

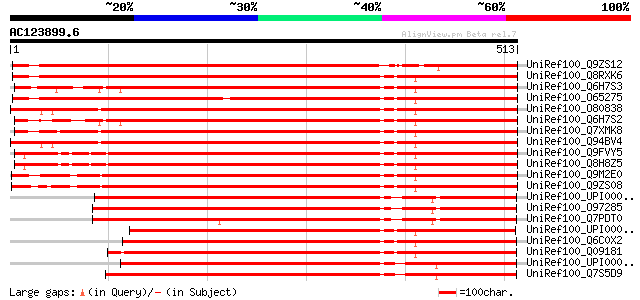
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123899.6 + phase: 0 /pseudo
(513 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZS12 RNA helicase [Arabidopsis thaliana] 707 0.0
UniRef100_Q8RXK6 Putative RNA helicase [Arabidopsis thaliana] 706 0.0
UniRef100_Q6H7S3 Putative RNA helicase [Oryza sativa] 684 0.0
UniRef100_O65275 F6N23.6 protein [Arabidopsis thaliana] 677 0.0
UniRef100_O80838 Putative ATP-dependent RNA helicase [Arabidopsi... 666 0.0
UniRef100_Q6H7S2 Putative RNA helicase [Oryza sativa] 665 0.0
UniRef100_Q7XMK8 OSJNBb0039L24.10 protein [Oryza sativa] 664 0.0
UniRef100_Q94BV4 At2g45810/F4I18.21 [Arabidopsis thaliana] 664 0.0
UniRef100_Q9FVY5 Putative RNA helicase [Oryza sativa] 659 0.0
UniRef100_Q8H8Z5 Putative RNA helicase [Oryza sativa] 654 0.0
UniRef100_Q9M2E0 DEAD box RNA helicase RH12 [Arabidopsis thaliana] 647 0.0
UniRef100_Q9ZS08 RNA helicase [Arabidopsis thaliana] 640 0.0
UniRef100_UPI00004688D2 UPI00004688D2 UniRef100 entry 501 e-140
UniRef100_O97285 Putative ATP-dependent RNA Helicase [Plasmodium... 497 e-139
UniRef100_Q7PDT0 ATP-dependent RNA helicase [Plasmodium yoelii y... 495 e-138
UniRef100_UPI00003C1C9D UPI00003C1C9D UniRef100 entry 494 e-138
UniRef100_Q6C0X2 YlDHH1 protein [Yarrowia lipolytica] 491 e-137
UniRef100_Q09181 Putative ATP-dependent RNA helicase ste13 [Schi... 486 e-136
UniRef100_UPI000023DF32 UPI000023DF32 UniRef100 entry 483 e-135
UniRef100_Q7S5D9 Hypothetical protein [Neurospora crassa] 481 e-134
>UniRef100_Q9ZS12 RNA helicase [Arabidopsis thaliana]
Length = 505
Score = 707 bits (1826), Expect = 0.0
Identities = 384/528 (72%), Positives = 410/528 (76%), Gaps = 43/528 (8%)
Query: 4 NNRGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQR-HMMQNQHQQH 62
NNRGRYPPGIG GRG N N +Q R +Q QYVQR + QN QQ
Sbjct: 2 NNRGRYPPGIGAGRGAF---------NPNPNYQSRSGYQQHPPPQYVQRGNYAQNHQQQF 52
Query: 63 YQNQQQNQQQNQQQQQQQQWLRRNQL-GGGTDTNVVEEVEKTVQSEANDSSSQDWKARLK 121
Q Q Q QQQQQQQQWLRR Q+ GG ++ + V EVEKTVQSE D +S+DWKARLK
Sbjct: 53 QQAPSQPHQYQQQQQQQQQWLRRGQIPGGNSNGDAVVEVEKTVQSEVIDPNSEDWKARLK 112
Query: 122 LPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDIL 181
LP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTG DIL
Sbjct: 113 LPAPDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGRDIL 172
Query: 182 ARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMV 241
ARAKNGTGKTAAF IP LEKIDQDNN+IQ VI+VPTRELALQTSQVCKELGKHL+IQVMV
Sbjct: 173 ARAKNGTGKTAAFCIPVLEKIDQDNNVIQAVIIVPTRELALQTSQVCKELGKHLKIQVMV 232
Query: 242 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQP 301
TTGGTSLKDDIMRLYQPVHLLVGTPGRILDL KKGVCVLKDCS+LVMDEADKLLS EFQP
Sbjct: 233 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLTKKGVCVLKDCSVLVMDEADKLLSQEFQP 292
Query: 302 SIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEE 361
S+E LI FLP +RQILMFSATFPVTVKDFKDR+L PY+INLMDELTLKGITQFYAFVEE
Sbjct: 293 SVEHLISFLPESRQILMFSATFPVTVKDFKDRFLTNPYVINLMDELTLKGITQFYAFVEE 352
Query: 362 RQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFP 421
RQK+HCLNTLFSK LQ +S C N RV + ++ S
Sbjct: 353 RQKIHCLNTLFSK---------LQINQSI-IFC--NSVNRVELLAKKITELGY----SCS 396
Query: 422 RLPQWCMQESR--------------LYC--LFTRGIDIQAVNVVINFDFPKNSETYLHRV 465
+ +Q+ R L C LFTRGIDIQAVNVVINFDFPKN+ETYLHRV
Sbjct: 397 YIHAKMLQDHRNRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKNAETYLHRV 456
Query: 466 GRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
GRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPP IDQAIYC
Sbjct: 457 GRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPHIDQAIYC 504
>UniRef100_Q8RXK6 Putative RNA helicase [Arabidopsis thaliana]
Length = 505
Score = 706 bits (1823), Expect = 0.0
Identities = 381/517 (73%), Positives = 406/517 (77%), Gaps = 21/517 (4%)
Query: 4 NNRGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQR-HMMQNQHQQH 62
NNRGRYPPGIG GRG N N +Q R +Q QYVQR + QN QQ
Sbjct: 2 NNRGRYPPGIGAGRGAF---------NPNPNYQSRSGYQQHPPPQYVQRGNYAQNHQQQF 52
Query: 63 YQNQQQNQQQNQQQQQQQQWLRRNQL-GGGTDTNVVEEVEKTVQSEANDSSSQDWKARLK 121
Q Q Q QQQQQQQQWLRR Q+ GG ++ + V EVEKTVQSE D +S+DWKARLK
Sbjct: 53 QQAPSQPHQYQQQQQQQQQWLRRGQIPGGNSNGDAVVEVEKTVQSEVIDPNSEDWKARLK 112
Query: 122 LPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDIL 181
LP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTG DIL
Sbjct: 113 LPAPDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGRDIL 172
Query: 182 ARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMV 241
ARAKNGTGKTAAF IP LEKIDQDNN+IQ VI+VPTRELALQTSQVCKELGKHL+IQVMV
Sbjct: 173 ARAKNGTGKTAAFCIPVLEKIDQDNNVIQAVIIVPTRELALQTSQVCKELGKHLKIQVMV 232
Query: 242 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQP 301
TTGGTSLKDDIMRLYQPVHLLVGTPGRILDL KKGVCVLKDCS+LVMDEADKLLS EFQP
Sbjct: 233 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLTKKGVCVLKDCSVLVMDEADKLLSQEFQP 292
Query: 302 SIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEE 361
S+E LI FLP +RQILMFSATFPVTVKDFKDR+L PY+INLMDELTLKGITQFYAFVEE
Sbjct: 293 SVEHLISFLPESRQILMFSATFPVTVKDFKDRFLTNPYVINLMDELTLKGITQFYAFVEE 352
Query: 362 RQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP 416
RQK+HCLNTLFSK I+ + F S E ++ YSC K +
Sbjct: 353 RQKIHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMLQDHR 407
Query: 417 **SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGL 476
F + LFTRGIDIQAVNVVINFDFPKN+ETYLHRVGRSGRFGHLGL
Sbjct: 408 NRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKNAETYLHRVGRSGRFGHLGL 467
Query: 477 AVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
AVNLITYEDRFNLYRIEQELGTEIKQIPP IDQAIYC
Sbjct: 468 AVNLITYEDRFNLYRIEQELGTEIKQIPPHIDQAIYC 504
>UniRef100_Q6H7S3 Putative RNA helicase [Oryza sativa]
Length = 508
Score = 684 bits (1765), Expect = 0.0
Identities = 377/525 (71%), Positives = 405/525 (76%), Gaps = 38/525 (7%)
Query: 6 RGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQ------QQYVQRHMMQNQH 59
R RYPPGIG GRGG+ N QQ+ +H QQQ QQYVQR Q+ H
Sbjct: 4 RARYPPGIGNGRGGNPN-----YYNRGPPLQQQHNHHQQQQTSAPHHQQYVQRQPQQHHH 58
Query: 60 QQHYQNQQQNQQQNQQQQQQQQWLRRNQLG---GGTDTNVVEEVEKTVQSEAND---SSS 113
H+Q Q QQQQQQWLRRNQ+ GTD N E + QS A D SSS
Sbjct: 59 HNHHQ---------QHQQQQQQWLRRNQIAREAAGTDRN--SEPKAVAQSPAVDGIDSSS 107
Query: 114 QDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPI 173
QDWKA+LKLPP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPI
Sbjct: 108 QDWKAQLKLPPQDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPI 167
Query: 174 ALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGK 233
ALTGSDILARAKNGTGKTAAF IPALEKIDQ+ N IQVVILVPTRELALQTSQVCKELGK
Sbjct: 168 ALTGSDILARAKNGTGKTAAFCIPALEKIDQEKNAIQVVILVPTRELALQTSQVCKELGK 227
Query: 234 HLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADK 293
HL+IQVMVTTGGTSLKDDI+RLYQPVHLLVGTPGRILDL KKG+C+LKDCSML+MDEADK
Sbjct: 228 HLKIQVMVTTGGTSLKDDIIRLYQPVHLLVGTPGRILDLTKKGICILKDCSMLIMDEADK 287
Query: 294 LLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGIT 353
LLSPEFQPS+EQLI++LP +RQILMFSATFPVTVK+FKD+YL KPY+INLMDELTLKGIT
Sbjct: 288 LLSPEFQPSVEQLIRYLPASRQILMFSATFPVTVKEFKDKYLPKPYVINLMDELTLKGIT 347
Query: 354 QFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC---- 409
QFYAFVEERQKVHCLNTLFSK I+ + F S E ++ YSC
Sbjct: 348 QFYAFVEERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIH 402
Query: 410 -KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRS 468
K + F + LFTRGIDIQAVNVVINFDFPK +ETYLHRVGRS
Sbjct: 403 AKMLQDHRNRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKTAETYLHRVGRS 462
Query: 469 GRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
GRFGHLGLAVNLITYEDRFNLYRIEQELGTEIK IPP IDQAIYC
Sbjct: 463 GRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKPIPPQIDQAIYC 507
>UniRef100_O65275 F6N23.6 protein [Arabidopsis thaliana]
Length = 499
Score = 677 bits (1747), Expect = 0.0
Identities = 370/517 (71%), Positives = 396/517 (76%), Gaps = 27/517 (5%)
Query: 4 NNRGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQR-HMMQNQHQQH 62
NNRGRYPPGIG GRG N N +Q R +Q QYVQR + QN QQ
Sbjct: 2 NNRGRYPPGIGAGRGAF---------NPNPNYQSRSGYQQHPPPQYVQRGNYAQNHQQQF 52
Query: 63 YQNQQQNQQQNQQQQQQQQWLRRNQL-GGGTDTNVVEEVEKTVQSEANDSSSQDWKARLK 121
Q Q Q QQQQQQQQWLRR Q+ GG ++ + V EVEKTVQSE D +S+DWKARLK
Sbjct: 53 QQAPSQPHQYQQQQQQQQQWLRRGQIPGGNSNGDAVVEVEKTVQSEVIDPNSEDWKARLK 112
Query: 122 LPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDIL 181
LP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTG DIL
Sbjct: 113 LPAPDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGRDIL 172
Query: 182 ARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMV 241
ARAKNGTGKTAAF IP LEKIDQDNN+IQ ++ TSQVCKELGKHL+IQVMV
Sbjct: 173 ARAKNGTGKTAAFCIPVLEKIDQDNNVIQGNCVLD------HTSQVCKELGKHLKIQVMV 226
Query: 242 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQP 301
TTGGTSLKDDIMRLYQPVHLLVGTPGRILDL KKGVCVLKDCS+LVMDEADKLLS EFQP
Sbjct: 227 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLTKKGVCVLKDCSVLVMDEADKLLSQEFQP 286
Query: 302 SIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEE 361
S+E LI FLP +RQILMFSATFPVTVKDFKDR+L PY+INLMDELTLKGITQFYAFVEE
Sbjct: 287 SVEHLISFLPESRQILMFSATFPVTVKDFKDRFLTNPYVINLMDELTLKGITQFYAFVEE 346
Query: 362 RQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP 416
RQK+HCLNTLFSK I+ + F S E ++ YSC K +
Sbjct: 347 RQKIHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMLQDHR 401
Query: 417 **SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGL 476
F + LFTRGIDIQAVNVVINFDFPKN+ETYLHRVGRSGRFGHLGL
Sbjct: 402 NRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKNAETYLHRVGRSGRFGHLGL 461
Query: 477 AVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
AVNLITYEDRFNLYRIEQELGTEIKQIPP IDQAIYC
Sbjct: 462 AVNLITYEDRFNLYRIEQELGTEIKQIPPHIDQAIYC 498
>UniRef100_O80838 Putative ATP-dependent RNA helicase [Arabidopsis thaliana]
Length = 528
Score = 666 bits (1718), Expect = 0.0
Identities = 365/531 (68%), Positives = 406/531 (75%), Gaps = 26/531 (4%)
Query: 2 NNNNRGRYPPGIGLGRGGSGGGGGGLTSN---------SNTGFQQRPHHQ---YQQQQQY 49
NNNNRGR+PPGIG G N S T F Q+P Q Y Q Q
Sbjct: 4 NNNNRGRFPPGIGAAGPGPDPNFQSRNPNPPQPQQYLQSRTPFPQQPQPQPPQYLQSQSD 63
Query: 50 VQRHMMQNQHQQHYQNQQ-QNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEA 108
Q+++ + QQ Q QQ Q QQQ QQQQQ+QQW RR QL G D + ++EVEKTVQSEA
Sbjct: 64 AQQYVQRGYPQQIQQQQQLQQQQQQQQQQQEQQWSRRAQLPG--DPSYIDEVEKTVQSEA 121
Query: 109 -NDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQ 167
+DS+++DWKA LKLPP D RY+TEDVTATKGNEFEDYFLKR+LL GIYEKGFE+PSPIQ
Sbjct: 122 ISDSNNEDWKATLKLPPRDNRYQTEDVTATKGNEFEDYFLKRDLLRGIYEKGFEKPSPIQ 181
Query: 168 EESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQV 227
EESIPIALTGSDILARAKNGTGKT AF IP LEKID +NN+IQ VILVPTRELALQTSQV
Sbjct: 182 EESIPIALTGSDILARAKNGTGKTGAFCIPTLEKIDPENNVIQAVILVPTRELALQTSQV 241
Query: 228 CKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLV 287
CKEL K+L+I+VMVTTGGTSL+DDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDC+MLV
Sbjct: 242 CKELSKYLKIEVMVTTGGTSLRDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCAMLV 301
Query: 288 MDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDEL 347
MDEADKLLS EFQPSIE+LIQFLP +RQILMFSATFPVTVK FKDRYL+KPYIINLMD+L
Sbjct: 302 MDEADKLLSVEFQPSIEELIQFLPESRQILMFSATFPVTVKSFKDRYLKKPYIINLMDQL 361
Query: 348 TLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLY 407
TL G+TQ+YAFVEERQKVHCLNTLFSK I+ + F S E ++ Y
Sbjct: 362 TLMGVTQYYAFVEERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGY 416
Query: 408 SC-----KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYL 462
SC K V F + LFTRGIDIQAVNVVINFDFP+ SE+YL
Sbjct: 417 SCFYIHAKMVQDHRNRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPRTSESYL 476
Query: 463 HRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
HRVGRSGRFGHLGLAVNL+TYEDRF +Y+ EQELGTEIK IP ID+AIYC
Sbjct: 477 HRVGRSGRFGHLGLAVNLVTYEDRFKMYQTEQELGTEIKPIPSLIDKAIYC 527
>UniRef100_Q6H7S2 Putative RNA helicase [Oryza sativa]
Length = 483
Score = 665 bits (1715), Expect = 0.0
Identities = 369/519 (71%), Positives = 395/519 (76%), Gaps = 51/519 (9%)
Query: 6 RGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQRHMMQNQHQQHYQN 65
R RYPPGIG GRGG N N Y + +Q+H N HQQH
Sbjct: 4 RARYPPGIGNGRGG----------NPN----------YYNRGPPLQQHHHHNHHQQH--- 40
Query: 66 QQQNQQQNQQQQQQQQWLRRNQLG---GGTDTNVVEEVEKTVQSEAND---SSSQDWKAR 119
QQQQQQWLRRNQ+ GTD N E + QS A D SSSQDWKA+
Sbjct: 41 ----------QQQQQQWLRRNQIAREAAGTDRN--SEPKAVAQSPAVDGIDSSSQDWKAQ 88
Query: 120 LKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSD 179
LKLPP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSD
Sbjct: 89 LKLPPQDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSD 148
Query: 180 ILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQV 239
ILARAKNGTGKTAAF IPALEKIDQ+ N IQVVILVPTRELALQTSQVCKELGKHL+IQV
Sbjct: 149 ILARAKNGTGKTAAFCIPALEKIDQEKNAIQVVILVPTRELALQTSQVCKELGKHLKIQV 208
Query: 240 MVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEF 299
MVTTGGTSLKDDI+RLYQPVHLLVGTPGRILDL KKG+C+LKDCSML+MDEADKLLSPEF
Sbjct: 209 MVTTGGTSLKDDIIRLYQPVHLLVGTPGRILDLTKKGICILKDCSMLIMDEADKLLSPEF 268
Query: 300 QPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFV 359
QPS+EQLI++LP +RQILMFSATFPVTVK+FKD+YL KPY+INLMDELTLKGITQFYAFV
Sbjct: 269 QPSVEQLIRYLPASRQILMFSATFPVTVKEFKDKYLPKPYVINLMDELTLKGITQFYAFV 328
Query: 360 EERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVAR 414
EERQKVHCLNTLFSK I+ + F S E ++ YSC K +
Sbjct: 329 EERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMLQD 383
Query: 415 PP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHL 474
F + LFTRGIDIQAVNVVINFDFPK +ETYLHRVGRSGRFGHL
Sbjct: 384 HRNRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKTAETYLHRVGRSGRFGHL 443
Query: 475 GLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
GLAVNLITYEDRFNLYRIEQELGTEIK IPP IDQAIYC
Sbjct: 444 GLAVNLITYEDRFNLYRIEQELGTEIKPIPPQIDQAIYC 482
>UniRef100_Q7XMK8 OSJNBb0039L24.10 protein [Oryza sativa]
Length = 498
Score = 664 bits (1713), Expect = 0.0
Identities = 363/513 (70%), Positives = 392/513 (75%), Gaps = 24/513 (4%)
Query: 6 RGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQRHMMQNQHQQHYQN 65
R RYPPGIG GRGG N N + P Q+QQ Q Q + H Y
Sbjct: 4 RARYPPGIGNGRGG----------NPNYYGRGPPPSQHQQHQH--QHQQPPHPHHHQYVQ 51
Query: 66 QQQNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPA 125
+Q QQ Q QQWLRRNQ+ + E + ++ DSSSQDWKA+LKLPP
Sbjct: 52 RQPQPQQTPHNSQHQQWLRRNQIAA--EAAGASEQKAPPVADGIDSSSQDWKAQLKLPPQ 109
Query: 126 DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAK 185
DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAK
Sbjct: 110 DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAK 169
Query: 186 NGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGG 245
NGTGKTAAF IPALEKIDQD N IQVVILVPTRELALQTSQVCKELGKHL+IQVMVTTGG
Sbjct: 170 NGTGKTAAFCIPALEKIDQDKNAIQVVILVPTRELALQTSQVCKELGKHLKIQVMVTTGG 229
Query: 246 TSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQ 305
TSLKDDI+RLYQPVHLLVGTPGRILDL KKGVCVLK+CSMLVMDEADKLLSPEFQPSI++
Sbjct: 230 TSLKDDIVRLYQPVHLLVGTPGRILDLTKKGVCVLKNCSMLVMDEADKLLSPEFQPSIQE 289
Query: 306 LIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKV 365
LI++LP RQILMFSATFPVTVK+FKD+YL KPY+INLMDELTLKGITQFYAFVEERQKV
Sbjct: 290 LIRYLPSNRQILMFSATFPVTVKEFKDKYLPKPYVINLMDELTLKGITQFYAFVEERQKV 349
Query: 366 HCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP**SF 420
HCLNTLFSK I+ + F S E ++ YSC K + F
Sbjct: 350 HCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMLQDHRNRVF 404
Query: 421 PRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNL 480
+ LFTRGIDIQAVNVVINFDFPK++ETYLHRVGRSGRFGHLGLAVNL
Sbjct: 405 HDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKSAETYLHRVGRSGRFGHLGLAVNL 464
Query: 481 ITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
ITYEDRFNLYRIEQELGTEIK IPP ID+AIYC
Sbjct: 465 ITYEDRFNLYRIEQELGTEIKPIPPQIDRAIYC 497
>UniRef100_Q94BV4 At2g45810/F4I18.21 [Arabidopsis thaliana]
Length = 528
Score = 664 bits (1712), Expect = 0.0
Identities = 364/531 (68%), Positives = 405/531 (75%), Gaps = 26/531 (4%)
Query: 2 NNNNRGRYPPGIGLGRGGSGGGGGGLTSN---------SNTGFQQRPHHQ---YQQQQQY 49
NNNNRGR+PPGIG G N S T F Q+P Q Y Q Q
Sbjct: 4 NNNNRGRFPPGIGAAGPGPDPNFQSRNPNPPQPQQYLQSRTPFPQQPQPQPPQYLQSQSD 63
Query: 50 VQRHMMQNQHQQHYQNQQ-QNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEA 108
Q+++ + QQ Q QQ Q QQQ QQQQQ+QQW RR QL G D + ++EVEKTVQSEA
Sbjct: 64 AQQYVQRGYPQQIQQQQQLQQQQQQQQQQQEQQWSRRAQLPG--DPSYIDEVEKTVQSEA 121
Query: 109 -NDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQ 167
+DS+++DWKA LKLPP D RY+TEDVTATKGNEFEDY LKR+LL GIYEKGFE+PSPIQ
Sbjct: 122 ISDSNNEDWKATLKLPPRDNRYQTEDVTATKGNEFEDYLLKRDLLRGIYEKGFEKPSPIQ 181
Query: 168 EESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQV 227
EESIPIALTGSDILARAKNGTGKT AF IP LEKID +NN+IQ VILVPTRELALQTSQV
Sbjct: 182 EESIPIALTGSDILARAKNGTGKTGAFCIPTLEKIDPENNVIQAVILVPTRELALQTSQV 241
Query: 228 CKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLV 287
CKEL K+L+I+VMVTTGGTSL+DDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDC+MLV
Sbjct: 242 CKELSKYLKIEVMVTTGGTSLRDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCAMLV 301
Query: 288 MDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDEL 347
MDEADKLLS EFQPSIE+LIQFLP +RQILMFSATFPVTVK FKDRYL+KPYIINLMD+L
Sbjct: 302 MDEADKLLSVEFQPSIEELIQFLPESRQILMFSATFPVTVKSFKDRYLKKPYIINLMDQL 361
Query: 348 TLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLY 407
TL G+TQ+YAFVEERQKVHCLNTLFSK I+ + F S E ++ Y
Sbjct: 362 TLMGVTQYYAFVEERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGY 416
Query: 408 SC-----KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYL 462
SC K V F + LFTRGIDIQAVNVVINFDFP+ SE+YL
Sbjct: 417 SCFYIHAKMVQDHRNRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPRTSESYL 476
Query: 463 HRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
HRVGRSGRFGHLGLAVNL+TYEDRF +Y+ EQELGTEIK IP ID+AIYC
Sbjct: 477 HRVGRSGRFGHLGLAVNLVTYEDRFKMYQTEQELGTEIKPIPSLIDKAIYC 527
>UniRef100_Q9FVY5 Putative RNA helicase [Oryza sativa]
Length = 521
Score = 659 bits (1701), Expect = 0.0
Identities = 365/527 (69%), Positives = 392/527 (74%), Gaps = 30/527 (5%)
Query: 6 RGRYPPGI--------------GLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQ 51
R RYPPG G G GG G GGGG N G+ R QQQ Y
Sbjct: 5 RARYPPGYTSGGGGGGGGGGGGGRGNGGGGFGGGGGGGGGNHGYYGRGPQPQPQQQHY-- 62
Query: 52 RHMMQNQHQQHYQNQQQNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDS 111
H Q HQ +Q QQQ+ Q+N QQQQ WLRR+Q + V Q EA DS
Sbjct: 63 HHQAQQLHQ--HQQQQQHAQRNSSSQQQQ-WLRRDQATAAAASGEVA-ARTAAQLEAVDS 118
Query: 112 SSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESI 171
SS+DWKA+L LP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESI
Sbjct: 119 SSEDWKAQLNLPAPDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESI 178
Query: 172 PIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKEL 231
PIALTGSDILARAKNGTGKTAAF IPALEKID + N IQVVILVPTRELALQTSQVCKEL
Sbjct: 179 PIALTGSDILARAKNGTGKTAAFCIPALEKIDPEKNAIQVVILVPTRELALQTSQVCKEL 238
Query: 232 GKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEA 291
GK+L IQVMV+TGGTSLKDDIMRLYQPVHLLVGTPGRILDL +KG+CVLKDCSMLVMDEA
Sbjct: 239 GKYLNIQVMVSTGGTSLKDDIMRLYQPVHLLVGTPGRILDLTRKGICVLKDCSMLVMDEA 298
Query: 292 DKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKG 351
DKLL+PEFQPSIEQLI FLP RQ+LMFSATFPVTVKDFK++YL +PY+INLMDELTLKG
Sbjct: 299 DKLLAPEFQPSIEQLIHFLPANRQLLMFSATFPVTVKDFKEKYLPRPYVINLMDELTLKG 358
Query: 352 ITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-- 409
ITQ+YAFVEERQKVHCLNTLFSK I+ + F S E ++ YSC
Sbjct: 359 ITQYYAFVEERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFY 413
Query: 410 ---KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVG 466
K + F + LFTRGIDIQAVNVVINFDFPK SETYLHRVG
Sbjct: 414 IHAKMLQDHRNRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKTSETYLHRVG 473
Query: 467 RSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
RSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIK IPP ID A+YC
Sbjct: 474 RSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKTIPPQIDLAVYC 520
>UniRef100_Q8H8Z5 Putative RNA helicase [Oryza sativa]
Length = 521
Score = 654 bits (1688), Expect = 0.0
Identities = 363/527 (68%), Positives = 390/527 (73%), Gaps = 30/527 (5%)
Query: 6 RGRYPPGI--------------GLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQ 51
R RYPPG G G GG G GGGG N G+ R QQQ Y
Sbjct: 5 RARYPPGYTSGGGGGGGGGGGGGRGNGGGGFGGGGGGGGGNHGYYGRGPQPQPQQQHY-- 62
Query: 52 RHMMQNQHQQHYQNQQQNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDS 111
H Q HQ +Q QQQ+ Q+N QQQQ WLRR+Q + V Q EA DS
Sbjct: 63 HHQAQQLHQ--HQQQQQHAQRNSSSQQQQ-WLRRDQATAAAASGEVA-ARTAAQLEAVDS 118
Query: 112 SSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESI 171
SS+DWKA+L LP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESI
Sbjct: 119 SSEDWKAQLNLPAPDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESI 178
Query: 172 PIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKEL 231
PIALTGSDILARAKNGTGKTAAF IPALEKID + N IQVVILVPTRELALQTSQVCKEL
Sbjct: 179 PIALTGSDILARAKNGTGKTAAFCIPALEKIDPEKNAIQVVILVPTRELALQTSQVCKEL 238
Query: 232 GKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEA 291
GK+L IQVMV+TGGTSLKDDIMRLYQPVHLLVGTPGRILDL +KG+CVLKDCSMLVMDEA
Sbjct: 239 GKYLNIQVMVSTGGTSLKDDIMRLYQPVHLLVGTPGRILDLTRKGICVLKDCSMLVMDEA 298
Query: 292 DKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKG 351
DKLL+PEFQPSIEQLI FLP RQ+LMFSATFPVTVKDFK++YL +PY+INLMDELTLKG
Sbjct: 299 DKLLAPEFQPSIEQLIHFLPANRQLLMFSATFPVTVKDFKEKYLPRPYVINLMDELTLKG 358
Query: 352 ITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-- 409
ITQ+YAFVEERQKVHCLNTLF K I+ + F S E ++ YSC
Sbjct: 359 ITQYYAFVEERQKVHCLNTLFLKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFY 413
Query: 410 ---KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVG 466
K + F + LFTRGI IQAVNVVINFDFPK SETYLHRVG
Sbjct: 414 IHAKMLQDHRNRVFHDFRNGACRNLVCTDLFTRGIGIQAVNVVINFDFPKTSETYLHRVG 473
Query: 467 RSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
RSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIK IPP ID A+YC
Sbjct: 474 RSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKTIPPQIDLAVYC 520
>UniRef100_Q9M2E0 DEAD box RNA helicase RH12 [Arabidopsis thaliana]
Length = 498
Score = 647 bits (1668), Expect = 0.0
Identities = 354/518 (68%), Positives = 391/518 (75%), Gaps = 29/518 (5%)
Query: 3 NNNRGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQ-QQQQYVQRHMMQNQHQQ 61
N NRGRYPPG+G GRG N + Q Q Q QQYVQR QN Q
Sbjct: 2 NTNRGRYPPGVGTGRGAP----------PNPDYHQSYRQQQPPQDQQYVQRGYSQNPQQM 51
Query: 62 HYQNQQQNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEAN-DSSSQDWKARL 120
Q QQQ+QQQQQQQQW RR QL G +N E V++T Q EA+ D++ QDWKA L
Sbjct: 52 ------QLQQQHQQQQQQQQWSRRPQLPGNA-SNANEVVQQTTQPEASSDANGQDWKATL 104
Query: 121 KLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDI 180
+LPP DTRY+T DVTATKGNEFEDYFLKR+LL GIYEKGFE+PSPIQEESIPIALTGSDI
Sbjct: 105 RLPPPDTRYQTADVTATKGNEFEDYFLKRDLLKGIYEKGFEKPSPIQEESIPIALTGSDI 164
Query: 181 LARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVM 240
LARAKNGTGKT AF IP LEKID +NN+IQ +ILVPTRELALQTSQVCKEL K+L IQVM
Sbjct: 165 LARAKNGTGKTGAFCIPVLEKIDPNNNVIQAMILVPTRELALQTSQVCKELSKYLNIQVM 224
Query: 241 VTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQ 300
VTTGGTSL+DDIMRL+QPVHLLVGTPGRILDL KKGVCVLKDC+MLVMDEADKLLS EFQ
Sbjct: 225 VTTGGTSLRDDIMRLHQPVHLLVGTPGRILDLTKKGVCVLKDCAMLVMDEADKLLSAEFQ 284
Query: 301 PSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVE 360
PS+E+LIQFLP RQ LMFSATFPVTVK FKDR+LRKPY+INLMD+LTL G+TQ+YAFVE
Sbjct: 285 PSLEELIQFLPQNRQFLMFSATFPVTVKAFKDRHLRKPYVINLMDQLTLMGVTQYYAFVE 344
Query: 361 ERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARP 415
ERQKVHCLNTLFSK I+ + F S E ++ YSC K V
Sbjct: 345 ERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMVQDH 399
Query: 416 P**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLG 475
F + LFTRGIDIQAVNVVINFDFP+ SE+YLHRVGRSGRFGHLG
Sbjct: 400 RNRVFHEFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPRTSESYLHRVGRSGRFGHLG 459
Query: 476 LAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
LAVNL+TYEDRF +Y+ EQELGTEIK IP IDQAIYC
Sbjct: 460 LAVNLVTYEDRFKMYQTEQELGTEIKPIPSNIDQAIYC 497
>UniRef100_Q9ZS08 RNA helicase [Arabidopsis thaliana]
Length = 498
Score = 640 bits (1651), Expect = 0.0
Identities = 350/517 (67%), Positives = 393/517 (75%), Gaps = 27/517 (5%)
Query: 3 NNNRGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQRHMMQNQHQQH 62
N NRGRYPPG+G GRG N + ++Q+ Q Q QQYVQR QN Q
Sbjct: 2 NTNRGRYPPGVGTGRGAPPN------PNYHQSYRQQ---QPPQDQQYVQRGYSQNPQQM- 51
Query: 63 YQNQQQNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEAN-DSSSQDWKARLK 121
Q QQQ+QQQQQQQQW +R QL +N E V++T Q EA+ D++ Q+WKA L+
Sbjct: 52 -----QLQQQHQQQQQQQQWSKRPQLPENA-SNANEVVQQTTQPEASSDANGQNWKATLR 105
Query: 122 LPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDIL 181
LPP DTRY+T DVTATKGNEFE+YFLKR+LL GIYEKGFE+PSPIQEESIPIALTGSDIL
Sbjct: 106 LPPPDTRYQTADVTATKGNEFENYFLKRDLLKGIYEKGFEKPSPIQEESIPIALTGSDIL 165
Query: 182 ARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMV 241
ARAKNGTGKT AF IP LEKID +NN+IQ +ILVPTRELALQTSQVCKEL K+L IQVMV
Sbjct: 166 ARAKNGTGKTGAFCIPVLEKIDPNNNVIQAMILVPTRELALQTSQVCKELSKYLNIQVMV 225
Query: 242 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQP 301
TTGGTSL+DDIMRL+QPVHLLVGTPGRILDL KKGVCVLKDC+MLVMDEADKLLS EFQP
Sbjct: 226 TTGGTSLRDDIMRLHQPVHLLVGTPGRILDLTKKGVCVLKDCAMLVMDEADKLLSAEFQP 285
Query: 302 SIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEE 361
S+E+LIQFLP RQ LMFSATFPVTVK FKDR+LRKPY+INLMD+LTL G+TQ+YAFVEE
Sbjct: 286 SLEELIQFLPQNRQFLMFSATFPVTVKAFKDRHLRKPYVINLMDQLTLMGVTQYYAFVEE 345
Query: 362 RQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP 416
RQKVHCLNTLFSK I+ + F S E ++ YSC K V
Sbjct: 346 RQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMVQDHR 400
Query: 417 **SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGL 476
F + LFTRGIDIQAVNVVINFDFP+ SE+YLHRVGRSGRFGHLGL
Sbjct: 401 NRVFHEFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPRTSESYLHRVGRSGRFGHLGL 460
Query: 477 AVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
AVNL+TYEDRF +Y+ EQELGTEIK IP IDQAIYC
Sbjct: 461 AVNLVTYEDRFKMYQTEQELGTEIKPIPSNIDQAIYC 497
>UniRef100_UPI00004688D2 UPI00004688D2 UniRef100 entry
Length = 429
Score = 501 bits (1291), Expect = e-140
Identities = 271/442 (61%), Positives = 321/442 (72%), Gaps = 30/442 (6%)
Query: 86 NQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDY 145
N + +TN + + E N + WK ++ P D RY+TEDVT TKGNEFEDY
Sbjct: 2 NCVASNANTNALNNSNNLNKIEDNVIFDEAWKKKILEPLKDPRYKTEDVTKTKGNEFEDY 61
Query: 146 FLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQD 205
FLKRELLMGI+EKG+E+PSPIQEESIP+AL G +ILARAKNGTGKTAAF+IP LEK +
Sbjct: 62 FLKRELLMGIFEKGYEKPSPIQEESIPVALAGKNILARAKNGTGKTAAFAIPLLEKCNTH 121
Query: 206 NNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGT 265
N IQ +ILVPTRELALQTS + KELGKH++IQ MVTTGGTSL++DIMRLY VH+L GT
Sbjct: 122 KNFIQGLILVPTRELALQTSAMIKELGKHMKIQCMVTTGGTSLREDIMRLYNAVHILCGT 181
Query: 266 PGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPV 325
PGRILDLA K V L C ++VMDEADKLLSPEFQP +E+L++FLP +QILM+SATFPV
Sbjct: 182 PGRILDLANKDVANLSGCHIMVMDEADKLLSPEFQPIVEELMKFLPKEKQILMYSATFPV 241
Query: 326 TVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQ 385
TVK+F+ YL + INLMDELTLKGITQ+YAFV+ERQKVHCLNTLF+K I+ +
Sbjct: 242 TVKEFRQIYLSDAHEINLMDELTLKGITQYYAFVKERQKVHCLNTLFAKL---QINQAII 298
Query: 386 FGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQW---------------CMQE 430
F C N TRV + ++ R+ Q C+
Sbjct: 299 F-------C--NSITRVELLAKKITELGYSSFYIHARMSQTHRNRVFHDFRNGACRCLVS 349
Query: 431 SRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLY 490
S LFTRGIDIQ+VNVVINFDFPKNSETYLHR+GRSGR+GHLGLA+NLITYEDRFNLY
Sbjct: 350 SD---LFTRGIDIQSVNVVINFDFPKNSETYLHRIGRSGRYGHLGLAINLITYEDRFNLY 406
Query: 491 RIEQELGTEIKQIPPFIDQAIY 512
+IE ELGTEI+ IP ID +IY
Sbjct: 407 KIELELGTEIQPIPNEIDPSIY 428
>UniRef100_O97285 Putative ATP-dependent RNA Helicase [Plasmodium falciparum]
Length = 433
Score = 497 bits (1280), Expect = e-139
Identities = 267/444 (60%), Positives = 322/444 (72%), Gaps = 30/444 (6%)
Query: 84 RRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFE 143
+ N +TN + + + N ++WK ++ P D RY+TEDVT TKGNEFE
Sbjct: 4 KTNCTNSNANTNTLNSSSNYNKIDDNIILDEEWKKKILEPLKDLRYKTEDVTKTKGNEFE 63
Query: 144 DYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKID 203
DYFLKRELLMGI+EKG+E+PSPIQEESIP+AL G +ILARAKNGTGKTAAF+IP LEK +
Sbjct: 64 DYFLKRELLMGIFEKGYEKPSPIQEESIPVALAGKNILARAKNGTGKTAAFAIPLLEKCN 123
Query: 204 QDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLV 263
N IQ +ILVPTRELALQTS + KELGKH+++Q MVTTGGTSL++DIMRLY VH+L
Sbjct: 124 THKNFIQGLILVPTRELALQTSAMIKELGKHMKVQCMVTTGGTSLREDIMRLYNVVHILC 183
Query: 264 GTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATF 323
GTPGRILDLA K V L C ++VMDEADKLLSPEFQP +E+L++FLP +QILM+SATF
Sbjct: 184 GTPGRILDLANKDVANLSGCHIMVMDEADKLLSPEFQPIVEELMKFLPKEKQILMYSATF 243
Query: 324 PVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHL 383
PVTVK+F+ YL + INLMDELTLKGITQ+YAFV+ERQKVHCLNTLF+K I+
Sbjct: 244 PVTVKEFRAIYLSDAHEINLMDELTLKGITQYYAFVKERQKVHCLNTLFAKL---QINQA 300
Query: 384 LQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQW---------------CM 428
+ F C N TRV + ++ R+ Q C+
Sbjct: 301 IIF-------C--NSITRVELLAKKITELGYSSFYIHARMSQTHRNRVFHDFRNGACRCL 351
Query: 429 QESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFN 488
S LFTRGIDIQ+VNVVINFDFPKNSETYLHR+GRSGR+GHLGLA+NLIT+EDRFN
Sbjct: 352 VSSD---LFTRGIDIQSVNVVINFDFPKNSETYLHRIGRSGRYGHLGLAINLITFEDRFN 408
Query: 489 LYRIEQELGTEIKQIPPFIDQAIY 512
LY+IE ELGTEI+ IP ID ++Y
Sbjct: 409 LYKIEVELGTEIQPIPNEIDPSLY 432
>UniRef100_Q7PDT0 ATP-dependent RNA helicase [Plasmodium yoelii yoelii]
Length = 477
Score = 495 bits (1275), Expect = e-138
Identities = 271/451 (60%), Positives = 322/451 (71%), Gaps = 37/451 (8%)
Query: 84 RRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFE 143
+ N + +TN + + E N + WK ++ P D RY+TEDVT TKGNEFE
Sbjct: 41 KTNCVASNANTNALNNSNNLNKIEDNVIFDEAWKKKILEPLKDPRYKTEDVTKTKGNEFE 100
Query: 144 DYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKID 203
DYFLKRELLMGI+EKG+E+PSPIQEESIP+AL G +ILARAKNGTGKTAAF+IP LEK +
Sbjct: 101 DYFLKRELLMGIFEKGYEKPSPIQEESIPVALAGKNILARAKNGTGKTAAFAIPLLEKCN 160
Query: 204 QDNNIIQ-------VVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLY 256
N IQ +ILVPTRELALQTS + KELGKH++IQ MVTTGGTSL++DIMRLY
Sbjct: 161 THKNFIQGKTIQKISLILVPTRELALQTSAMIKELGKHMKIQCMVTTGGTSLREDIMRLY 220
Query: 257 QPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQI 316
VH+L GTPGRILDLA K V L C ++VMDEADKLLSPEFQP +E+L++FLP +QI
Sbjct: 221 NAVHILCGTPGRILDLANKDVANLSGCHIMVMDEADKLLSPEFQPIVEELMKFLPKEKQI 280
Query: 317 LMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAA 376
LM+SATFPVTVK+F+ YL + INLMDELTLKGITQ+YAFV+ERQKVHCLNTLF+K
Sbjct: 281 LMYSATFPVTVKEFRQIYLSDAHEINLMDELTLKGITQYYAFVKERQKVHCLNTLFAKL- 339
Query: 377 NKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQW---------- 426
I+ + F C N TRV + ++ R+ Q
Sbjct: 340 --QINQAIIF-------C--NSITRVELLAKKITELGYSSFYIHARMSQTHRNRVFHDFR 388
Query: 427 -----CMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLI 481
C+ S LFTRGIDIQ+VNVVINFDFPKNSETYLHR+GRSGR+GHLGLA+NLI
Sbjct: 389 NGACRCLVSSD---LFTRGIDIQSVNVVINFDFPKNSETYLHRIGRSGRYGHLGLAINLI 445
Query: 482 TYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
TYEDRFNLY+IE ELGTEI+ IP ID +IY
Sbjct: 446 TYEDRFNLYKIELELGTEIQPIPNEIDPSIY 476
>UniRef100_UPI00003C1C9D UPI00003C1C9D UniRef100 entry
Length = 534
Score = 494 bits (1271), Expect = e-138
Identities = 268/395 (67%), Positives = 304/395 (76%), Gaps = 10/395 (2%)
Query: 122 LPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDIL 181
L P R +TEDV TKGNEFEDYFLKRELLMGI+E GFERPSPIQEE+IPIALTG DIL
Sbjct: 129 LLPHHERPQTEDVLNTKGNEFEDYFLKRELLMGIFEAGFERPSPIQEEAIPIALTGRDIL 188
Query: 182 ARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMV 241
ARAKNGTGKTAA+ IP+LEK++ N IQ V+LVPTRELALQTSQV K LGKHL ++VMV
Sbjct: 189 ARAKNGTGKTAAYVIPSLEKLNTKKNKIQAVLLVPTRELALQTSQVAKTLGKHLGVEVMV 248
Query: 242 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQP 301
TTGGT+L+DDI+RL Q VHLLVGTPGRILDLA KGV L C+ VMDEADKLLSPEF P
Sbjct: 249 TTGGTTLRDDILRLGQTVHLLVGTPGRILDLAGKGVADLSQCTTFVMDEADKLLSPEFTP 308
Query: 302 SIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEE 361
+EQL+ FLP RQ+++FSATFP+ VKDFKDR + KPY INLMDELTL+G+TQ+YAFVEE
Sbjct: 309 VMEQLLSFLPKERQVMLFSATFPLIVKDFKDRNMVKPYEINLMDELTLRGVTQYYAFVEE 368
Query: 362 RQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP 416
RQKVHCLNTLFSK I+ + F S T E ++ YSC K +
Sbjct: 369 RQKVHCLNTLFSKL---QINQSIIFCNS--TNRVELLAKKITELGYSCFYSHAKMLQAHR 423
Query: 417 **SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGL 476
F + L TRGIDIQAVNVVINFDFPKN+ETYLHR+GRSGRFGHLGL
Sbjct: 424 NRVFHDFRNGACRNLVCSDLLTRGIDIQAVNVVINFDFPKNAETYLHRIGRSGRFGHLGL 483
Query: 477 AVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAI 511
A+NLITYEDRFNLYRIEQELGTEI+ IP ID+ +
Sbjct: 484 AINLITYEDRFNLYRIEQELGTEIQPIPSNIDKRL 518
>UniRef100_Q6C0X2 YlDHH1 protein [Yarrowia lipolytica]
Length = 522
Score = 491 bits (1263), Expect = e-137
Identities = 261/403 (64%), Positives = 306/403 (75%), Gaps = 10/403 (2%)
Query: 115 DWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIA 174
+WK L +P DTR++TEDVTATKG FED+FLKRELLMGI+E GFE PSPIQEE+IPIA
Sbjct: 3 EWKESLNVPKKDTRHKTEDVTATKGTGFEDFFLKRELLMGIFEAGFENPSPIQEEAIPIA 62
Query: 175 LTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKH 234
L G DILARAKNGTGKTAAF IPAL++++ N IQ +I+VPTRELALQTSQVCK LGKH
Sbjct: 63 LAGRDILARAKNGTGKTAAFVIPALQQVNPKVNKIQALIMVPTRELALQTSQVCKTLGKH 122
Query: 235 LQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKL 294
L I+VMVTTGGT+L+DDIMRL VH+LVGTPGR+LDLA KGV L + M +MDEADKL
Sbjct: 123 LGIKVMVTTGGTNLRDDIMRLEDTVHVLVGTPGRVLDLAGKGVADLSESPMFIMDEADKL 182
Query: 295 LSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQ 354
LSP+F P IEQ++ F P RQIL+FSATFP+TVK F DR L KPY INLMDELTL+GITQ
Sbjct: 183 LSPDFTPIIEQVLHFFPEDRQILLFSATFPLTVKAFMDRNLHKPYEINLMDELTLRGITQ 242
Query: 355 FYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC----- 409
+YAFV+E+QK+HCLNTLFSK I+ + F S T E ++ YSC
Sbjct: 243 YYAFVDEKQKLHCLNTLFSKL---DINQSIIFCNS--TVRVELLARKITELGYSCYYSHA 297
Query: 410 KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSG 469
K + F + L TRGIDIQAVNVVINFDFPKN+ETYLHR+GRSG
Sbjct: 298 KMIQSHRNRVFHEFRNGTCRNLVCSDLLTRGIDIQAVNVVINFDFPKNAETYLHRIGRSG 357
Query: 470 RFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
RFGHLG+A+NLI + DR+NLY+IEQELGTEIK IP ID+ +Y
Sbjct: 358 RFGHLGIAINLINWNDRYNLYKIEQELGTEIKPIPAQIDKNLY 400
>UniRef100_Q09181 Putative ATP-dependent RNA helicase ste13 [Schizosaccharomyces
pombe]
Length = 485
Score = 486 bits (1252), Expect = e-136
Identities = 258/418 (61%), Positives = 311/418 (73%), Gaps = 12/418 (2%)
Query: 100 VEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKG 159
++K + ND S +K ++K P D R +TEDVT T+G EFEDY+LKRELLMGI+E G
Sbjct: 6 IQKLENANLNDRES--FKGQMKAQPVDMRPKTEDVTKTRGTEFEDYYLKRELLMGIFEAG 63
Query: 160 FERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRE 219
FERPSPIQEESIPIAL+G DILARAKNGTGKTAAF IP+LEK+D + IQ +ILVPTRE
Sbjct: 64 FERPSPIQEESIPIALSGRDILARAKNGTGKTAAFVIPSLEKVDTKKSKIQTLILVPTRE 123
Query: 220 LALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCV 279
LALQTSQVCK LGKH+ ++VMVTTGGT+L+DDI+RL VH++VGTPGR+LDLA KGV
Sbjct: 124 LALQTSQVCKTLGKHMNVKVMVTTGGTTLRDDIIRLNDTVHIVVGTPGRVLDLAGKGVAD 183
Query: 280 LKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPY 339
+C+ VMDEADKLLSPEF P IEQL+ + P RQI ++SATFP+ VK+F D++L KPY
Sbjct: 184 FSECTTFVMDEADKLLSPEFTPIIEQLLSYFPKNRQISLYSATFPLIVKNFMDKHLNKPY 243
Query: 340 IINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH* 399
INLMDELTL+G+TQ+YAFV+E QKVHCLNTLFSK I+ + F S T E
Sbjct: 244 EINLMDELTLRGVTQYYAFVDESQKVHCLNTLFSKL---QINQSIIFCNS--TNRVELLA 298
Query: 400 TRVFMFLYSC-----KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDF 454
++ YSC K + F + L TRGIDIQAVNVVINFDF
Sbjct: 299 KKITELGYSCFYSHAKMLQSHRNRVFHNFRNGVCRNLVCSDLLTRGIDIQAVNVVINFDF 358
Query: 455 PKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
PKN+ETYLHR+GRSGRFGH GLA++ I++ DRFNLYRIE ELGTEI+ IPP ID ++Y
Sbjct: 359 PKNAETYLHRIGRSGRFGHRGLAISFISWADRFNLYRIENELGTEIQPIPPSIDPSLY 416
>UniRef100_UPI000023DF32 UPI000023DF32 UniRef100 entry
Length = 486
Score = 483 bits (1244), Expect = e-135
Identities = 256/412 (62%), Positives = 306/412 (74%), Gaps = 24/412 (5%)
Query: 113 SQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIP 172
S +WK L LP D R +TEDVT TKG EFE++ LKR+LLMGI+E GFE+PSPIQEESIP
Sbjct: 15 SDEWKKNLNLPAKDNRQQTEDVTNTKGLEFENFALKRDLLMGIFEAGFEKPSPIQEESIP 74
Query: 173 IALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELG 232
+ALTG DILARAKNGTGKTAAF IP LE+I+ + IQ +ILVPTRELA+QTSQVCK LG
Sbjct: 75 VALTGRDILARAKNGTGKTAAFVIPTLERINPKISKIQCLILVPTRELAMQTSQVCKTLG 134
Query: 233 KHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEAD 292
KHL I VMVTTGGT L+DDI+RL PVH++VGTPGRILDLA K V L +C M +MDEAD
Sbjct: 135 KHLGINVMVTTGGTGLRDDIIRLQDPVHIVVGTPGRILDLAGKNVADLSECPMFIMDEAD 194
Query: 293 KLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGI 352
KLLS EF P IEQL+QF P RQ+++FSATFP++VKDF D+ + PY INLMDELTL+GI
Sbjct: 195 KLLSIEFTPVIEQLLQFHPKDRQVMLFSATFPLSVKDFSDKNMVSPYEINLMDELTLRGI 254
Query: 353 TQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDV 412
TQ+YAFVEE+QKVHCLNTLFSK I+ + F S+ RV + ++
Sbjct: 255 TQYYAFVEEKQKVHCLNTLFSKL---QINQSIIFCNST---------NRVELLAKKITEL 302
Query: 413 ARPP**SFPRLPQWCMQE----------SRLYC--LFTRGIDIQAVNVVINFDFPKNSET 460
S ++ Q L C L TRGIDIQAVNVVINFDFPKN+ET
Sbjct: 303 GYSCFYSHAKMQQHARNRVFHDFRNGVCRNLVCSDLLTRGIDIQAVNVVINFDFPKNAET 362
Query: 461 YLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
YLHR+GRSGR+GHLGLA+NLI ++DRFNLY IE++LGTEI+ IP ID+++Y
Sbjct: 363 YLHRIGRSGRYGHLGLAINLINWDDRFNLYNIERDLGTEIQPIPASIDKSLY 414
>UniRef100_Q7S5D9 Hypothetical protein [Neurospora crassa]
Length = 569
Score = 481 bits (1238), Expect = e-134
Identities = 254/427 (59%), Positives = 313/427 (72%), Gaps = 24/427 (5%)
Query: 98 EEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYE 157
+++ +++ + S +DWK L LP DTR +TEDVT T+G ++ED+ R+LLMGI+E
Sbjct: 3 DQLADQLKATSLSSGPEDWKKGLNLPARDTRQQTEDVTNTRGLDWEDFIHDRDLLMGIFE 62
Query: 158 KGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPT 217
GFE+PSPIQEE+IP+ALTG DILARAKNGTGKTAAF IPAL KI+ + IQ +ILVPT
Sbjct: 63 AGFEKPSPIQEEAIPVALTGRDILARAKNGTGKTAAFVIPALNKINPKVSKIQCLILVPT 122
Query: 218 RELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGV 277
RELA+QTSQVCK LGKHL I VMVTTGGT L+DDI+RL PVH++VGTPGRILDLA K V
Sbjct: 123 RELAMQTSQVCKTLGKHLGINVMVTTGGTGLRDDIVRLQDPVHIVVGTPGRILDLAGKQV 182
Query: 278 CVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRK 337
L +C M +MDEADKLLS EF P IEQL+QF P RQ+++FSATFP++VKDF D+ +
Sbjct: 183 ADLSECPMFIMDEADKLLSQEFTPVIEQLLQFHPKDRQVMLFSATFPLSVKDFSDKNMTS 242
Query: 338 PYIINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*EN 397
PY INLMDELTL+GITQ+YAFVEE+QKVHCLNTLFSK I+ + F S+
Sbjct: 243 PYEINLMDELTLRGITQYYAFVEEKQKVHCLNTLFSKL---QINQSIIFCNST------- 292
Query: 398 H*TRVFMFLYSCKDVARPP**SFPRLPQWCMQE----------SRLYC--LFTRGIDIQA 445
RV + ++ S ++ Q L C L TRGIDIQA
Sbjct: 293 --NRVELLAKKITELGYSCFYSHAKMAQQARNRVFHDFRNGVCRNLVCSDLLTRGIDIQA 350
Query: 446 VNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPP 505
VNVVINFDFPKN+ETYLHR+GRSGR+GHLGLA+NLI ++DRFNLY IE++LGTEI+ IP
Sbjct: 351 VNVVINFDFPKNAETYLHRIGRSGRYGHLGLAINLINWDDRFNLYNIERDLGTEIQPIPQ 410
Query: 506 FIDQAIY 512
ID+++Y
Sbjct: 411 TIDKSLY 417
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 863,359,583
Number of Sequences: 2790947
Number of extensions: 37453129
Number of successful extensions: 641561
Number of sequences better than 10.0: 7940
Number of HSP's better than 10.0 without gapping: 6635
Number of HSP's successfully gapped in prelim test: 1428
Number of HSP's that attempted gapping in prelim test: 380994
Number of HSP's gapped (non-prelim): 76805
length of query: 513
length of database: 848,049,833
effective HSP length: 132
effective length of query: 381
effective length of database: 479,644,829
effective search space: 182744679849
effective search space used: 182744679849
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC123899.6


