

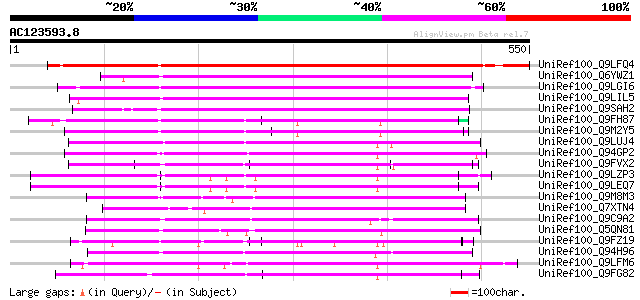
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.8 + phase: 0
(550 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LFQ4 Hypothetical protein F2G14_130 [Arabidopsis tha... 612 e-173
UniRef100_Q6YWZ1 Pentatricopeptide (PPR) repeat-containing prote... 286 8e-76
UniRef100_Q9LGI6 Hypothetical protein P0009G03.25 [Oryza sativa] 271 4e-71
UniRef100_Q9LIL5 Gb|AAF27119.1 [Arabidopsis thaliana] 266 9e-70
UniRef100_Q9SAH2 F23A5.24 protein [Arabidopsis thaliana] 265 3e-69
UniRef100_Q9FH87 Similarity to salt-inducible protein [Arabidops... 233 1e-59
UniRef100_Q9M2Y5 Hypothetical protein T16K5.80 [Arabidopsis thal... 219 2e-55
UniRef100_Q9LUJ4 Gb|AAF26800.1 [Arabidopsis thaliana] 218 3e-55
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 217 8e-55
UniRef100_Q9FVX2 Hypothetical protein F2P24.7 [Arabidopsis thali... 211 6e-53
UniRef100_Q9LZP3 Hypothetical protein T12C14_170 [Arabidopsis th... 209 2e-52
UniRef100_Q9LEQ7 Hypothetical protein T9L3_120 [Arabidopsis thal... 207 5e-52
UniRef100_Q9M8M3 Hypothetical protein T21F11.12 [Arabidopsis tha... 206 2e-51
UniRef100_Q7XTN4 OSJNBa0033G05.8 protein [Oryza sativa] 204 7e-51
UniRef100_Q9C9A2 Hypothetical protein F23N20.5 [Arabidopsis thal... 202 2e-50
UniRef100_Q5QN81 Pentatricopeptide (PPR) repeat-containing prote... 199 2e-49
UniRef100_Q9FZ19 T6A9.11 protein [Arabidopsis thaliana] 199 2e-49
UniRef100_Q94H96 Hypothetical protein OSJNBb0048A17.10 [Oryza sa... 195 3e-48
UniRef100_Q9LFM6 Hypothetical protein F2I11_200 [Arabidopsis tha... 185 3e-45
UniRef100_Q9FG82 Emb|CAB67677.1 [Arabidopsis thaliana] 177 7e-43
>UniRef100_Q9LFQ4 Hypothetical protein F2G14_130 [Arabidopsis thaliana]
Length = 532
Score = 612 bits (1577), Expect = e-173
Identities = 301/511 (58%), Positives = 382/511 (73%), Gaps = 14/511 (2%)
Query: 41 EDDGDDDHDKNSSQLHLRDDGFVKDVKTILDIIHKPGSGPYEIKQKLEDCNVKASSELVV 100
E D + D D S + D+ +DV I ++ GS E++ KLE+C+VK S+ELVV
Sbjct: 35 EKDDESDIDLGCS---ISDELVSEDVGKISKLVKDCGSDRKELRNKLEECDVKPSNELVV 91
Query: 101 EVLSRVRNDWEAAFTFFLWAGKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGK 160
E+LSRVRNDWE AFTFF+WAGKQ GY SVREYHSMIS+LGKMR+FDTAW L++EMR K
Sbjct: 92 EILSRVRNDWETAFTFFVWAGKQQGYVRSVREYHSMISILGKMRKFDTAWTLIDEMR--K 149
Query: 161 TGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQ 220
S+V QTLLIMIRKYCAVHDVG+AINTF+A+KRF ++G+ +FQ LLSALCRYKNV
Sbjct: 150 FSPSLVNSQTLLIMIRKYCAVHDVGKAINTFHAYKRFKLEMGIDDFQSLLSALCRYKNVS 209
Query: 221 DAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASII 280
DA HL+FCNK+ +P D KSFNI+LNGWCN+I S R AER+W EM ++HDVVSY+S+I
Sbjct: 210 DAGHLIFCNKDKYPFDAKSFNIVLNGWCNVIGSPREAERVWMEMGNVGVKHDVVSYSSMI 269
Query: 281 SCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKM-EDNNV 339
SCYSK L +VL+LF++MKK I PDRKVYNAV+ +LAK V EA NL+ M E+ +
Sbjct: 270 SCYSKGGSLNKVLKLFDRMKKECIEPDRKVYNAVVHALAKASFVSEARNLMKTMEEEKGI 329
Query: 340 TPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVKEEVFELLD 399
P+ +TYNSLIKPLCKARK +EAK++F+ MLE+G+ P+IRT+HAF RILR EEVFELL
Sbjct: 330 EPNVVTYNSLIKPLCKARKTEEAKQVFDEMLEKGLFPTIRTYHAFMRILRTGEEVFELLA 389
Query: 400 KMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNC 459
KM+++GC PT+ETYIMLIRK CRWR D V +W+ M+E +G D SSYIV+IHGLFLN
Sbjct: 390 KMRKMGCEPTVETYIMLIRKLCRWRDFDNVLLLWDEMKEKTVGPDLSSYIVMIHGLFLNG 449
Query: 460 KVEEAYKYYIEMQEKGFLPEPKTESMLQAWLSGRQVTDSQATDLEHNQLEHGGLKKSVKP 519
K+EEAY YY EM++KG P E M+Q+W SG+Q + + TD ++ G + K
Sbjct: 450 KIEEAYGYYKEMKDKGMRPNENVEDMIQSWFSGKQYAEQRITD-SKGEVNKGAIVK---- 504
Query: 520 IQSKFDREKDFLREPETRSVSREGGFSFWEK 550
K +REK+FL++PE R V RE G+SFW++
Sbjct: 505 ---KSEREKNFLQQPEVRKVVREHGYSFWDE 532
>UniRef100_Q6YWZ1 Pentatricopeptide (PPR) repeat-containing protein-like [Oryza
sativa]
Length = 528
Score = 286 bits (733), Expect = 8e-76
Identities = 150/405 (37%), Positives = 231/405 (57%), Gaps = 16/405 (3%)
Query: 97 ELVVEVLSRVRNDWEAAFTFFLW-----------AGKQPGYDHSVREYHSMISVLGKMRR 145
+L+V L +R D +AA W AG P +H I GK RR
Sbjct: 99 DLLVRALWELRRDPDAAALALRWGEEGCAAAGERAGPPPPPPPPAEAWHLTIWAAGKARR 158
Query: 146 FDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYE 205
FD AWA+V M R ++T + ++I++ +Y A ++V +AI TF A ++F +
Sbjct: 159 FDLAWAVVRRMLR----RGVLTSRAMVIVMERYAAANEVNKAIKTFDAMEKFKTEADQTV 214
Query: 206 FQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMS 265
F LL ALC+ KN++DAE LL K FPL + FNIIL+GWCN+I A+RIW EMS
Sbjct: 215 FYSLLRALCKNKNIEDAEELLLVRKKFFPLTAEGFNIILDGWCNVITDIAEAKRIWREMS 274
Query: 266 KRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVK 325
I D SY ++SC++K L+ L+++++MKKR TP VYN++I+ L K +K
Sbjct: 275 NYCITPDGTSYTLMVSCFAKVGNLFDTLRVYDEMKKRGWTPSIAVYNSLIYVLTKENCMK 334
Query: 326 EAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFF 385
+ N+ ++ D + P+ TYNS+I PLC++RK+DEA+ + M+ +GI P+I T+H F
Sbjct: 335 DVQNIFTRIIDEGLQPNVKTYNSMIVPLCESRKLDEARMVLEDMMLKGIVPTILTYHTFL 394
Query: 386 RILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDR 445
R + +E + L KMK+ GC P +T++MLI +F + + ++WN M+ I
Sbjct: 395 RQENI-DETLKFLKKMKDDGCGPKSDTFLMLIDRFFQLNEPGHALKLWNEMKRYDIRPSY 453
Query: 446 SSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQAWL 490
S Y+ ++ GL + ++ A +YY EM+E GF +PK E + +L
Sbjct: 454 SHYMSVVQGLIKHGCMQRALEYYDEMKENGFASDPKLEKEFRTFL 498
>UniRef100_Q9LGI6 Hypothetical protein P0009G03.25 [Oryza sativa]
Length = 534
Score = 271 bits (693), Expect = 4e-71
Identities = 144/452 (31%), Positives = 254/452 (55%), Gaps = 9/452 (1%)
Query: 51 NSSQLHLRDDGFVKDVKTILDIIHKPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDW 110
+S+ L D V ++ IL K G E+ + L + ++V++VL + R++W
Sbjct: 71 HSTALDGSKDVHVSEIVKIL----KSRDGDSELAEVLNQFADEMDEDVVLKVLQKQRSNW 126
Query: 111 EAAFTFFLWAGKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQT 170
+ A +FF WA P Y+H R Y M+ +LG+M++ L +E+ ++ +S+VT +
Sbjct: 127 KVALSFFKWAAGLPQYNHGSRAYTEMLDILGRMKKVRLMRQLFDEIPV-ESRQSVVTNRM 185
Query: 171 LLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNK 230
+++ +Y H V AI+ FY K + F++ L FQ LL +LCRYK+V++AE L K
Sbjct: 186 FAVLLNRYAGAHKVQEAIDMFYKRKDYGFELDLVGFQILLMSLCRYKHVEEAEALFLQKK 245
Query: 231 NVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLY 290
+ FP KS+NIILNGWC + S +A+R+W E+ +++ D+ +Y + I+ +KS KL
Sbjct: 246 DEFPPVIKSWNIILNGWC-VKGSLADAKRVWNEIIASKLKPDLFTYGTFINSLTKSGKLS 304
Query: 291 RVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLI 350
++LF M ++ I PD + N +I L + + EA+ + +M D D TYN+LI
Sbjct: 305 TAVKLFTSMWEKGINPDVAICNCIIDQLCFKKRIPEALEIFGEMNDRGCQADVATYNTLI 364
Query: 351 KPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVKEEVFELLDKMKELGCNPTI 410
K CK ++++ E+ + M +G+SP+ T+ + ++V L+ +M++ GC
Sbjct: 365 KHFCKINRMEKVYELLDDMEVKGVSPNNMTYSYILKTTEKPKDVISLMQRMEKSGCRLDS 424
Query: 411 ETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIE 470
+TY +++ + W V+ +W+ M +G G D+ S+ +++HGL + K++EA YY
Sbjct: 425 DTYNLILNLYVSWDYEKGVQLVWDEMERNGSGPDQRSFTIMVHGLHSHDKLDEALHYYRT 484
Query: 471 MQEKGFLPEPKTESMLQAWLSGRQVTDSQATD 502
M+ +G PEP+T+ +++A R D AT+
Sbjct: 485 MESRGMTPEPRTKLLVKAI---RMKKDEPATE 513
>UniRef100_Q9LIL5 Gb|AAF27119.1 [Arabidopsis thaliana]
Length = 523
Score = 266 bits (681), Expect = 9e-70
Identities = 146/429 (34%), Positives = 241/429 (56%), Gaps = 10/429 (2%)
Query: 64 KDVKTILD----IIHKPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLW 119
KD ++ LD I H GS P +IK+ L+ C + + ELV+EV++R R+DW+ A+
Sbjct: 73 KDKQSALDVHNIIKHHRGSSPEKIKRILDKCGIDLTEELVLEVVNRNRSDWKPAYILSQL 132
Query: 120 AGKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYC 179
KQ + S Y+ ++ VLGKMRRF+ + +EM + + V +T +++ +Y
Sbjct: 133 VVKQSVHLSSSMLYNEILDVLGKMRRFEEFHQVFDEMSKR---DGFVNEKTYEVLLNRYA 189
Query: 180 AVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKS 239
A H V A+ F K F L F GLL LCRYK+V+ AE L + F D K+
Sbjct: 190 AAHKVDEAVGVFERRKEFGIDDDLVAFHGLLMWLCRYKHVEFAETLFCSRRREFGCDIKA 249
Query: 240 FNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQM 299
N+ILNGWC ++ + A+R W+++ + + DVVSY ++I+ +K KL + ++L+ M
Sbjct: 250 MNMILNGWC-VLGNVHEAKRFWKDIIASKCRPDVVSYGTMINALTKKGKLGKAMELYRAM 308
Query: 300 KKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKI 359
PD K+ N VI +L + + EA+ + ++ + P+ +TYNSL+K LCK R+
Sbjct: 309 WDTRRNPDVKICNNVIDALCFKKRIPEALEVFREISEKGPDPNVVTYNSLLKHLCKIRRT 368
Query: 360 DEAKEIFNVMLERG--ISPSIRTFHAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLI 417
++ E+ M +G SP+ TF + + ++V +L++M + C T + Y ++
Sbjct: 369 EKVWELVEEMELKGGSCSPNDVTFSYLLKYSQRSKDVDIVLERMAKNKCEMTSDLYNLMF 428
Query: 418 RKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFL 477
R + +W + ++V+ IW+ M G+G D+ +Y + IHGL K+ EA Y+ EM KG +
Sbjct: 429 RLYVQWDKEEKVREIWSEMERSGLGPDQRTYTIRIHGLHTKGKIGEALSYFQEMMSKGMV 488
Query: 478 PEPKTESML 486
PEP+TE +L
Sbjct: 489 PEPRTEMLL 497
>UniRef100_Q9SAH2 F23A5.24 protein [Arabidopsis thaliana]
Length = 540
Score = 265 bits (677), Expect = 3e-69
Identities = 141/423 (33%), Positives = 239/423 (56%), Gaps = 9/423 (2%)
Query: 67 KTILDIIHKPGS--GPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQP 124
K ++D+I + + LED + + + ++ +R++W AF F W G++
Sbjct: 92 KGLIDLIRQVSELESEADAMASLEDSSFDLNHDSFYSLIWELRDEWRLAFLAFKW-GEKR 150
Query: 125 GYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDV 184
G D + MI VLG ++F+ AW L+ +M S T + + +M+ +Y A +D
Sbjct: 151 GCDDQ-KSCDLMIWVLGNHQKFNIAWCLIRDM----FNVSKDTRKAMFLMMDRYAAANDT 205
Query: 185 GRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIIL 244
+AI TF +F FQGLL ALCR+ +++ AE + +K +FP+D + FN+IL
Sbjct: 206 SQAIRTFDIMDKFKHTPYDEAFQGLLCALCRHGHIEKAEEFMLASKKLFPVDVEGFNVIL 265
Query: 245 NGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNI 304
NGWCN+ A+RIW EM I + SY+ +ISC+SK L+ L+L+++MKKR +
Sbjct: 266 NGWCNIWTDVTEAKRIWREMGNYCITPNKDSYSHMISCFSKVGNLFDSLRLYDEMKKRGL 325
Query: 305 TPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKE 364
P +VYN++++ L + EA+ L+ K+ + + PD++TYNS+I+PLC+A K+D A+
Sbjct: 326 APGIEVYNSLVYVLTREDCFDEAMKLMKKLNEEGLKPDSVTYNSMIRPLCEAGKLDVARN 385
Query: 365 IFNVMLERGISPSIRTFHAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWR 424
+ M+ +SP++ TFHAF + E+ E+L +MK PT ET+++++ K + +
Sbjct: 386 VLATMISENLSPTVDTFHAFLEAVNF-EKTLEVLGQMKISDLGPTEETFLLILGKLFKGK 444
Query: 425 QLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTES 484
Q + +IW M I + + Y+ I GL +E+A + Y EM+ KGF+ P +
Sbjct: 445 QPENALKIWAEMDRFEIVANPALYLATIQGLLSCGWLEKAREIYSEMKSKGFVGNPMLQK 504
Query: 485 MLQ 487
+L+
Sbjct: 505 LLE 507
>UniRef100_Q9FH87 Similarity to salt-inducible protein [Arabidopsis thaliana]
Length = 637
Score = 233 bits (594), Expect = 1e-59
Identities = 133/467 (28%), Positives = 240/467 (50%), Gaps = 20/467 (4%)
Query: 21 KFSFSVRFFSAESSSDDDYNEDDG---------DDDHDKNSSQLHLRDDGFVKDVKTILD 71
+F R F + +D++ +G +D KNS D F DV+
Sbjct: 34 RFDLIHRSFHVSRALEDNFRRSNGIGLVCLEKSHNDRTKNSKY-----DEFASDVEKSYR 88
Query: 72 IIHKPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQPGYDHSVR 131
I+ K S +++ L + V+ L+ VL+R + + FF+WA KQP Y HS+
Sbjct: 89 ILRKFHSRVPKLELALNESGVELRPGLIERVLNRCGDAGNLGYRFFVWAAKQPRYCHSIE 148
Query: 132 EYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTF 191
Y SM+ +L KMR+F W L+EEMR K ++ P+ +++++++ + V +AI
Sbjct: 149 VYKSMVKILSKMRQFGAVWGLIEEMR--KENPQLIEPELFVVLVQRFASADMVKKAIEVL 206
Query: 192 YAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLI 251
+F F+ Y F LL ALC++ +V+DA L + FP++ + F +L GWC +
Sbjct: 207 DEMPKFGFEPDEYVFGCLLDALCKHGSVKDAAKLFEDMRMRFPVNLRYFTSLLYGWCR-V 265
Query: 252 VSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVY 311
A+ + +M++ + D+V Y +++S Y+ + K+ L M++R P+ Y
Sbjct: 266 GKMMEAKYVLVQMNEAGFEPDIVDYTNLLSGYANAGKMADAYDLLRDMRRRGFEPNANCY 325
Query: 312 NAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLE 371
+I +L K ++EA+ + ++ME D +TY +L+ CK KID+ + + M++
Sbjct: 326 TVLIQALCKVDRMEEAMKVFVEMERYECEADVVTYTALVSGFCKWGKIDKCYIVLDDMIK 385
Query: 372 RGISPSIRTFHAFFRILRVK---EEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDE 428
+G+ PS T+ K EE EL++KM+++ +P I Y ++IR C+ ++ E
Sbjct: 386 KGLMPSELTYMHIMVAHEKKESFEECLELMEKMRQIEYHPDIGIYNVVIRLACKLGEVKE 445
Query: 429 VKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKG 475
R+WN M E+G+ +++++I+GL + EA ++ EM +G
Sbjct: 446 AVRLWNEMEENGLSPGVDTFVIMINGLASQGCLLEASDHFKEMVTRG 492
Score = 56.2 bits (134), Expect = 2e-06
Identities = 51/222 (22%), Positives = 89/222 (39%), Gaps = 38/222 (17%)
Query: 268 RIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRN---ITPDRKVYNAVIFSLAKNRMV 324
R H + Y S++ SK + V L E+M+K N I P+ ++ ++ A MV
Sbjct: 142 RYCHSIEVYKSMVKILSKMRQFGAVWGLIEEMRKENPQLIEPE--LFVVLVQRFASADMV 199
Query: 325 KEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAF 384
K+A+ ++ +M PD + L+ LCK + +A ++F M R
Sbjct: 200 KKAIEVLDEMPKFGFEPDEYVFGCLLDALCKHGSVKDAAKLFEDMRMR------------ 247
Query: 385 FRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHD 444
F + + + L+ +CR ++ E K + M E G D
Sbjct: 248 FPV---------------------NLRYFTSLLYGWCRVGKMMEAKYVLVQMNEAGFEPD 286
Query: 445 RSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
Y L+ G K+ +AY +M+ +GF P ++L
Sbjct: 287 IVDYTNLLSGYANAGKMADAYDLLRDMRRRGFEPNANCYTVL 328
>UniRef100_Q9M2Y5 Hypothetical protein T16K5.80 [Arabidopsis thaliana]
Length = 1184
Score = 219 bits (557), Expect = 2e-55
Identities = 121/427 (28%), Positives = 220/427 (51%), Gaps = 7/427 (1%)
Query: 59 DDGFVKDVKTILDIIHKPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFL 118
+D F +V+ I I+ S +++ L + + L++ VLSR + + FFL
Sbjct: 60 EDEFAGEVEKIYRILRNHHSRVPKLELALNESGIDLRPGLIIRVLSRCGDAGNLGYRFFL 119
Query: 119 WAGKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKY 178
WA KQPGY HS SM+ +L KMR+F W L+EEMR KT ++ P+ ++++R++
Sbjct: 120 WATKQPGYFHSYEVCKSMVMILSKMRQFGAVWGLIEEMR--KTNPELIEPELFVVLMRRF 177
Query: 179 CAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTK 238
+ + V +A+ ++ + Y F LL ALC+ +V++A + + FP + +
Sbjct: 178 ASANMVKKAVEVLDEMPKYGLEPDEYVFGCLLDALCKNGSVKEASKVFEDMREKFPPNLR 237
Query: 239 SFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQ 298
F +L GWC A+ + +M + ++ D+V + +++S Y+ + K+ L
Sbjct: 238 YFTSLLYGWCR-EGKLMEAKEVLVQMKEAGLEPDIVVFTNLLSGYAHAGKMADAYDLMND 296
Query: 299 MKKRNITPDRKVYNAVIFSLAK-NRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKAR 357
M+KR P+ Y +I +L + + + EA+ + ++ME D +TY +LI CK
Sbjct: 297 MRKRGFEPNVNCYTVLIQALCRTEKRMDEAMRVFVEMERYGCEADIVTYTALISGFCKWG 356
Query: 358 KIDEAKEIFNVMLERGISPSIRTFHAFFRILRVK---EEVFELLDKMKELGCNPTIETYI 414
ID+ + + M ++G+ PS T+ K EE EL++KMK GC+P + Y
Sbjct: 357 MIDKGYSVLDDMRKKGVMPSQVTYMQIMVAHEKKEQFEECLELIEKMKRRGCHPDLLIYN 416
Query: 415 MLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEK 474
++IR C+ ++ E R+WN M +G+ +++++I+G + EA ++ EM +
Sbjct: 417 VVIRLACKLGEVKEAVRLWNEMEANGLSPGVDTFVIMINGFTSQGFLIEACNHFKEMVSR 476
Query: 475 GFLPEPK 481
G P+
Sbjct: 477 GIFSAPQ 483
Score = 59.3 bits (142), Expect = 3e-07
Identities = 52/212 (24%), Positives = 92/212 (42%), Gaps = 38/212 (17%)
Query: 278 SIISCYSKSSKLYRVLQLFEQMKKRN---ITPDRKVYNAVIFSLAKNRMVKEAVNLIIKM 334
S++ SK + V L E+M+K N I P+ ++ ++ A MVK+AV ++ +M
Sbjct: 136 SMVMILSKMRQFGAVWGLIEEMRKTNPELIEPE--LFVVLMRRFASANMVKKAVEVLDEM 193
Query: 335 EDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVKEEV 394
+ PD + L+ LCK + EA ++F M E+ P++R F +
Sbjct: 194 PKYGLEPDEYVFGCLLDALCKNGSVKEASKVFEDMREK-FPPNLRYFTS----------- 241
Query: 395 FELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHG 454
L+ +CR +L E K + M+E G+ D + L+ G
Sbjct: 242 ---------------------LLYGWCREGKLMEAKEVLVQMKEAGLEPDIVVFTNLLSG 280
Query: 455 LFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
K+ +AY +M+++GF P ++L
Sbjct: 281 YAHAGKMADAYDLMNDMRKRGFEPNVNCYTVL 312
>UniRef100_Q9LUJ4 Gb|AAF26800.1 [Arabidopsis thaliana]
Length = 562
Score = 218 bits (556), Expect = 3e-55
Identities = 127/444 (28%), Positives = 227/444 (50%), Gaps = 9/444 (2%)
Query: 63 VKDVKTILDIIHKPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGK 122
V+D+ + D ++K + ++ ++L C+V + LV++VL R N W A+ FF+WA
Sbjct: 99 VEDIDKVCDFLNKKDTSHEDVVKELSKCDVVVTESLVLQVLRRFSNGWNQAYGFFIWANS 158
Query: 123 QPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVH 182
Q GY HS Y++M+ VLGK R FD W LV EM + + + +VT T+ ++R+
Sbjct: 159 QTGYVHSGHTYNAMVDVLGKCRNFDLMWELVNEMNKNEESK-LVTLDTMSKVMRRLAKSG 217
Query: 183 DVGRAINTFYAF-KRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFN 241
+A++ F K + + L+ AL + +++ A + + D ++FN
Sbjct: 218 KYNKAVDAFLEMEKSYGVKTDTIAMNSLMDALVKENSIEHAHEVFLKLFDTIKPDARTFN 277
Query: 242 IILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKK 301
I+++G+C +A + + M DVV+Y S + Y K RV ++ E+M++
Sbjct: 278 ILIHGFCK-ARKFDDARAMMDLMKVTEFTPDVVTYTSFVEAYCKEGDFRRVNEMLEEMRE 336
Query: 302 RNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDE 361
P+ Y V+ SL K++ V EA+ + KM+++ PDA Y+SLI L K + +
Sbjct: 337 NGCNPNVVTYTIVMHSLGKSKQVAEALGVYEKMKEDGCVPDAKFYSSLIHILSKTGRFKD 396
Query: 362 AKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKE---LGCNPTIETYIM 415
A EIF M +G+ + ++ E LL +M++ C+P +ETY
Sbjct: 397 AAEIFEDMTNQGVRRDVLVYNTMISAALHHSRDEMALRLLKRMEDEEGESCSPNVETYAP 456
Query: 416 LIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKG 475
L++ C +++ + + + M ++ + D S+YI+LI GL ++ KVEEA ++ E KG
Sbjct: 457 LLKMCCHKKKMKLLGILLHHMVKNDVSIDVSTYILLIRGLCMSGKVEEACLFFEEAVRKG 516
Query: 476 FLPEPKTESMLQAWLSGRQVTDSQ 499
+P T ML L + + +++
Sbjct: 517 MVPRDSTCKMLVDELEKKNMAEAK 540
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 217 bits (552), Expect = 8e-55
Identities = 136/459 (29%), Positives = 227/459 (48%), Gaps = 16/459 (3%)
Query: 59 DDGFVKDVKTILDIIHKPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFL 118
D F D+ + I+ + P I ++ C+V+ + LV ++L+R NDW AAF FF+
Sbjct: 1374 DAEFEADIDEVSRILSARFASPEAIMIAMDCCSVRVTGCLVDKILTRFSNDWVAAFGFFM 1433
Query: 119 WAGKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKY 178
W G Q GY H Y M+ +LGK ++FD W L+ +M + G +++ T+ ++R+
Sbjct: 1434 WVGTQGGYCHCADSYDLMVDILGKFKQFDLMWGLINQM--VEVG-GLMSLMTMTKVMRRL 1490
Query: 179 CAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTK 238
AI+ F+ RF LL LC+ ++V+ A + + P D
Sbjct: 1491 AGASRWTEAIDAFHKMDRFGVVKDTKAMNVLLDTLCKERSVKRARGVFQELRGTIPPDEN 1550
Query: 239 SFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQ 298
SFN +++GWC + + A EEM + VV+Y S++ Y V L ++
Sbjct: 1551 SFNTLVHGWCKARM-LKEALDTMEEMKQHGFSPSVVTYTSLVEAYCMEKDFQTVYALLDE 1609
Query: 299 MKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARK 358
M+KR P+ Y ++ +L K +EA++ K++++ V PDA YNSLI L +A +
Sbjct: 1610 MRKRRCPPNVVTYTILMHALGKAGRTREALDTFDKLKEDGVAPDASFYNSLIYILGRAGR 1669
Query: 359 IDEAKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKELGCNPTIETYIM 415
+++A + M GI+P++ TF+ E +LL KM+E CNP I+TY
Sbjct: 1670 LEDAYSVVEEMRTTGIAPNVTTFNTLISAACDHSQAENALKLLVKMEEQSCNPDIKTYTP 1729
Query: 416 LIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKG 475
L++ C+ + + + + M I D S+Y +L+ L N KV ++ + EM KG
Sbjct: 1730 LLKLCCKRQWVKILLFLVCHMFRKDISPDFSTYTLLVSWLCRNGKVAQSCLFLEEMVSKG 1789
Query: 476 FLPEPKTESMLQAWLSGR---------QVTDSQATDLEH 505
F P+ +T ++ L R QV +Q T+L+H
Sbjct: 1790 FAPKQETFDLVMEKLEKRNLQSVYKKIQVLRTQVTNLKH 1828
>UniRef100_Q9FVX2 Hypothetical protein F2P24.7 [Arabidopsis thaliana]
Length = 481
Score = 211 bits (536), Expect = 6e-53
Identities = 125/433 (28%), Positives = 219/433 (49%), Gaps = 11/433 (2%)
Query: 63 VKDVKTILDIIHKP--GSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWA 120
V+DV + I K S + L+ ++ S E+V +VL+R RN + FF W+
Sbjct: 29 VRDVADVAKNISKVLMSSPQLVLDSALDQSGLRVSQEVVEDVLNRFRNAGLLTYRFFQWS 88
Query: 121 GKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCA 180
KQ Y+HSVR YH MI K+R++ W L+ MR+ K ++ +T I++RKY
Sbjct: 89 EKQRHYEHSVRAYHMMIESTAKIRQYKLMWDLINAMRKKK----MLNVETFCIVMRKYAR 144
Query: 181 VHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSF 240
V AI F ++++ L F GLLSALC+ KNV+ A+ + ++ F D+K++
Sbjct: 145 AQKVDEAIYAFNVMEKYDLPPNLVAFNGLLSALCKSKNVRKAQEVFENMRDRFTPDSKTY 204
Query: 241 NIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMK 300
+I+L GW + A ++ EM D+V+Y+ ++ K+ ++ L + M
Sbjct: 205 SILLEGW-GKEPNLPKAREVFREMIDAGCHPDIVTYSIMVDILCKAGRVDEALGIVRSMD 263
Query: 301 KRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKID 360
P +Y+ ++ + ++EAV+ ++ME + + D +NSLI CKA ++
Sbjct: 264 PSICKPTTFIYSVLVHTYGTENRLEEAVDTFLEMERSGMKADVAVFNSLIGAFCKANRMK 323
Query: 361 EAKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKELGCNPTIETYIMLI 417
+ M +G++P+ ++ + R L K+E F++ KM ++ C P +TY M+I
Sbjct: 324 NVYRVLKEMKSKGVTPNSKSCNIILRHLIERGEKDEAFDVFRKMIKV-CEPDADTYTMVI 382
Query: 418 RKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFL 477
+ FC ++++ ++W MR+ G+ ++ VLI+GL ++A EM E G
Sbjct: 383 KMFCEKKEMETADKVWKYMRKKGVFPSMHTFSVLINGLCEERTTQKACVLLEEMIEMGIR 442
Query: 478 PEPKTESMLQAWL 490
P T L+ L
Sbjct: 443 PSGVTFGRLRQLL 455
Score = 84.3 bits (207), Expect = 8e-15
Identities = 59/248 (23%), Positives = 120/248 (47%), Gaps = 6/248 (2%)
Query: 255 RNAERIWEEMSKRRIQH--DVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYN 312
R + +W+ ++ R + +V ++ ++ Y+++ K+ + F M+K ++ P+ +N
Sbjct: 112 RQYKLMWDLINAMRKKKMLNVETFCIVMRKYARAQKVDEAIYAFNVMEKYDLPPNLVAFN 171
Query: 313 AVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLER 372
++ +L K++ V++A + M D TPD+ TY+ L++ K + +A+E+F M++
Sbjct: 172 GLLSALCKSKNVRKAQEVFENMRDR-FTPDSKTYSILLEGWGKEPNLPKAREVFREMIDA 230
Query: 373 GISPSIRTFHAFFRILRVKEEVFELLDKMKELG---CNPTIETYIMLIRKFCRWRQLDEV 429
G P I T+ IL V E L ++ + C PT Y +L+ + +L+E
Sbjct: 231 GCHPDIVTYSIMVDILCKAGRVDEALGIVRSMDPSICKPTTFIYSVLVHTYGTENRLEEA 290
Query: 430 KRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQAW 489
+ M G+ D + + LI +++ Y+ EM+ KG P K+ +++
Sbjct: 291 VDTFLEMERSGMKADVAVFNSLIGAFCKANRMKNVYRVLKEMKSKGVTPNSKSCNIILRH 350
Query: 490 LSGRQVTD 497
L R D
Sbjct: 351 LIERGEKD 358
Score = 59.3 bits (142), Expect = 3e-07
Identities = 64/273 (23%), Positives = 105/273 (38%), Gaps = 44/273 (16%)
Query: 133 YHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLL--IMIRKYCAVHDVGRAINT 190
Y M+ +L K R D A +V M SI P T + +++ Y + + A++T
Sbjct: 239 YSIMVDILCKAGRVDEALGIVRSM-----DPSICKPTTFIYSVLVHTYGTENRLEEAVDT 293
Query: 191 FYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNL 250
F +R + + F L+ A FC N
Sbjct: 294 FLEMERSGMKADVAVFNSLIGA--------------FCKAN------------------- 320
Query: 251 IVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKV 310
+N R+ +EM + + + S I+ + + +F +M K PD
Sbjct: 321 --RMKNVYRVLKEMKSKGVTPNSKSCNIILRHLIERGEKDEAFDVFRKMIKV-CEPDADT 377
Query: 311 YNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVML 370
Y VI + + ++ A + M V P T++ LI LC+ R +A + M+
Sbjct: 378 YTMVIKMFCEKKEMETADKVWKYMRKKGVFPSMHTFSVLINGLCEERTTQKACVLLEEMI 437
Query: 371 ERGISPSIRTFHAFFRILRVKEEVFELLDKMKE 403
E GI PS TF R L +KEE ++L + E
Sbjct: 438 EMGIRPSGVTF-GRLRQLLIKEEREDVLKFLNE 469
>UniRef100_Q9LZP3 Hypothetical protein T12C14_170 [Arabidopsis thaliana]
Length = 599
Score = 209 bits (531), Expect = 2e-52
Identities = 131/458 (28%), Positives = 239/458 (51%), Gaps = 10/458 (2%)
Query: 23 SFSVRFFSAESS--SDDDYNEDDGDDDHDKNSSQLHLRDDGFVKDVKTILDIIHKPGSGP 80
S R FS+ SS SD E + + D+D+ + + ++V+ + +I + +
Sbjct: 86 SLGCRGFSSGSSNVSDGCDEEVESECDNDEETGVSCVESSTNPEEVERVCKVIDELFALD 145
Query: 81 YEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQPGYDHSVREYHSMISVL 140
++ L++ + S +L+VEVL R R+ + AF FF WA ++ G+ H R Y+SM+S+L
Sbjct: 146 RNMEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMSIL 205
Query: 141 GKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQ 200
K R+F+T +++EEM G G ++T +T I ++ + A + +A+ F K++ F+
Sbjct: 206 AKTRQFETMVSVLEEM--GTKG--LLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFK 261
Query: 201 VGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSARNAERI 260
+G+ LL +L R K ++A+ L K F + ++ ++LNGWC + + A RI
Sbjct: 262 IGVETINCLLDSLGRAKLGKEAQVLFDKLKERFTPNMMTYTVLLNGWCR-VRNLIEAARI 320
Query: 261 WEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAK 320
W +M + ++ D+V++ ++ +S K ++LF MK + P+ + Y +I K
Sbjct: 321 WNDMIDQGLKPDIVAHNVMLEGLLRSRKKSDAIKLFHVMKSKGPCPNVRSYTIMIRDFCK 380
Query: 321 NRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRT 380
++ A+ M D+ + PDA Y LI +K+D E+ M E+G P +T
Sbjct: 381 QSSMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKT 440
Query: 381 FHAFFRIL---RVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMR 437
++A +++ ++ E + +KM + P+I T+ M+++ + R + + +W M
Sbjct: 441 YNALIKLMANQKMPEHATRIYNKMIQNEIEPSIHTFNMIMKSYFMARNYEMGRAVWEEMI 500
Query: 438 EDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKG 475
+ GI D +SY VLI GL K EA +Y EM +KG
Sbjct: 501 KKGICPDDNSYTVLIRGLIGEGKSREACRYLEEMLDKG 538
Score = 108 bits (269), Expect = 5e-22
Identities = 89/366 (24%), Positives = 174/366 (47%), Gaps = 29/366 (7%)
Query: 160 KTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLS------AL 213
+TG S V T + + C V I+ +A R N + L E + LS L
Sbjct: 116 ETGVSCVESSTNPEEVERVCKV------IDELFALDR-NMEAVLDEMKLDLSHDLIVEVL 168
Query: 214 CRYKNVQDAEHLLFC---NKNVFPLDTKSFNIILNGWCNLIVSARNAER---IWEEMSKR 267
R+++ + FC + F D++++N +++ ++ R E + EEM +
Sbjct: 169 ERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMS----ILAKTRQFETMVSVLEEMGTK 224
Query: 268 RIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEA 327
+ + ++ + ++ + + + + +FE MKK + N ++ SL + ++ KEA
Sbjct: 225 GLL-TMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVETINCLLDSLGRAKLGKEA 283
Query: 328 VNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRI 387
L K+++ TP+ +TY L+ C+ R + EA I+N M+++G+ P I +
Sbjct: 284 QVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWNDMIDQGLKPDIVAHNVMLEG 342
Query: 388 L---RVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHD 444
L R K + +L MK G P + +Y ++IR FC+ ++ ++ M + G+ D
Sbjct: 343 LLRSRKKSDAIKLFHVMKSKGPCPNVRSYTIMIRDFCKQSSMETAIEYFDDMVDSGLQPD 402
Query: 445 RSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQAWLSGRQVTDSQATDLE 504
+ Y LI G K++ Y+ EMQEKG P+ KT + L ++ +++ + AT +
Sbjct: 403 AAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYNALIKLMANQKMPE-HATRIY 461
Query: 505 HNQLEH 510
+ +++
Sbjct: 462 NKMIQN 467
>UniRef100_Q9LEQ7 Hypothetical protein T9L3_120 [Arabidopsis thaliana]
Length = 598
Score = 207 bits (528), Expect = 5e-52
Identities = 131/458 (28%), Positives = 239/458 (51%), Gaps = 10/458 (2%)
Query: 23 SFSVRFFSAESS--SDDDYNEDDGDDDHDKNSSQLHLRDDGFVKDVKTILDIIHKPGSGP 80
S R FS+ SS SD E + + D+D+ + + ++V+ + +I + +
Sbjct: 85 SLGCRGFSSGSSNVSDGCDEEVESECDNDEETGVSCVESSTNPEEVERVCKVIDELFALD 144
Query: 81 YEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQPGYDHSVREYHSMISVL 140
++ L++ + S +L+VEVL R R+ + AF FF WA ++ G+ H R Y+SM+S+L
Sbjct: 145 RNMEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMSIL 204
Query: 141 GKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQ 200
K R+F+T +++EEM G G ++T +T I ++ + A + +A+ F K++ F+
Sbjct: 205 AKTRQFETMVSVLEEM--GTKG--LLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFK 260
Query: 201 VGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSARNAERI 260
+G+ LL +L R K ++A+ L K F + ++ ++LNGWC + + A RI
Sbjct: 261 IGVETINCLLDSLGRAKLGKEAQVLFDKLKERFTPNMMTYTVLLNGWCR-VRNLIEAARI 319
Query: 261 WEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAK 320
W +M ++ D+V++ ++ +S K ++LF MK + P+ + Y +I K
Sbjct: 320 WNDMIDHGLKPDIVAHNVMLEGLLRSMKKSDAIKLFHVMKSKGPCPNVRSYTIMIRDFCK 379
Query: 321 NRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRT 380
++ A+ M D+ + PDA Y LI +K+D E+ M E+G P +T
Sbjct: 380 QSSMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKT 439
Query: 381 FHAFFRIL---RVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMR 437
++A +++ ++ E + +KM + P+I T+ M+++ + R + + +W+ M
Sbjct: 440 YNALIKLMANQKMPEHGTRIYNKMIQNEIEPSIHTFNMIMKSYFVARNYEMGRAVWDEMI 499
Query: 438 EDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKG 475
+ GI D +SY VLI GL K EA +Y EM +KG
Sbjct: 500 KKGICPDDNSYTVLIRGLISEGKSREACRYLEEMLDKG 537
Score = 105 bits (262), Expect = 3e-21
Identities = 87/353 (24%), Positives = 167/353 (46%), Gaps = 28/353 (7%)
Query: 160 KTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLS------AL 213
+TG S V T + + C V I+ +A R N + L E + LS L
Sbjct: 115 ETGVSCVESSTNPEEVERVCKV------IDELFALDR-NMEAVLDEMKLDLSHDLIVEVL 167
Query: 214 CRYKNVQDAEHLLFC---NKNVFPLDTKSFNIILNGWCNLIVSARNAER---IWEEMSKR 267
R+++ + FC + F D++++N +++ ++ R E + EEM +
Sbjct: 168 ERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMS----ILAKTRQFETMVSVLEEMGTK 223
Query: 268 RIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEA 327
+ + ++ + ++ + + + + +FE MKK + N ++ SL + ++ KEA
Sbjct: 224 GLL-TMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVETINCLLDSLGRAKLGKEA 282
Query: 328 VNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFR- 386
L K+++ TP+ +TY L+ C+ R + EA I+N M++ G+ P I +
Sbjct: 283 QVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWNDMIDHGLKPDIVAHNVMLEG 341
Query: 387 ILRV--KEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHD 444
+LR K + +L MK G P + +Y ++IR FC+ ++ ++ M + G+ D
Sbjct: 342 LLRSMKKSDAIKLFHVMKSKGPCPNVRSYTIMIRDFCKQSSMETAIEYFDDMVDSGLQPD 401
Query: 445 RSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQAWLSGRQVTD 497
+ Y LI G K++ Y+ EMQEKG P+ KT + L ++ +++ +
Sbjct: 402 AAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYNALIKLMANQKMPE 454
Score = 45.4 bits (106), Expect = 0.004
Identities = 39/176 (22%), Positives = 71/176 (40%), Gaps = 2/176 (1%)
Query: 173 IMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLF-CNKN 231
IMIR +C + AI F Q + L++ K + LL +
Sbjct: 372 IMIRDFCKQSSMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEK 431
Query: 232 VFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYR 291
P D K++N ++ N + + RI+ +M + I+ + ++ I+ Y +
Sbjct: 432 GHPPDGKTYNALIKLMANQKMP-EHGTRIYNKMIQNEIEPSIHTFNMIMKSYFVARNYEM 490
Query: 292 VLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYN 347
++++M K+ I PD Y +I L +EA + +M D + I YN
Sbjct: 491 GRAVWDEMIKKGICPDDNSYTVLIRGLISEGKSREACRYLEEMLDKGMKTPLIDYN 546
>UniRef100_Q9M8M3 Hypothetical protein T21F11.12 [Arabidopsis thaliana]
Length = 448
Score = 206 bits (523), Expect = 2e-51
Identities = 124/407 (30%), Positives = 207/407 (50%), Gaps = 10/407 (2%)
Query: 82 EIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQPGYDHSVREYHSMISVLG 141
+ + + E+ + V E L+ NDW+ A FF W ++ G+ H+ ++ +I +LG
Sbjct: 33 KFRSQEEEDQSSYDQKTVCEALTCYSNDWQKALEFFNWVERESGFRHTTETFNRVIDILG 92
Query: 142 KMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQV 201
K F+ +WAL+ M G T ES+ T I+ ++Y H V AI+ + FN +
Sbjct: 93 KYFEFEISWALINRMI-GNT-ESVPNHVTFRIVFKRYVTAHLVQEAIDAYDKLDDFNLRD 150
Query: 202 GLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFP-----LDTKSFNIILNGWCNLIVSARN 256
F L+ ALC +K+V +AE L F KNV +TK N+IL GW L +
Sbjct: 151 ET-SFYNLVDALCEHKHVVEAEELCF-GKNVIGNGFSVSNTKIHNLILRGWSKLGWWGKC 208
Query: 257 AERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIF 316
E W++M + D+ SY+ + KS K ++ ++L+++MK R + D YN VI
Sbjct: 209 KE-YWKKMDTEGVTKDLFSYSIYMDIMCKSGKPWKAVKLYKEMKSRRMKLDVVAYNTVIR 267
Query: 317 SLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISP 376
++ ++ V+ + + +M + P+ T+N++IK LC+ ++ +A + + M +RG P
Sbjct: 268 AIGASQGVEFGIRVFREMRERGCEPNVATHNTIIKLLCEDGRMRDAYRMLDEMPKRGCQP 327
Query: 377 SIRTFHAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAM 436
T+ F L E+ L +M G P ++TY+ML+RKF RW L V +W M
Sbjct: 328 DSITYMCLFSRLEKPSEILSLFGRMIRSGVRPKMDTYVMLMRKFERWGFLQPVLYVWKTM 387
Query: 437 REDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTE 483
+E G D ++Y +I L ++ A +Y EM E+G P + E
Sbjct: 388 KESGDTPDSAAYNAVIDALIQKGMLDMAREYEEEMIERGLSPRRRPE 434
Score = 41.6 bits (96), Expect = 0.061
Identities = 41/189 (21%), Positives = 83/189 (43%), Gaps = 11/189 (5%)
Query: 306 PDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEI 365
P+ + V +V+EA++ K++D N+ + YN L+ LC+ + + EA+E+
Sbjct: 115 PNHVTFRIVFKRYVTAHLVQEAIDAYDKLDDFNLRDETSFYN-LVDALCEHKHVVEAEEL 173
Query: 366 F--NVMLERGISPSIRTFHAFFRILRVKEEVF------ELLDKMKELGCNPTIETYIMLI 417
++ G S S H ILR ++ E KM G + +Y + +
Sbjct: 174 CFGKNVIGNGFSVSNTKIHNL--ILRGWSKLGWWGKCKEYWKKMDTEGVTKDLFSYSIYM 231
Query: 418 RKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFL 477
C+ + + +++ M+ + D +Y +I + + VE + + EM+E+G
Sbjct: 232 DIMCKSGKPWKAVKLYKEMKSRRMKLDVVAYNTVIRAIGASQGVEFGIRVFREMRERGCE 291
Query: 478 PEPKTESML 486
P T + +
Sbjct: 292 PNVATHNTI 300
>UniRef100_Q7XTN4 OSJNBa0033G05.8 protein [Oryza sativa]
Length = 442
Score = 204 bits (518), Expect = 7e-51
Identities = 130/392 (33%), Positives = 200/392 (50%), Gaps = 12/392 (3%)
Query: 99 VVEVLSRVRNDWEAAFTFFLWAGKQPGYD--HSVREYHSMISVLGKMRRFDTAWAL-VEE 155
V+E LS NDW A FF W+ G + + + VLGK F A +L V
Sbjct: 36 VLETLSLYANDWRRALEFFHWSASPDGANVPPTPATVARAVDVLGKHFEFPLATSLLVSH 95
Query: 156 MRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYE---FQGLLSA 212
G+ S + P L ++ + A + + AI AF +GL + F L+ A
Sbjct: 96 HDPGRADPSFLRP-ALRSLLNRLAAANLIDDAIR---AFDSTAGSIGLRDEASFHALVDA 151
Query: 213 LCRYKNVQDAEHLLFC-NKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQH 271
LC ++ V +A HL F + FP TK++N++L GW AR ++W +M R +
Sbjct: 152 LCDHRRVDEAHHLCFGKDPPPFPPVTKTYNLLLRGWAKTRAWAR-LRQLWFDMDSRGVAK 210
Query: 272 DVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLI 331
D+ SY+ + +KS K ++ ++F++MK++ + D YN I S+ + V A+ L
Sbjct: 211 DLHSYSIYMDALAKSGKPWKAFKVFKEMKQKGMAIDVVAYNTAIHSVGLAQGVDFAIRLY 270
Query: 332 IKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRILRVK 391
+M D P+A T+N+++K LCK + E M + GI P++ T+H FF+ L
Sbjct: 271 RQMVDAGCKPNASTFNTIVKLLCKEGRFKEGYAFVQQMHKFGIEPNVLTYHCFFQYLSRP 330
Query: 392 EEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVL 451
+EV L +KM E GC P ++TY+MLI++F RW L V +W AM + G+ D +Y L
Sbjct: 331 QEVLGLFEKMLERGCRPRMDTYVMLIKRFGRWGFLRPVFIVWKAMEKQGLSPDAFAYNSL 390
Query: 452 IHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTE 483
I L V+ A KY EM KG P+P+ E
Sbjct: 391 IDALLQKGMVDLARKYDEEMLSKGLSPKPRKE 422
>UniRef100_Q9C9A2 Hypothetical protein F23N20.5 [Arabidopsis thaliana]
Length = 510
Score = 202 bits (514), Expect = 2e-50
Identities = 123/425 (28%), Positives = 222/425 (51%), Gaps = 18/425 (4%)
Query: 82 EIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQPGYDHSVREYHSMISVLG 141
+++ L + +VK S L+ EVL ++ N A + F WA Q G+ H+ Y+++I LG
Sbjct: 80 KVETLLNEASVKLSPALIEEVLKKLSNAGVLALSVFKWAENQKGFKHTTSNYNALIESLG 139
Query: 142 KMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQV 201
K+++F W+LV++M+ K +++ +T ++ R+Y V AI F+ + F F++
Sbjct: 140 KIKQFKLIWSLVDDMKAKK----LLSKETFALISRRYARARKVKEAIGAFHKMEEFGFKM 195
Query: 202 GLYEFQGLLSALCRYKNVQDAEHLLF-CNKNVFPLDTKSFNIILNGWCNLIVSARNAERI 260
+F +L L + +NV DA+ + K F D KS+ I+L GW + R + +
Sbjct: 196 ESSDFNRMLDTLSKSRNVGDAQKVFDKMKKKRFEPDIKSYTILLEGWGQELNLLR-VDEV 254
Query: 261 WEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAK 320
EM + DVV+Y II+ + K+ K ++ F +M++RN P ++ ++I L
Sbjct: 255 NREMKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNEMEQRNCKPSPHIFCSLINGLGS 314
Query: 321 NRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRT 380
+ + +A+ + + + +A TYN+L+ C ++++++A + + M +G+ P+ RT
Sbjct: 315 EKKLNDALEFFERSKSSGFPLEAPTYNALVGAYCWSQRMEDAYKTVDEMRLKGVGPNART 374
Query: 381 F----HAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAM 436
+ H R+ R KE +E+ M C PT+ TY +++R FC +LD +IW+ M
Sbjct: 375 YDIILHHLIRMQRSKE-AYEVYQTMS---CEPTVSTYEIMVRMFCNKERLDMAIKIWDEM 430
Query: 437 REDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQAWL--SGR- 493
+ G+ + LI L K++EA +Y+ EM + G P S L+ L GR
Sbjct: 431 KGKGVLPGMHMFSSLITALCHENKLDEACEYFNEMLDVGIRPPGHMFSRLKQTLLDEGRK 490
Query: 494 -QVTD 497
+VTD
Sbjct: 491 DKVTD 495
Score = 38.5 bits (88), Expect = 0.51
Identities = 53/286 (18%), Positives = 111/286 (38%), Gaps = 49/286 (17%)
Query: 57 LRDDGFVKDVKTILDIIH-----KPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWE 111
++D+GF DV II+ K ++E N K S + +++ + ++ +
Sbjct: 258 MKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNEMEQRNCKPSPHIFCSLINGLGSEKK 317
Query: 112 A--AFTFFLWAGKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQ 169
A FF K G+ Y++++ +R + A+ V+EMR G + T
Sbjct: 318 LNDALEFFE-RSKSSGFPLEAPTYNALVGAYCWSQRMEDAYKTVDEMRLKGVGPNARTYD 376
Query: 170 TLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLFCN 229
+L H + R + A++ +Y+ + Y+ + +FCN
Sbjct: 377 IIL---------HHLIRMQRSKEAYE-------VYQTMSCEPTVSTYEIMVR----MFCN 416
Query: 230 KNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKL 289
K + A +IW+EM + + + ++S+I+ +KL
Sbjct: 417 KERLDM---------------------AIKIWDEMKGKGVLPGMHMFSSLITALCHENKL 455
Query: 290 YRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKME 335
+ F +M I P +++ + +L + +L++KM+
Sbjct: 456 DEACEYFNEMLDVGIRPPGHMFSRLKQTLLDEGRKDKVTDLVVKMD 501
>UniRef100_Q5QN81 Pentatricopeptide (PPR) repeat-containing protein-like [Oryza
sativa]
Length = 540
Score = 199 bits (505), Expect = 2e-49
Identities = 126/430 (29%), Positives = 215/430 (49%), Gaps = 21/430 (4%)
Query: 81 YEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQPGYDHSVREYHSMISVL 140
+ I L+ V S +LV EVL + N A FF WA +Q G+ +S +H++I L
Sbjct: 105 HRIAPVLDALGVTVSPQLVAEVLKNLSNAGILALAFFRWAERQQGFRYSAEGFHNLIEAL 164
Query: 141 GKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQ 200
GK+++F W+LVE MR S ++ T I++R+Y V A+ TF F +
Sbjct: 165 GKIKQFRLVWSLVEAMRC----RSCLSKDTFKIIVRRYARARKVKEAVETFEKMSSFGLK 220
Query: 201 VGLYEFQGLLSALCRYKNVQDAEHLLFCN---KNVFPLDTKSFNIILNGWCN----LIVS 253
L ++ L+ L + K V+ A H +F K F D K++ +++ GW + L++
Sbjct: 221 TDLSDYNWLIDILSKSKQVKKA-HAIFKEMKRKGRFIPDLKTYTVLMEGWGHEKDLLMLK 279
Query: 254 ARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNA 313
A +++EM I+ DVV+Y +IS + KS K +++F +M++ P VY
Sbjct: 280 A-----VYQEMLDAGIKPDVVAYGMLISAFCKSGKCDEAIKVFHEMEESGCMPSPHVYCM 334
Query: 314 VIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERG 373
+I L + EA+ +++ + T N++I C+A + A ++ + M + G
Sbjct: 335 LINGLGSMERLDEALKYFQLSKESGFPMEVPTCNAVIGAYCRALEFHHAFKMVDEMRKSG 394
Query: 374 ISPSIRTFHAFFRILRVKE---EVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVK 430
I P+ RT+ L E E + L +M+ GC P + TY M++ FC ++D
Sbjct: 395 IGPNTRTYDIILNHLIKSEKIEEAYNLFQRMERDGCEPELNTYTMMVGMFCSNERVDMAL 454
Query: 431 RIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESML-QAW 489
++W M+E G+ + LI+GL ++EEA Y+ EM +KG P + S L +A
Sbjct: 455 KVWKQMKEKGVLPCMHMFSALINGLCFENRLEEACVYFQEMLDKGIRPPGQLFSNLKEAL 514
Query: 490 LSGRQVTDSQ 499
+ G ++T +Q
Sbjct: 515 VEGGRITLAQ 524
>UniRef100_Q9FZ19 T6A9.11 protein [Arabidopsis thaliana]
Length = 490
Score = 199 bits (505), Expect = 2e-49
Identities = 129/424 (30%), Positives = 224/424 (52%), Gaps = 20/424 (4%)
Query: 65 DVKTILDIIHKPGSG-PYEIKQKLEDCNVKASSELVVEVLSRVR---NDWEAAFTFFLWA 120
D +T+ +I+ GS E+K+ L + S +L+ VL RVR + F+ +A
Sbjct: 40 DAETVFRMIN--GSNLQVELKESLSSSGIHLSKDLIDRVLKRVRFSHGNPIQTLEFYRYA 97
Query: 121 GKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCA 180
G+ HS +M+ +LG+ R+FD W L+ E +R S+++P+T+ +++ +
Sbjct: 98 SAIRGFYHSSFSLDTMLYILGRNRKFDQIWELLIETKR--KDRSLISPRTMQVVLGRVAK 155
Query: 181 VHDVGRAINTFYAFKRFN---FQVGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDT 237
+ V + + +F+ FKR F F LL LC+ K++ DA ++ K+ F D
Sbjct: 156 LCSVRQTVESFWKFKRLVPDFFDTAC--FNALLRTLCQEKSMTDARNVYHSLKHQFQPDL 213
Query: 238 KSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFE 297
++FNI+L+GW S+ AE +EEM + ++ DVV+Y S+I Y K ++ + +L +
Sbjct: 214 QTFNILLSGW----KSSEEAEAFFEEMKGKGLKPDVVTYNSLIDVYCKDREIEKAYKLID 269
Query: 298 QMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKAR 357
+M++ TPD Y VI L +A ++ +M++ PD YN+ I+ C AR
Sbjct: 270 KMREEEETPDVITYTTVIGGLGLIGQPDKAREVLKEMKEYGCYPDVAAYNAAIRNFCIAR 329
Query: 358 KIDEAKEIFNVMLERGISPSIRTFHAFFRILRVKEEV---FELLDKMKELGCNPTIETYI 414
++ +A ++ + M+++G+SP+ T++ FFR+L + ++ +EL +M C P ++ +
Sbjct: 330 RLGDADKLVDEMVKKGLSPNATTYNLFFRVLSLANDLGRSWELYVRMLGNECLPNTQSCM 389
Query: 415 MLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEK 474
LI+ F R ++D R+W M G G VL+ L KVEEA K +EM EK
Sbjct: 390 FLIKMFKRHEKVDMAMRLWEDMVVKGFGSYSLVSDVLLDLLCDLAKVEEAEKCLLEMVEK 449
Query: 475 GFLP 478
G P
Sbjct: 450 GHRP 453
Score = 73.2 bits (178), Expect = 2e-11
Identities = 52/219 (23%), Positives = 110/219 (49%), Gaps = 11/219 (5%)
Query: 267 RRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRN---ITPDRKVYNAVIFSLAKNRM 323
R H S +++ ++ K ++ +L + K+++ I+P + V+ +AK
Sbjct: 101 RGFYHSSFSLDTMLYILGRNRKFDQIWELLIETKRKDRSLISP--RTMQVVLGRVAKLCS 158
Query: 324 VKEAVNLIIKMEDNNVTPD---AITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRT 380
V++ V K + + PD +N+L++ LC+ + + +A+ +++ L+ P ++T
Sbjct: 159 VRQTVESFWKFK--RLVPDFFDTACFNALLRTLCQEKSMTDARNVYH-SLKHQFQPDLQT 215
Query: 381 FHAFFRILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDG 440
F+ + EE ++MK G P + TY LI +C+ R++++ ++ + MRE+
Sbjct: 216 FNILLSGWKSSEEAEAFFEEMKGKGLKPDVVTYNSLIDVYCKDREIEKAYKLIDKMREEE 275
Query: 441 IGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPE 479
D +Y +I GL L + ++A + EM+E G P+
Sbjct: 276 ETPDVITYTTVIGGLGLIGQPDKAREVLKEMKEYGCYPD 314
Score = 71.6 bits (174), Expect = 5e-11
Identities = 60/245 (24%), Positives = 117/245 (47%), Gaps = 12/245 (4%)
Query: 255 RNAERIWEEMSK-RRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKK-RNITPD---RK 309
R ++IWE + + +R ++S ++ + +KL V Q E K + + PD
Sbjct: 121 RKFDQIWELLIETKRKDRSLISPRTMQVVLGRVAKLCSVRQTVESFWKFKRLVPDFFDTA 180
Query: 310 VYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVM 369
+NA++ +L + + + +A N+ ++ + PD T+N L+ + +EA+ F M
Sbjct: 181 CFNALLRTLCQEKSMTDARNVYHSLK-HQFQPDLQTFNILLSGW---KSSEEAEAFFEEM 236
Query: 370 LERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQL 426
+G+ P + T+++ + R E+ ++L+DKM+E P + TY +I Q
Sbjct: 237 KGKGLKPDVVTYNSLIDVYCKDREIEKAYKLIDKMREEEETPDVITYTTVIGGLGLIGQP 296
Query: 427 DEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESML 486
D+ + + M+E G D ++Y I + ++ +A K EM +KG P T ++
Sbjct: 297 DKAREVLKEMKEYGCYPDVAAYNAAIRNFCIARRLGDADKLVDEMVKKGLSPNATTYNLF 356
Query: 487 QAWLS 491
LS
Sbjct: 357 FRVLS 361
>UniRef100_Q94H96 Hypothetical protein OSJNBb0048A17.10 [Oryza sativa]
Length = 484
Score = 195 bits (495), Expect = 3e-48
Identities = 114/413 (27%), Positives = 207/413 (49%), Gaps = 11/413 (2%)
Query: 83 IKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWAGKQP--GYDHSVREYHSMISVL 140
++ +L+ V+ + ++ VL R+ N A+ FF WA +Q G H+VR YH++++ L
Sbjct: 52 VEHELDHSGVRVTPDVAERVLERLDNAGMLAYRFFEWARRQKRGGCAHTVRSYHTVVASL 111
Query: 141 GKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQ 200
K+R++ W +V MRR E V +T I++RKY V A+ TF +++
Sbjct: 112 AKIRQYQLMWDVVAVMRR----EGAVNVETFGIIMRKYARAQKVDEAVYTFNVMEKYGVV 167
Query: 201 VGLYEFQGLLSALCRYKNVQDAEHLLFCNKNVFPLDTKSFNIILNGWCNLIVSARNAERI 260
L F LL ALC+ KNV+ A+ + + F D K+++I+L GW + +
Sbjct: 168 PNLAAFNSLLGALCKSKNVRKAQEIFDKMNSRFSPDAKTYSILLEGW-GRAPNLPKMREV 226
Query: 261 WEEMSKRRIQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAK 320
+ EM + D+V+Y ++ K+ ++ +++ + M R P +Y+ ++ +
Sbjct: 227 YSEMLDAGCEPDIVTYGIMVDSLCKTGRVEEAVRVVQDMTSRGCQPTTYIYSVLVHTYGV 286
Query: 321 NRMVKEAVNLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRT 380
+++AV + ME + + PD + YN+L+ CKA+K + A + N M GI+ + RT
Sbjct: 287 EMRIEDAVATFLDMEKDGIVPDIVVYNALVSAFCKAKKFENAFRVLNDMEGHGITTNSRT 346
Query: 381 FHAFFR---ILRVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMR 437
++ L +E +++ +M + C P +TY M+I+ FC +++ ++W MR
Sbjct: 347 WNIILNHLISLGRDDEAYKVFRRMIKC-CQPDCDTYTMMIKMFCENDKVEMALKVWKYMR 405
Query: 438 EDGIGHDRSSYIVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKTESMLQAWL 490
++ VLI+GL +V +A +M EKG P T L+ L
Sbjct: 406 LKQFLPSMHTFSVLINGLCDKREVSQACVLLEDMIEKGIRPPGSTFGKLRQLL 458
>UniRef100_Q9LFM6 Hypothetical protein F2I11_200 [Arabidopsis thaliana]
Length = 602
Score = 185 bits (469), Expect = 3e-45
Identities = 134/496 (27%), Positives = 240/496 (48%), Gaps = 28/496 (5%)
Query: 65 DVKTILDIIHK----PGSGPYEIKQKLEDCNVKASSELVVEVLSRVRNDWEAAFTFFLWA 120
D+ TI +++ PGS ++ L++ ++ S ELV + R+ + + F WA
Sbjct: 69 DLSTISNLLENTDVVPGSS---LESALDETGIEPSVELVHALFDRLSSSPMLLHSVFKWA 125
Query: 121 GKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTPQTLLIMIRKYCA 180
+PG+ S + S+++ L K R F+ AW+LV + R G ++V+ T +++IR+Y
Sbjct: 126 EMKPGFTLSPSLFDSVVNSLCKAREFEIAWSLVFDRVRSDEGSNLVSADTFIVLIRRYAR 185
Query: 181 VHDVGRAINTFYAFKRFN----FQVGLYEFQGLLSALCRYKNVQDAEHLL-----FCNKN 231
V +AI F + + L + LL ALC+ +V++A L + N
Sbjct: 186 AGMVQQAIRAFEFARSYEPVCKSATELRLLEVLLDALCKEGHVREASMYLERIGGTMDSN 245
Query: 232 VFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSSKLYR 291
P + FNI+LNGW + AE++WEEM ++ VV+Y ++I Y + ++
Sbjct: 246 WVP-SVRIFNILLNGWFRSR-KLKQAEKLWEEMKAMNVKPTVVTYGTLIEGYCRMRRVQI 303
Query: 292 VLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYNSLIK 351
+++ E+MK + + V+N +I L + + EA+ ++ + P +TYNSL+K
Sbjct: 304 AMEVLEEMKMAEMEINFMVFNPIIDGLGEAGRLSEALGMMERFFVCESGPTIVTYNSLVK 363
Query: 352 PLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKELGCNP 408
CKA + A +I +M+ RG+ P+ T++ FF+ EE L K+ E G +P
Sbjct: 364 NFCKAGDLPGASKILKMMMTRGVDPTTTTYNHFFKYFSKHNKTEEGMNLYFKLIEAGHSP 423
Query: 409 TIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEAYKYY 468
TY ++++ C +L ++ M+ GI D + +LIH L +EEA++ +
Sbjct: 424 DRLTYHLILKMLCEDGKLSLAMQVNKEMKNRGIDPDLLTTTMLIHLLCRLEMLEEAFEEF 483
Query: 469 IEMQEKGFLPEPKTESMLQAWLSGRQVTD------SQATDLEHNQLEHGGLKKSVKPIQS 522
+G +P+ T M+ L + ++D S + L H++ +++V
Sbjct: 484 DNAVRRGIIPQYITFKMIDNGLRSKGMSDMAKRLSSLMSSLPHSKKLPNTYREAVDAPPD 543
Query: 523 KFDREKDFLREPETRS 538
K DR K L E S
Sbjct: 544 K-DRRKSILHRAEAMS 558
>UniRef100_Q9FG82 Emb|CAB67677.1 [Arabidopsis thaliana]
Length = 680
Score = 177 bits (449), Expect = 7e-43
Identities = 114/434 (26%), Positives = 203/434 (46%), Gaps = 9/434 (2%)
Query: 49 DKNSSQLHLRDDGFVKDVKTILDIIHKPGSGPYEIKQKLEDCNVKASSELVVEVLSRVRN 108
+ +SS+ + D F+ + + + G I++ L + S ++V +VL+R
Sbjct: 70 EDSSSKNQVAIDSFLSAEDKLRGVFLQKLKGKSAIQKSLSSLGIGLSIDIVADVLNRGNL 129
Query: 109 DWEAAFTFFLWAGKQPGYDHSVREYHSMISVLGKMRRFDTAWALVEEMRRGKTGESIVTP 168
EA TFF WA ++PG V Y ++ LG+ + F + + ++ +G E +
Sbjct: 130 SGEAMVTFFDWAVREPGVTKDVGSYSVILRALGRRKLF----SFMMDVLKGMVCEGVNPD 185
Query: 169 -QTLLIMIRKYCAVHDVGRAINTFYAFKRFNFQVGLYEFQGLLSALCRYKNVQDAEHLLF 227
+ L I + + VH V RAI F + F + F LL LC +V A+ +
Sbjct: 186 LECLTIAMDSFVRVHYVRRAIELFEESESFGVKCSTESFNALLRCLCERSHVSAAKSVFN 245
Query: 228 CNKNVFPLDTKSFNIILNGWCNLIVSARNAERIWEEMSKRRIQHDVVSYASIISCYSKSS 287
K P D+ S+NI+++GW L E++ +EM + D +SY+ +I ++
Sbjct: 246 AKKGNIPFDSCSYNIMISGWSKL-GEVEEMEKVLKEMVESGFGPDCLSYSHLIEGLGRTG 304
Query: 288 KLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAVNLIIKMEDNNVTPDAITYN 347
++ +++F+ +K + PD VYNA+I + R E++ +M D P+ TY+
Sbjct: 305 RINDSVEIFDNIKHKGNVPDANVYNAMICNFISARDFDESMRYYRRMLDEECEPNLETYS 364
Query: 348 SLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL---RVKEEVFELLDKMKEL 404
L+ L K RK+ +A EIF ML RG+ P+ +F + L + K ++
Sbjct: 365 KLVSGLIKGRKVSDALEIFEEMLSRGVLPTTGLVTSFLKPLCSYGPPHAAMVIYQKSRKA 424
Query: 405 GCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSYIVLIHGLFLNCKVEEA 464
GC + Y +L+++ R+ + + +W+ M+E G D Y ++ GL + +E A
Sbjct: 425 GCRISESAYKLLLKRLSRFGKCGMLLNVWDEMQESGYPSDVEVYEYIVDGLCIIGHLENA 484
Query: 465 YKYYIEMQEKGFLP 478
E KGF P
Sbjct: 485 VLVMEEAMRKGFCP 498
Score = 57.0 bits (136), Expect = 1e-06
Identities = 47/231 (20%), Positives = 100/231 (42%), Gaps = 34/231 (14%)
Query: 269 IQHDVVSYASIISCYSKSSKLYRVLQLFEQMKKRNITPDRKVYNAVIFSLAKNRMVKEAV 328
+ DV SY+ I+ + ++ + + M + PD + + S + V+ A+
Sbjct: 147 VTKDVGSYSVILRALGRRKLFSFMMDVLKGMVCEGVNPDLECLTIAMDSFVRVHYVRRAI 206
Query: 329 NLIIKMEDNNVTPDAITYNSLIKPLCKARKIDEAKEIFNVMLERGISPSIRTFHAFFRIL 388
L + E V ++N+L++ LC+ + AK +FN ++G P
Sbjct: 207 ELFEESESFGVKCSTESFNALLRCLCERSHVSAAKSVFNA--KKGNIP------------ 252
Query: 389 RVKEEVFELLDKMKELGCNPTIETYIMLIRKFCRWRQLDEVKRIWNAMREDGIGHDRSSY 448
F+ C+ Y ++I + + +++E++++ M E G G D SY
Sbjct: 253 ------FD--------SCS-----YNIMISGWSKLGEVEEMEKVLKEMVESGFGPDCLSY 293
Query: 449 IVLIHGLFLNCKVEEAYKYYIEMQEKGFLPEPKT-ESMLQAWLSGRQVTDS 498
LI GL ++ ++ + + ++ KG +P+ +M+ ++S R +S
Sbjct: 294 SHLIEGLGRTGRINDSVEIFDNIKHKGNVPDANVYNAMICNFISARDFDES 344
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 921,750,693
Number of Sequences: 2790947
Number of extensions: 38400468
Number of successful extensions: 167694
Number of sequences better than 10.0: 1171
Number of HSP's better than 10.0 without gapping: 754
Number of HSP's successfully gapped in prelim test: 421
Number of HSP's that attempted gapping in prelim test: 153467
Number of HSP's gapped (non-prelim): 5856
length of query: 550
length of database: 848,049,833
effective HSP length: 132
effective length of query: 418
effective length of database: 479,644,829
effective search space: 200491538522
effective search space used: 200491538522
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC123593.8


