGSVKDESQVFEAHGRHQFRWEAEY 309
Query: 158 ---FPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGFYD 214
F +N TV + + L + +W+ + ++ R+D+ + E K A D
Sbjct: 310 SFVFKDYNCTVEFFASPFLVQ-EWEVTEKNGTKKETL----RLDLVGKSSEQYKGA---D 361
Query: 215 VLLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEF-PRNT 273
+L+FNTGHWW H+K K + +Y+ G + P L++ + + L +++K P+ +
Sbjct: 362 ILVFNTGHWWTHEKTSKGED--YYQEGSTVHPKLDVDEAFRKALTTWGRWVDKNVNPKKS 419
Query: 274 LKFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNNGVNKEARQMNF-VIEKV 332
L F+R SP HF GG WN G+C D EP N + ++E+V
Sbjct: 420 LVFFRGYSPSHFSGGQWNAGGAC-----------DDETEPIKNETYLTPYMLKMEILERV 468
Query: 333 LQG--TNIHVVDFTHLSEFRADAHPAIWLGRKDAVA-----IWGQDCMHWCLPGVPDTWV 385
L+G T + ++ T L+++R DAHP+I+ +K + + QDC HWCLPGVPD+W
Sbjct: 469 LRGMKTPVTYLNITRLTDYRKDAHPSIYRKQKLSAEESKSPLLYQDCSHWCLPGVPDSWN 528
Query: 386 DI 387
+I
Sbjct: 529 EI 530
>UniRef100_Q8VYR3 Hypothetical protein At1g60790 [Arabidopsis thaliana]
Length = 541
Score = 163 bits (412), Expect = 9e-39
Identities = 107/358 (29%), Positives = 175/358 (47%), Gaps = 36/358 (10%)
Query: 48 CNLFKGTWVFDPNHTPLY--DDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRID 105
C+++ G+WV + T Y +CP+ +NC N R + + + W+W P GCD+ R++
Sbjct: 190 CDIYDGSWVRADDETMPYYPPGSCPYIDRDFNCHANGRPDDAYVK-WRWQPNGCDIPRLN 248
Query: 106 PFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLR---------VADGGARKWKKKGAWRGA 156
FL +R K + VGDS+N N+ S +C LR G R++KKKG +
Sbjct: 249 GTDFLEKLRGKKLVFVGDSINRNMWESLICILRHSLKDKKRVYEISGRREFKKKGFY-AF 307
Query: 157 YFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGFYDVL 216
F +N TV + + + + S G+ + R+D+ D+ + D+L
Sbjct: 308 RFEDYNCTVDFVGSPFFVR-----ESSFKGVNGTTLETLRLDM---MDKTTSMYRDADIL 359
Query: 217 LFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEFPRN-TLK 275
+FNTGHWW HDK + +Y+ G + P L++L+ K L +++K R+ T
Sbjct: 360 IFNTGHWWTHDKTKLGEN--YYQEGNVVYPRLKVLEAYKRALITWAKWVDKNIDRSQTHI 417
Query: 276 FWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNNGVNKEARQMNFVIEKVLQG 335
+R S HF GG WN G C N L P + + + +++ ++
Sbjct: 418 VFRGYSVTHFRGGPWNSGGQCHKETEPIFNTSYLAKYP------SKMKALEYILRDTMK- 470
Query: 336 TNIHVVDFTHLSEFRADAHPAIW-----LGRKDAVAIWGQDCMHWCLPGVPDTWVDIL 388
T + ++ + L++FR D HP+I+ ++ A+ QDC HWCLPGVPDTW +L
Sbjct: 471 TPVIYMNISRLTDFRKDGHPSIYRMVYRTEKEKREAVSHQDCSHWCLPGVPDTWNQLL 528
>UniRef100_Q6Z7L5 Hypothetical protein OJ1448_G06.4 [Oryza sativa]
Length = 700
Score = 160 bits (406), Expect = 5e-38
Identities = 110/363 (30%), Positives = 181/363 (49%), Gaps = 43/363 (11%)
Query: 45 ISTCNLFKGTWVFDPNHTPLYDD-TCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNR 103
+++C++F G WV D ++ PLY + +CP ++NC N R + + +W P GC + R
Sbjct: 348 MASCDMFYGNWVRDDSY-PLYPEGSCPHIDESFNCPLNGRPD-NAYQRLRWQPSGCSIPR 405
Query: 104 IDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLR--VAD-------GGARKWKKKGAWR 154
++P L +R K + VGDSLN N+ S +C LR V D G ++++ +G++
Sbjct: 406 LNPSDMLERLRGKRLVFVGDSLNRNMWESLVCILRNSVKDKRKVFEVSGRQQFRAEGSY- 464
Query: 155 GAYFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGFYD 214
F +N +V + R+ L + +W+ G+ + + + P + A D
Sbjct: 465 SFLFQDYNCSVEFFRSPFLVQ-EWE-FPVRKGLTKETLRLDMISNSFPRYKDA------D 516
Query: 215 VLLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEF-PRNT 273
+++FNTGHWW H+K K +Y+ G + L + D + L +++ P+ T
Sbjct: 517 IIIFNTGHWWTHEKTSLGKD--YYQEGNRVYSELNVDDAFQKALITWAKWVDSSVNPKKT 574
Query: 274 LKFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNNGVNKEARQMNFVIEKVL 333
F+R S HF GG WN GSC NE L PR ++E VL
Sbjct: 575 TVFFRGYSSSHFSGGQWNSGGSCDKETEPITNEKFLTPYPRKMS----------ILEDVL 624
Query: 334 QGTNIHVV--DFTHLSEFRADAHPAIWLGR------KDAVAIWGQDCMHWCLPGVPDTWV 385
G VV + T ++++R +AHP+++ + K + I+ QDC HWCLPGVPD+W
Sbjct: 625 SGMKTPVVYLNITRMTDYRKEAHPSVYRKQKLTEEEKKSPQIY-QDCSHWCLPGVPDSWN 683
Query: 386 DIL 388
++L
Sbjct: 684 ELL 686
>UniRef100_Q9LZQ1 Hypothetical protein T12C14_90 [Arabidopsis thaliana]
Length = 475
Score = 160 bits (405), Expect = 6e-38
Identities = 111/358 (31%), Positives = 176/358 (49%), Gaps = 47/358 (13%)
Query: 45 ISTCNLFKGTWVFDPNHTPLYDD-TCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNR 103
I C++ KG WV+D ++ PLY + +CPF + C N R +L+ +N W+W P+ C R
Sbjct: 135 IEECDVTKGKWVYDSDY-PLYTNASCPFIDEGFGCQSNGRLDLNYMN-WRWEPQDCHAPR 192
Query: 104 IDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTL--------RVADGGARKWKKKGAWRG 155
+ + L +R K + VGDS+N N S LC L RV + R+ K+
Sbjct: 193 FNATKMLEMIRGKRLVFVGDSINRNQWESMLCLLFQAVKDPKRVYETHNRRITKEKGNYS 252
Query: 156 AYFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGFYDV 215
F + TV ++ L + E + G + + +D ++ G ++
Sbjct: 253 FRFVDYKCTVEFYVTHFLVR--------EGRARIGKKRRETLRIDAMDRTSSRWKG-ANI 303
Query: 216 LLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEF-PRNTL 274
L+FNT HWW+H + + + +Y+ G I P L++ K L ++++K P+ T
Sbjct: 304 LVFNTAHWWSH--YKTKSGVNYYQEGDLIHPKLDVSTAFKKALQTWSSWVDKNVDPKKTR 361
Query: 275 KFWRLQSPRHFYGGDWNQNGSCL-FNKPLEENELDLWFEPRNNGVNKEARQMNFVIEKVL 333
F+R +P HF GG+WN G C N PL + F+P + ++E VL
Sbjct: 362 VFFRSAAPSHFSGGEWNSGGHCREANMPLNQT-----FKPSYSSKKS-------IVEDVL 409
Query: 334 Q--GTNIHVVDFTHLSEFRADAHPAIW-----LGRKDAVAIWGQDCMHWCLPGVPDTW 384
+ T + +++ + LS++R DAHP+I+ R AV QDC HWCLPGVPDTW
Sbjct: 410 KQMRTPVTLLNVSGLSQYRIDAHPSIYGTKPENRRSRAV----QDCSHWCLPGVPDTW 463
>UniRef100_Q9FG35 Emb|CAB82953.1 [Arabidopsis thaliana]
Length = 608
Score = 160 bits (404), Expect = 8e-38
Identities = 113/372 (30%), Positives = 185/372 (49%), Gaps = 44/372 (11%)
Query: 42 SLTISTCNLFKGTWVFDPNHTPLYDD-TCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCD 100
S ++ C F G W+ D ++ PLY +C +NCI N R + KW P+ C
Sbjct: 249 SESLKNCEFFDGEWIKDDSY-PLYKPGSCNLIDEQFNCITNGRPDKDF-QKLKWKPKKCS 306
Query: 101 MNRIDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLR--------VADGGARKWKKKGA 152
+ R++ L +R + + VGDSLN N+ S +C L+ V + R + A
Sbjct: 307 LPRLNGAILLEMLRGRRLVFVGDSLNRNMWESLVCILKGSVKDETKVYEARGRHHFRGEA 366
Query: 153 WRGAYFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGF 212
F +N TV + + L + +W+ + ++ R+D+ + E K A
Sbjct: 367 EYSFVFQDYNCTVEFFVSPFLVQ-EWEIVDKKGTKKETL----RLDLVGKSSEQYKGA-- 419
Query: 213 YDVLLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEF-PR 271
DV++FNTGHWW H+K K + +Y+ G + L +L+ + L ++EK P
Sbjct: 420 -DVIVFNTGHWWTHEKTSKGED--YYQEGSNVYHELAVLEAFRKALTTWGRWVEKNVNPA 476
Query: 272 NTLKFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNNGVNKEARQMNFVIEK 331
+L F+R S HF GG WN G+C + E + D + P + + V+EK
Sbjct: 477 KSLVFFRGYSASHFSGGQWNSGGAC--DSETEPIKNDTYLTPYPSKMK--------VLEK 526
Query: 332 VLQG--TNIHVVDFTHLSEFRADAHPAIWLGRKDAVA-------IWGQDCMHWCLPGVPD 382
VL+G T + ++ T L+++R D HP+++ RK +++ + QDC HWCLPGVPD
Sbjct: 527 VLRGMKTPVTYLNITRLTDYRKDGHPSVY--RKQSLSEKEKKSPLLYQDCSHWCLPGVPD 584
Query: 383 TWVDIL-SQLII 393
+W +IL ++LI+
Sbjct: 585 SWNEILYAELIV 596
>UniRef100_Q69T67 Lustrin A-like [Oryza sativa]
Length = 847
Score = 159 bits (402), Expect = 1e-37
Identities = 114/366 (31%), Positives = 183/366 (49%), Gaps = 38/366 (10%)
Query: 45 ISTCNLFKGTWVFDPNHTPLYDD-TCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNR 103
+++C++F G WV D ++ PLY + +CP ++C N R + + +W P C++ R
Sbjct: 495 MASCDMFHGNWVRDESY-PLYPEGSCPHIDEPFDCYLNGRPDRAY-QKLRWQPSSCNIPR 552
Query: 104 IDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLR--VAD-------GGARKWKKKGAWR 154
++P L +R K + VGDSLN N+ S +C LR V D G ++K +G++
Sbjct: 553 LNPTDMLERLRGKRLVFVGDSLNRNMWESLVCILRNSVKDKRKVFEASGRHEFKTEGSY- 611
Query: 155 GAYFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGFYD 214
F +N +V + R+ L + +W+ K S ++ R+D+ + K A D
Sbjct: 612 SFLFTDYNCSVEFFRSPFLVQ-EWEMKVSNGKKKE----TLRLDIVEQSSPKYKDA---D 663
Query: 215 VLLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYIEKEF-PRNT 273
L+FNTGHWW H+K K +Y+ G + L ++D L +I+ P+ T
Sbjct: 664 FLIFNTGHWWTHEKTSLGKD--YYQEGNHVYSELNVVDAFHKALVTWSRWIDANVNPKKT 721
Query: 274 LKFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEPRNNGVNKEARQMNFVIEKVL 333
+R S HF GG WN GSC NE L P + ++ VI K+
Sbjct: 722 TVLFRGYSASHFSGGQWNSGGSCDKETEPIRNEQYLSTYPPKMSILED------VIHKM- 774
Query: 334 QGTNIHVVDFTHLSEFRADAHPAIWLGR-----KDAVAIWGQDCMHWCLPGVPDTWVDIL 388
T + ++ T ++++R DAHP+I+ R + QDC HWCLPGVPD+W ++L
Sbjct: 775 -KTPVVYLNITRMTDYRKDAHPSIYRKRNLTEDERRSPERYQDCSHWCLPGVPDSWNELL 833
Query: 389 -SQLII 393
+QL+I
Sbjct: 834 YAQLLI 839
>UniRef100_Q93ZW1 Hypothetical protein At5g20680 [Arabidopsis thaliana]
Length = 551
Score = 155 bits (391), Expect = 2e-36
Identities = 119/396 (30%), Positives = 177/396 (44%), Gaps = 61/396 (15%)
Query: 25 SSFISLNTLSSSSSTSPSLTISTCNLFKGTWVFDPNHTPLYDDT-CP-FHRNAWNCIRNQ 82
S+ ++ + + +++ +T CN KG WV D NH PLY + C + + W C Q
Sbjct: 189 SNILATDEERTDGTSTARITNQACNYAKGKWVVD-NHRPLYSGSQCKQWLASMWACRLMQ 247
Query: 83 RQNLSLINSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLR---- 138
R + + S +W P+ C M + +FL M+NK + VGDSL S +C +
Sbjct: 248 RTDFAF-ESLRWQPKDCSMEEFEGSKFLRRMKNKTLAFVGDSLGRQQFQSMMCMISGGKE 306
Query: 139 --------------VADGGARKWKKKGAWRGAY-FPKFNVTVAYHRAVLLSKYKWQPKQS 183
+GGAR G W AY FP+ N TV YH + L +P
Sbjct: 307 RLDVLDVGPEFGFITPEGGARP----GGW--AYRFPETNTTVLYHWSSTLCDI--EPLNI 358
Query: 184 ESGMQDGSEGIHRVDVDVPADEWAKIAGFYDVLLFNTGHWWNHDKFPKEKPLVFYKAGQP 243
+ H + +D P + DVL+ NTGH WN K K ++
Sbjct: 359 TDPATE-----HAMHLDRPPAFLRQYLQKIDVLVMNTGHHWNRGKLNGNKWVMHVNGVPN 413
Query: 244 IVPPLEMLDGLK-VVLGNMITYIEKEFPRNT-LK-FWRLQSPRHFYGGDWNQNGSCLFNK 300
L L K + + ++++ + P + LK F+R SPRHF GG+WN GSC
Sbjct: 414 TNRKLAALGNAKNFTIHSTVSWVNSQLPLHPGLKAFYRSLSPRHFVGGEWNTGGSCNNTT 473
Query: 301 PLEENELDLWFEPRNNGVNKEARQ---MNFVIEKVLQGTNIHVVDFTHLSEFRADAHPAI 357
P+ + KE Q ++ + ++GT + ++D T LS R + H
Sbjct: 474 PMS--------------IGKEVLQEESSDYSAGRAVKGTGVKLLDITALSHIRDEGH--- 516
Query: 358 WLGRKDAVAIWG-QDCMHWCLPGVPDTWVDILSQLI 392
+ R A G QDC+HWCLPGVPDTW +IL +I
Sbjct: 517 -ISRFSISASRGVQDCLHWCLPGVPDTWNEILFAMI 551
>UniRef100_Q9LDG2 Hypothetical protein F24F17.6 [Arabidopsis thaliana]
Length = 469
Score = 153 bits (387), Expect = 7e-36
Identities = 114/398 (28%), Positives = 181/398 (44%), Gaps = 48/398 (12%)
Query: 22 YYFSSFISLNTLSSSSSTSPSL------TISTCNLFKGTWVFDPNHTPLYDDT-CPFHRN 74
+ + F ++++ SSSSS S + S C++F G WV+D ++ PLY C F
Sbjct: 71 FVWLKFDNISSSSSSSSNSSKRVGFLEESGSGCDVFDGDWVWDESY-PLYQSKDCRFLDE 129
Query: 75 AWNCIRNQRQNLSLINSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSLNENLLVSFL 134
+ C R +L W+W PR C++ R D L +R+K + VGDS+ N S L
Sbjct: 130 GFRCSDFGRSDL-FYTQWRWQPRHCNLPRFDAKLMLEKLRDKRLVFVGDSIGRNQWESLL 188
Query: 135 CTLRVA---------DGGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLSKYKWQPKQSES 185
C L A G+ K KG + F ++N TV Y+R+ L P
Sbjct: 189 CLLSSAVKNESLIYEINGSPITKHKG-FLVFKFEEYNCTVEYYRSPFLVPQSRPP----- 242
Query: 186 GMQDGSEGIHRVDVDVPADEWAKIA-GFYDVLLFNTGHWWNHDKFPKEKPLVFYKAGQPI 244
GS G + + + +W DVL+ NTGHWWN K + +++ G+ +
Sbjct: 243 ---IGSPGKVKTSLKLDTMDWTSSKWRDADVLVLNTGHWWNEGKTTRTG--CYFQEGEEV 297
Query: 245 VPPLEMLDGLKVVLGNMITYIEKEFPRN-TLKFWRLQSPRHFYGGDWNQNGSCLFNKPLE 303
+ + D K L ++ +I E N T F+R +P HF GGDW G+C
Sbjct: 298 KLKMNVDDAYKRALNTVVKWIHTELDSNKTQVFFRTFAPVHFRGGDWKTGGTC------- 350
Query: 304 ENELDLWFEPRNNGVNKEARQMNFVIEKVL-------QGTNIHVVDFTHLSEFRADAHPA 356
++ E + + E + ++ VL + + +++ T ++ R D HP+
Sbjct: 351 --HMETLPEIGTSLASSETWEQLKILRDVLSHNSNRSETVKVKLLNITAMAAQRKDGHPS 408
Query: 357 I-WLGRKDAVAIWGQDCMHWCLPGVPDTWVDILSQLII 393
+ +LG + QDC HWCLPGVPDTW ++ L +
Sbjct: 409 LYYLGPHGPAPLHRQDCSHWCLPGVPDTWNELFYALFM 446
>UniRef100_Q9FFZ4 Gb|AAF00670.1 [Arabidopsis thaliana]
Length = 413
Score = 153 bits (387), Expect = 7e-36
Identities = 125/410 (30%), Positives = 190/410 (45%), Gaps = 50/410 (12%)
Query: 4 KLPSHLFPFLIILTLASLYYFSSFI-----SLNTLSSSSSTSPSLTISTCNLFKGTWVFD 58
K+ HLF L +L SL FS+ + SL SSS+S ++T C+ KG WV
Sbjct: 17 KIKKHLFVSLFLL---SLLIFSTVVVDVMPSLRIGLLSSSSSQTVT-KECDYSKGKWVRR 72
Query: 59 PNHTP------LYDDTCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRIDPFRFLGS 112
+ + Y + C F + + C ++ R++ ++ W+W P GCD+ R + L
Sbjct: 73 ASSSSSSVNGLFYGEECRFLDSGFRCHKHGRKDSGYLD-WRWQPHGCDLPRFNASDLLER 131
Query: 113 MRNKNVGIVGDSLNENLLVSFLCTLRVAD---------GGARKWKKKGAWRGAYFPKFNV 163
RN + VGDS+ N S +C L A G K KG + FP+ N+
Sbjct: 132 SRNGRIVFVGDSIGRNQWESLMCMLSQAIPNKSEIYEVNGNPITKHKG-FLSMRFPRENL 190
Query: 164 TVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDVPADEW-AKIAGFYDVLLFNTGH 222
TV YHR+ L P +S + + V V W +K DVL+FN+GH
Sbjct: 191 TVEYHRSPFLVVIGRPPDKSPKEI--------KTTVRVDEFNWQSKRWVGSDVLVFNSGH 242
Query: 223 WWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGNMITYI-EKEFPRNTLKFWRLQS 281
WWN DK +++ G+ + + +++ L +++ EK P + F+R S
Sbjct: 243 WWNEDKTVLTG--CYFEEGRKVNKTMGVMEAFGKSLKTWKSWVLEKLDPDKSYVFFRSYS 300
Query: 282 PRHFYGGDWNQNGSCLFNKPLEENELDLWFEP---RNNGVNKEARQMNFVIEKVLQGTNI 338
P H+ G WN G C E ++ L EP N + K +M + KV
Sbjct: 301 PVHYRNGTWNTGGLCDAEIEPETDKRKL--EPDASHNEYIYKVIEEMRYRHSKV------ 352
Query: 339 HVVDFTHLSEFRADAHPAIWLGRKDAVAIWGQDCMHWCLPGVPDTWVDIL 388
++ T+L+EFR D H + + + +V + QDC HWCLPGVPDTW +IL
Sbjct: 353 KFLNITYLTEFRKDGHISRYREQGTSVDV-PQDCSHWCLPGVPDTWNEIL 401
>UniRef100_Q9CAX1 Hypothetical protein F24K9.24 [Arabidopsis thaliana]
Length = 427
Score = 152 bits (385), Expect = 1e-35
Identities = 111/373 (29%), Positives = 172/373 (45%), Gaps = 38/373 (10%)
Query: 33 LSSSSSTSPSLTISTCNLFKGTWVF---DPNHTPLYDDTCPFHRNAWNCIRNQRQNLSLI 89
LS + T C+ G WV D + T Y + C F + C+ N R++ S
Sbjct: 64 LSPRTLTKERGNDDVCDYSYGRWVRRRRDVDETSYYGEECRFLDPGFRCLNNGRKD-SGF 122
Query: 90 NSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTL--------RVAD 141
W+W P GCD+ R + FL RN + VGDS+ N S LC L + +
Sbjct: 123 RQWRWQPHGCDLPRFNASDFLERSRNGRIVFVGDSIGRNQWESLLCMLSQAVSNKSEIYE 182
Query: 142 GGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVDV 201
K + FP+ N+TV YHR L P+ S + ++ V V
Sbjct: 183 VNGNPISKHKGFLSMRFPEQNLTVEYHRTPFLVVVGRPPENSPVDV--------KMTVRV 234
Query: 202 PADEW-AKIAGFYDVLLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLGN 260
W +K DVL+FNTGHWWN DK +++ G + + +++G + L
Sbjct: 235 DEFNWQSKKWVGSDVLVFNTGHWWNEDK--TFIAGCYFQEGGKLNKTMGVMEGFEKSLKT 292
Query: 261 MITYI-EKEFPRNTLKFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEP-RNNGV 318
+++ E+ + F+R SP H+ G WN G C + E + + +P NN +
Sbjct: 293 WKSWVLERLDSERSHVFFRSFSPVHYRNGTWNLGGLCDADTEPETDMKKMEPDPIHNNYI 352
Query: 319 NKEARQMNFVIEKVLQGTNIHVVDFTHLSEFRADAHPAIWL---GRKDAVAIWGQDCMHW 375
++ ++M + + + + ++ T+L+EFR DAHP+ + +DA QDC HW
Sbjct: 353 SQAIQEMRY------EHSKVKFLNITYLTEFRKDAHPSRYREPGTPEDA----PQDCSHW 402
Query: 376 CLPGVPDTWVDIL 388
CLPGVPDTW +IL
Sbjct: 403 CLPGVPDTWNEIL 415
>UniRef100_Q6YWS2 Leaf senescence protein-like [Oryza sativa]
Length = 516
Score = 151 bits (381), Expect = 4e-35
Identities = 119/419 (28%), Positives = 190/419 (44%), Gaps = 64/419 (15%)
Query: 13 LIILTLASLYYFSSF-----ISLNTLSSSSSTSPS-LTISTCNLFKGTWVFDPNHTPLYD 66
L+++ L SLY F+ I + SSSSS+SPS T+S CNL +G WV D P Y
Sbjct: 124 LLVVGLISLYDFTFADRYPNIDAASSSSSSSSSPSPATVSKCNLTRGEWVPD-GEAPYYT 182
Query: 67 D-TCPFHRNAWNCIRNQRQNLSLINSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSL 125
+ TCPF + NC++ + +L + SW+W P GC++ R D RFL +MR K++ VGDSL
Sbjct: 183 NLTCPFIDDHQNCMKFGKPSLEYV-SWRWKPDGCELPRFDAARFLEAMRGKSMAFVGDSL 241
Query: 126 NENLLVSFLCTLRVADGGARKWKKKGAWRGAYFPKFNVT-------VAYHRAVLLSKYKW 178
N S LC L A+ + GA P+ +VT Y + W
Sbjct: 242 ARNHFKSLLCLLSKV---AQPVELVGA-----APEIDVTGRAVRRDFRYDSHGFTASLFW 293
Query: 179 QPKQSESGMQDGSEGIHRVDVDVPADEWAKIAGFYDVLLFNTGHWWNHDKFPKEKPLVFY 238
P ++ + + + G+ + +D WA +D ++ + +W+ +P V+Y
Sbjct: 294 SPFLVKANLANATLGLWDLHLDTADARWAAHVAEFDYVVLSDTNWFL-------RPSVYY 346
Query: 239 KAGQP--------------IVPPLEMLDGLKVVLGNMI----TYIEKEFPRNTLKFWRLQ 280
+ G+ I P + LG + T+ K R+
Sbjct: 347 EGGRAVGRNGAAPVTNATEIAVPRAVRAAFSTALGALAAAPGTFRGKAILRSV------- 399
Query: 281 SPRHFYGGDWNQNGSCLFNKPLEENELDLW-FEPRNNGVNKEA-RQMNFVIEKVLQGTNI 338
+P HF G+WN G C+ +P +E L E V +A R+ + + G +
Sbjct: 400 TPAHFENGEWNTGGDCVRTRPFRRDERALGAVEAEYLAVQVDAVREAEAAVRR--NGGEL 457
Query: 339 HVVDFTHLSEFRADAHPAIWLGRKDAVAIWGQ---DCMHWCLPGVPDTWVDILSQLIID 394
++D T + R D HP+ + G ++ G DC+HWCLPG D W ++L +++D
Sbjct: 458 RLLDITEAMDLRPDGHPSRY-GHPPGGSVEGSFVVDCLHWCLPGPIDLWSELLFHMLVD 515
>UniRef100_Q9ASZ0 AT5g06230/MBL20_11 [Arabidopsis thaliana]
Length = 372
Score = 146 bits (369), Expect = 9e-34
Identities = 113/373 (30%), Positives = 174/373 (46%), Gaps = 42/373 (11%)
Query: 36 SSSTSPSLTISTCNLFKGTWVFDPNHTP------LYDDTCPFHRNAWNCIRNQRQNLSLI 89
SSS+S ++T C+ KG WV + + Y + C F + + C ++ R++ +
Sbjct: 10 SSSSSQTVT-KECDYSKGKWVRRASSSSSSVNGLFYGEECRFLDSGFRCHKHGRKDSGYL 68
Query: 90 NSWKWVPRGCDMNRIDPFRFLGSMRNKNVGIVGDSLNENLLVSFLCTLRVAD-------- 141
+ W+W P GCD+ R + L RN + VGDS+ N S +C L A
Sbjct: 69 D-WRWQPHGCDLPRFNASDLLERSRNGRIVFVGDSIGRNQWESLMCMLSQAIPNKSEIYE 127
Query: 142 -GGARKWKKKGAWRGAYFPKFNVTVAYHRAVLLSKYKWQPKQSESGMQDGSEGIHRVDVD 200
G K KG + FP+ N+TV YHR+ L P +S + + V
Sbjct: 128 VNGNPITKHKG-FLSMRFPRENLTVEYHRSPFLVVIGRPPDKSPKEI--------KTTVR 178
Query: 201 VPADEW-AKIAGFYDVLLFNTGHWWNHDKFPKEKPLVFYKAGQPIVPPLEMLDGLKVVLG 259
V W +K DVL+FN+GHWWN DK +++ G+ + + +++ L
Sbjct: 179 VDEFNWQSKRWVGSDVLVFNSGHWWNEDKTVLTG--CYFEEGRKVNKTMGVMEAFGKSLK 236
Query: 260 NMITYI-EKEFPRNTLKFWRLQSPRHFYGGDWNQNGSCLFNKPLEENELDLWFEP---RN 315
+++ EK P + F+R SP H+ G WN G C E ++ L EP N
Sbjct: 237 TWKSWVLEKLDPDKSYVFFRSYSPVHYRNGTWNTGGLCDAEIEPETDKRKL--EPDASHN 294
Query: 316 NGVNKEARQMNFVIEKVLQGTNIHVVDFTHLSEFRADAHPAIWLGRKDAVAIWGQDCMHW 375
+ K +M + KV ++ T+L+EFR D H + + + +V + QDC HW
Sbjct: 295 EYIYKVIEEMRYRHSKV------KFLNITYLTEFRKDGHISRYREQGTSVDV-PQDCSHW 347
Query: 376 CLPGVPDTWVDIL 388
CLPGVPDTW +IL
Sbjct: 348 CLPGVPDTWNEIL 360
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 759,482,656
Number of Sequences: 2790947
Number of extensions: 34139589
Number of successful extensions: 78197
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 122
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 77602
Number of HSP's gapped (non-prelim): 164
length of query: 402
length of database: 848,049,833
effective HSP length: 130
effective length of query: 272
effective length of database: 485,226,723
effective search space: 131981668656
effective search space used: 131981668656
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC124214.2


