

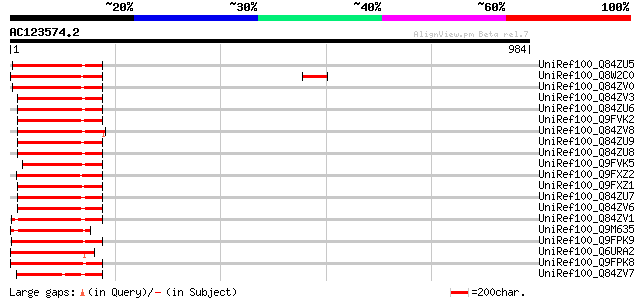
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.2 + phase: 0 /pseudo
(984 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84ZU5 R 8 protein [Glycine max] 209 3e-52
UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Gl... 206 2e-51
UniRef100_Q84ZV0 R 14 protein [Glycine max] 205 5e-51
UniRef100_Q84ZV3 R 4 protein [Glycine max] 204 8e-51
UniRef100_Q84ZU6 R 1 protein [Glycine max] 202 3e-50
UniRef100_Q9FVK2 Resistance protein MG13 [Glycine max] 202 4e-50
UniRef100_Q84ZV8 R 3 protein [Glycine max] 201 1e-49
UniRef100_Q84ZU9 R 13 protein [Glycine max] 200 2e-49
UniRef100_Q84ZU8 R 10 protein [Glycine max] 196 2e-48
UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max] 195 6e-48
UniRef100_Q9FXZ2 Resistance protein LM12 [Glycine max] 195 6e-48
UniRef100_Q9FXZ1 Resistance protein MG23 [Glycine max] 191 7e-47
UniRef100_Q84ZU7 R 5 protein [Glycine max] 187 1e-45
UniRef100_Q84ZV6 R 6 protein [Glycine max] 181 7e-44
UniRef100_Q84ZV1 R 9 protein [Glycine max] 180 2e-43
UniRef100_Q9M635 Hypothetical protein [Glycine max] 179 4e-43
UniRef100_Q9FPK9 Putative resistance protein [Glycine max] 178 6e-43
UniRef100_Q6URA2 TIR-NBS-LRR type R protein 7 [Malus baccata] 166 3e-39
UniRef100_Q9FPK8 Putative resistance protein [Glycine max] 166 4e-39
UniRef100_Q84ZV7 R 12 protein [Glycine max] 166 4e-39
>UniRef100_Q84ZU5 R 8 protein [Glycine max]
Length = 892
Score = 209 bits (532), Expect = 3e-52
Identities = 108/172 (62%), Positives = 131/172 (75%), Gaps = 6/172 (3%)
Query: 6 SSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDN 65
++++ S + Y VFLSF G DTR GFTG LYKAL D+GI+TFIDD EL+RGDEI P+L N
Sbjct: 2 AATTRSLAYNYDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSN 61
Query: 66 AIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYG 125
AI+ESRI I V S NYASSSFCLDELV I+H K G LV+PVF+ VDPSHVRH +GSYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYG 120
Query: 126 EALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
EA+AKH++RF+ N E+LQKW++AL Q A+LSG H G YEY+ G I
Sbjct: 121 EAMAKHQKRFKAN---KEKLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSI 169
Score = 41.6 bits (96), Expect = 0.12
Identities = 20/34 (58%), Positives = 21/34 (60%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
AF KM LK LII N FS G Y P LRVL+W
Sbjct: 554 AFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEW 587
>UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Glycine max]
Length = 1124
Score = 206 bits (525), Expect = 2e-51
Identities = 106/178 (59%), Positives = 132/178 (73%), Gaps = 4/178 (2%)
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
M SSSSS SY F VFLSFRG DTR GFTGNLYKAL+D+GIHTF+DD ++ RGD+IT
Sbjct: 1 MAKQSSSSSFSYRFSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKKIPRGDQIT 60
Query: 61 PSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHH 120
L+ AIEESRIFI V S NYASSSFCL+EL +I+ K G L+LPVF+ VDPS VR+H
Sbjct: 61 SGLEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKVDPSDVRNH 120
Query: 121 RGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH---RSPGYEYKLTGKI 175
GS+G+AL HE++F+ +T+ ME+L+ WK+AL + ANLSG H YEY+ +I
Sbjct: 121 TGSFGKALTNHEKKFK-STNDMEKLETWKMALNKVANLSGYHHFKHGEEYEYEFIQRI 177
Score = 54.3 bits (129), Expect = 2e-05
Identities = 28/48 (58%), Positives = 33/48 (68%)
Query: 555 YSSCFK*KHYQKGMAFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
YSS + + G AFKKM LKTLII +GHFS G K+ P SLRVL+W
Sbjct: 546 YSSFEEVEIQWDGDAFKKMKNLKTLIIRSGHFSKGPKHFPKSLRVLEW 593
Score = 42.7 bits (99), Expect = 0.054
Identities = 24/69 (34%), Positives = 38/69 (54%), Gaps = 7/69 (10%)
Query: 898 GIKYESEMDKALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGIHVLKEKNNAEEENVI 957
G + +D+ALLENEW E+ F + + G+HVLK+++N E+ +
Sbjct: 1001 GERVTDNLDEALLENEWNHAEVTCPGFTFTFAP-----TFIKTGLHVLKQESNMED--IR 1053
Query: 958 FTDPYRETK 966
F+DP R+TK
Sbjct: 1054 FSDPCRKTK 1062
>UniRef100_Q84ZV0 R 14 protein [Glycine max]
Length = 641
Score = 205 bits (522), Expect = 5e-51
Identities = 105/172 (61%), Positives = 128/172 (74%), Gaps = 6/172 (3%)
Query: 6 SSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDN 65
++++ S F Y VFLSFRG DTRYGFTGNLY+AL +KGIHTF D+ +L GDEITP+L
Sbjct: 2 AATTRSLPFIYDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSK 61
Query: 66 AIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYG 125
AI+ESRI I V S NYA SSFCLDELV I+H K G LV+PVF+ VDPS +RH +GSYG
Sbjct: 62 AIQESRIAITVLSQNYAFSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDLRHQKGSYG 120
Query: 126 EALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
EA+ KH++RF+ ME+LQKW++AL Q A+LSG H G YEYK G I
Sbjct: 121 EAMIKHQKRFE---SKMEKLQKWRMALKQVADLSGHHFKDGDAYEYKFIGSI 169
>UniRef100_Q84ZV3 R 4 protein [Glycine max]
Length = 895
Score = 204 bits (520), Expect = 8e-51
Identities = 107/162 (66%), Positives = 124/162 (76%), Gaps = 6/162 (3%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLYKAL D+GI+T IDD EL RGDEITP+L AI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S NYASSSFCLDELV I+H K G LV+PVF+ VDPS VRH +GSYGEA+AKH++RF
Sbjct: 72 VLSQNYASSSFCLDELVTILHC-KSEGLLVIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 130
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
+ E+LQKW++AL Q A+LSG H G YEYK G I
Sbjct: 131 KAK---KEKLQKWRMALKQVADLSGYHFEDGDAYEYKFIGSI 169
Score = 45.1 bits (105), Expect = 0.011
Identities = 21/34 (61%), Positives = 22/34 (63%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
AF KM LK LII NG FS G Y P LRVL+W
Sbjct: 553 AFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEW 586
>UniRef100_Q84ZU6 R 1 protein [Glycine max]
Length = 902
Score = 202 bits (515), Expect = 3e-50
Identities = 105/162 (64%), Positives = 123/162 (75%), Gaps = 6/162 (3%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFL+FRG DTRYGFTGNLYKAL DKGIHTF D+ +L GD+ITP+L AI+ESRI I
Sbjct: 12 YDVFLNFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGDDITPALSKAIQESRIAIT 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S NYASSSFCLDELV I+H K+ G LV+PVF VDPS VRH +GSYGEA+AKH++RF
Sbjct: 72 VLSQNYASSSFCLDELVTILHC-KREGLLVIPVFHNVDPSAVRHLKGSYGEAMAKHQKRF 130
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
+ E+LQKW++AL Q A+LSG H G YEYK G I
Sbjct: 131 KAK---KEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIGNI 169
Score = 42.7 bits (99), Expect = 0.054
Identities = 20/34 (58%), Positives = 21/34 (60%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
AF KM LK LII NG FS G Y P L VL+W
Sbjct: 556 AFMKMENLKILIIRNGKFSKGPNYFPEGLTVLEW 589
>UniRef100_Q9FVK2 Resistance protein MG13 [Glycine max]
Length = 344
Score = 202 bits (514), Expect = 4e-50
Identities = 102/162 (62%), Positives = 124/162 (75%), Gaps = 6/162 (3%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFLSFRG+DTRYGFTGNLYKAL DKG HTF D+ +L G+EITP+L AI++SR+ I
Sbjct: 12 YDVFLSFRGTDTRYGFTGNLYKALCDKGFHTFFDEDKLHSGEEITPALLKAIQDSRVAII 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S NYA SSFCLDELV I H K+ G LV+PVF+ VDPS+VRH +GSYGEA+ KH+ERF
Sbjct: 72 VLSENYAFSSFCLDELVTIFHC-KREGLLVIPVFYKVDPSYVRHQKGSYGEAMTKHQERF 130
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
+ D ME+LQ+W++AL Q A+LSG H G YEY+ G I
Sbjct: 131 K---DKMEKLQEWRMALKQVADLSGSHFKDGGSYEYEFIGSI 169
>UniRef100_Q84ZV8 R 3 protein [Glycine max]
Length = 897
Score = 201 bits (510), Expect = 1e-49
Identities = 108/174 (62%), Positives = 128/174 (73%), Gaps = 12/174 (6%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLYKAL D+GI+TFIDD EL RGDEITP+L AI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S NYASSSFCLDELV ++ L K+ G LV+PVF+ VDPS VR +GSYGEA+AKH++RF
Sbjct: 72 VLSQNYASSSFCLDELVTVL-LCKRKGLLVIPVFYNVDPSDVRQQKGSYGEAMAKHQKRF 130
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKIA------FNQTP 181
+ E+LQKW++AL Q A+LSG H G YEYK I N+TP
Sbjct: 131 KAK---KEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIQSIVEQVSREINRTP 181
Score = 45.1 bits (105), Expect = 0.011
Identities = 21/34 (61%), Positives = 22/34 (63%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
AF KM LK LII NG FS G Y P LRVL+W
Sbjct: 553 AFMKMENLKILIIRNGKFSKGPNYFPQGLRVLEW 586
>UniRef100_Q84ZU9 R 13 protein [Glycine max]
Length = 641
Score = 200 bits (509), Expect = 2e-49
Identities = 104/162 (64%), Positives = 122/162 (75%), Gaps = 6/162 (3%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFLSFRG DTR GFTGNLYKAL D+GI+TFIDD EL RGD+ITP+L NAI ESRI I
Sbjct: 12 YDVFLSFRGLDTRNGFTGNLYKALGDRGIYTFIDDQELPRGDKITPALSNAINESRIAIT 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S NYA SSFCLDELV I+H K G LV+PVF+ VDPS VRH +GSYGE + KH++RF
Sbjct: 72 VLSENYAFSSFCLDELVTILHC-KSEGLLVIPVFYKVDPSDVRHQKGSYGETMTKHQKRF 130
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
+ ME+L++W++AL Q A+LSG H G YEYK G I
Sbjct: 131 E---SKMEKLREWRMALQQVADLSGYHFKDGDSYEYKFIGNI 169
>UniRef100_Q84ZU8 R 10 protein [Glycine max]
Length = 901
Score = 196 bits (499), Expect = 2e-48
Identities = 102/162 (62%), Positives = 120/162 (73%), Gaps = 6/162 (3%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFL+FRG DTRYGFTGNLY+AL DKGIHTF D+ +L RG+EITP+L AI+ESRI I
Sbjct: 12 YDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLKAIQESRIAIT 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S NYASSSFCLDELV I+H K G LV+PVF+ VDPS VRH +GSYG +AKH++RF
Sbjct: 72 VLSKNYASSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDVRHQKGSYGVEMAKHQKRF 130
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
+ E+LQKW+IAL Q A+L G H G YEYK I
Sbjct: 131 KAK---KEKLQKWRIALKQVADLCGYHFKDGDAYEYKFIQSI 169
Score = 42.4 bits (98), Expect = 0.070
Identities = 20/34 (58%), Positives = 21/34 (60%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
AF KM LK LII N FS G Y P LRVL+W
Sbjct: 556 AFMKMENLKILIIRNDKFSKGPNYFPEGLRVLEW 589
>UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max]
Length = 863
Score = 195 bits (495), Expect = 6e-48
Identities = 101/154 (65%), Positives = 119/154 (76%), Gaps = 6/154 (3%)
Query: 24 GSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPVFSANYAS 83
G DTR GFTG LYKAL D+GI+TFIDD EL+RGDEI P+L NAI+ESRI I V S NYAS
Sbjct: 3 GQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAITVLSQNYAS 62
Query: 84 SSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERFQHNTDHME 143
SSFCLDELV I+H K G LV+PVF+ VDPSHVRH +GSYGEA+AKH++RF+ N E
Sbjct: 63 SSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFKAN---KE 118
Query: 144 RLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
+LQKW++AL Q A+LSG H G YEY+ G I
Sbjct: 119 KLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSI 152
Score = 43.5 bits (101), Expect = 0.031
Identities = 31/88 (35%), Positives = 38/88 (42%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKWKGCLLESLSSSILSKASEVTSFYNCK 628
AF KM LK LII N FS G Y P LRVL+W L S+ + +
Sbjct: 530 AFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSNCLPSNFDPINLVICKLPDSS 589
Query: 629 ITKRVFVSSLCLRRSSRI*KS*HWTIVN 656
IT F S S + K H T++N
Sbjct: 590 ITSFEFHGSSKASLKSSLQKLGHLTVLN 617
>UniRef100_Q9FXZ2 Resistance protein LM12 [Glycine max]
Length = 438
Score = 195 bits (495), Expect = 6e-48
Identities = 100/166 (60%), Positives = 124/166 (74%), Gaps = 6/166 (3%)
Query: 14 FKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIF 73
F Y VFLSFRG DTRYGFTG LY L ++GIHTFIDD E Q GDEIT +L+ AIE+S+IF
Sbjct: 6 FSYDVFLSFRGEDTRYGFTGYLYNVLRERGIHTFIDDDEPQEGDEITTALEAAIEKSKIF 65
Query: 74 IPVFSANYASSSFCLDELVHIIHLYKQNGR-LVLPVFFGVDPSHVRHHRGSYGEALAKHE 132
I V S NYASSSFCL+ L HI++ K+N LVLPVF+ V+PS VRHHRGS+GEALA HE
Sbjct: 66 IIVLSENYASSSFCLNSLTHILNFTKENNDVLVLPVFYRVNPSDVRHHRGSFGEALANHE 125
Query: 133 ERFQHNTDHMERLQKWKIALTQAANLSG---DHRSPGYEYKLTGKI 175
++ N+++ME+L+ WK+AL Q +N+SG H YEYK +I
Sbjct: 126 KK--SNSNNMEKLETWKMALHQVSNISGHHFQHDGNKYEYKFIKEI 169
>UniRef100_Q9FXZ1 Resistance protein MG23 [Glycine max]
Length = 435
Score = 191 bits (486), Expect = 7e-47
Identities = 98/164 (59%), Positives = 123/164 (74%), Gaps = 6/164 (3%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLY L ++GI TFIDD ELQ+G EIT +L+ AIE+S+IFI
Sbjct: 8 YDVFLSFRGEDTRHGFTGNLYNVLRERGIDTFIDDEELQKGHEITKALEEAIEKSKIFII 67
Query: 76 VFSANYASSSFCLDELVHIIHLYK-QNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEER 134
V S NYASSSFCL+EL HI++ K ++ R +LPVF+ VDPS VR+HRGS+GEALA HE++
Sbjct: 68 VLSENYASSSFCLNELTHILNFTKGKSDRSILPVFYKVDPSDVRYHRGSFGEALANHEKK 127
Query: 135 FQHNTDHMERLQKWKIALTQAANLSGDHRSPG---YEYKLTGKI 175
+ N +ME+LQ WK+AL Q +N SG H P YEY +I
Sbjct: 128 LKSN--YMEKLQIWKMALQQVSNFSGHHFQPDGDKYEYDFIKEI 169
>UniRef100_Q84ZU7 R 5 protein [Glycine max]
Length = 907
Score = 187 bits (476), Expect = 1e-45
Identities = 97/162 (59%), Positives = 118/162 (71%), Gaps = 8/162 (4%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFLSFRG DTRYGFTGNLYKAL DKGIHTF D+ +L G+EITP+L AI++SRI I
Sbjct: 12 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S ++ASSSFCLDEL I+ + NG +V+PVF+ V P VRH +G+YGEALAKH++RF
Sbjct: 72 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 131
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDH--RSPGYEYKLTGKI 175
++LQKW+ AL Q ANLSG H YEYK G+I
Sbjct: 132 P------DKLQKWERALRQVANLSGLHFKDRDEYEYKFIGRI 167
Score = 45.1 bits (105), Expect = 0.011
Identities = 21/34 (61%), Positives = 22/34 (63%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
AF KM LK LII NG FS G Y P LRVL+W
Sbjct: 552 AFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEW 585
>UniRef100_Q84ZV6 R 6 protein [Glycine max]
Length = 264
Score = 181 bits (460), Expect = 7e-44
Identities = 92/162 (56%), Positives = 117/162 (71%), Gaps = 8/162 (4%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VFL+FRG DTRYGFTGNLY+AL+DKGI TF D+ +L G+EITP+L AI++SRI I
Sbjct: 12 YDVFLNFRGEDTRYGFTGNLYRALSDKGIRTFFDEEKLHSGEEITPALLKAIKDSRIAIT 71
Query: 76 VFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEERF 135
V S ++ASSSFCLDEL I+H + NG +++PVF+ V PS VRH +G+YGEALAKH+ RF
Sbjct: 72 VLSEDFASSSFCLDELTSIVHCAQYNGMMIIPVFYKVYPSDVRHQKGTYGEALAKHKIRF 131
Query: 136 QHNTDHMERLQKWKIALTQAANLSGDH--RSPGYEYKLTGKI 175
E+ Q W++AL Q A+LSG H YEYK +I
Sbjct: 132 P------EKFQNWEMALRQVADLSGFHFKYRDEYEYKFIERI 167
>UniRef100_Q84ZV1 R 9 protein [Glycine max]
Length = 264
Score = 180 bits (456), Expect = 2e-43
Identities = 95/175 (54%), Positives = 123/175 (70%), Gaps = 13/175 (7%)
Query: 3 SSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPS 62
++SS +SI Y VFL+FRG DTRYGFT NLY+AL+DKGI TF D+ +L G+EITP+
Sbjct: 4 TTSSRASI-----YDVFLNFRGEDTRYGFTSNLYRALSDKGIRTFFDEEKLHSGEEITPA 58
Query: 63 LDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRG 122
L AI++SRI I V S ++ASSSFCLDEL I+H + NG +++PVF+ V PS VRH +G
Sbjct: 59 LLKAIKDSRIAITVLSEDFASSSFCLDELTSIVHCAQYNGMMIIPVFYKVYPSDVRHQKG 118
Query: 123 SYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH--RSPGYEYKLTGKI 175
+YGEALAKH+ RF E+ Q W++AL Q A+LSG H YEYK +I
Sbjct: 119 TYGEALAKHKIRFP------EKFQNWEMALRQVADLSGFHFKYRDEYEYKFIERI 167
>UniRef100_Q9M635 Hypothetical protein [Glycine max]
Length = 148
Score = 179 bits (454), Expect = 4e-43
Identities = 94/151 (62%), Positives = 113/151 (74%), Gaps = 9/151 (5%)
Query: 2 QSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITP 61
+SSSSSS+ Y VFLSFRG DTR FTG+LY L KGIHTFIDD +LQRG++ITP
Sbjct: 6 RSSSSSSN------YDVFLSFRGEDTRSAFTGHLYNTLQSKGIHTFIDDEKLQRGEQITP 59
Query: 62 SLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHR 121
+L AIE+SR+ I V S +YASSSFCLDEL I+H ++ LV+PVF+ VDPS VRH +
Sbjct: 60 ALMKAIEDSRVAITVLSEHYASSSFCLDELATILHCDQRKRLLVIPVFYKVDPSDVRHQK 119
Query: 122 GSYGEALAKHEERFQHNTDHMERLQKWKIAL 152
GSYGEALAK E RFQH+ E+LQ WK+AL
Sbjct: 120 GSYGEALAKLERRFQHDP---EKLQNWKMAL 147
>UniRef100_Q9FPK9 Putative resistance protein [Glycine max]
Length = 1093
Score = 178 bits (452), Expect = 6e-43
Identities = 96/174 (55%), Positives = 115/174 (65%), Gaps = 5/174 (2%)
Query: 4 SSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSL 63
S + S + + Y VFLSFRG DTR FTGNLY L +GIHTFI D + + G+EI SL
Sbjct: 2 SKAVSESTDIRVYDVFLSFRGEDTRRSFTGNLYNCLEKRGIHTFIGDYDFESGEEIKASL 61
Query: 64 DNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGS 123
AIE SR+F+ VFS NYASSS+CLD LV I+ + N R V+PVFF V+PSHVRH +G
Sbjct: 62 SEAIEHSRVFVIVFSENYASSSWCLDGLVRILDFTEDNHRPVIPVFFDVEPSHVRHQKGI 121
Query: 124 YGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSG--DHRSPGYEYKLTGKI 175
YGEALA HE R + ++ KW+ AL QAANLSG GYEYKL KI
Sbjct: 122 YGEALAMHERRLNPES---YKVMKWRNALRQAANLSGYAFKHGDGYEYKLIEKI 172
Score = 36.6 bits (83), Expect = 3.8
Identities = 22/71 (30%), Positives = 41/71 (56%), Gaps = 13/71 (18%)
Query: 899 IKYESEMDKALLENEW----IRVELELKSFNLSEEEKNEMLRSAQMGIHVLKEKNNAEEE 954
IK+E +D+ + EN+W + V+++ K +N +E + G+HV+K K++ E+
Sbjct: 1030 IKFEDNVDEVVSENDWNHVVVSVDVDFK-WNPTEP------LVVRTGLHVIKPKSSVED- 1081
Query: 955 NVIFTDPYRET 965
+ F DPY+ T
Sbjct: 1082 -IRFIDPYKPT 1091
Score = 35.8 bits (81), Expect = 6.6
Identities = 23/46 (50%), Positives = 28/46 (60%), Gaps = 2/46 (4%)
Query: 567 GMAFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKWKGCLLESLSS 612
GMAF KM L+TLII FS G K L++L+W GC +SL S
Sbjct: 552 GMAFVKMISLRTLIIRK-MFSKGPKNF-QILKMLEWWGCPSKSLPS 595
>UniRef100_Q6URA2 TIR-NBS-LRR type R protein 7 [Malus baccata]
Length = 1095
Score = 166 bits (420), Expect = 3e-39
Identities = 89/165 (53%), Positives = 111/165 (66%), Gaps = 6/165 (3%)
Query: 2 QSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITP 61
++SSSSSS+S ++ Y +FLSFRG DTR GFTG+L+ AL D+G ++D +L RG+EI
Sbjct: 9 EASSSSSSMSKLWNYDLFLSFRGEDTRNGFTGHLHAALKDRGYQAYMDQDDLNRGEEIKE 68
Query: 62 SLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHR 121
L AIE SRI I VFS YA SS+CLDELV I+ + GR VLP+F+ VDPSHVR
Sbjct: 69 ELFRAIEGSRISIIVFSKRYADSSWCLDELVKIMECRSKLGRHVLPIFYHVDPSHVRKQD 128
Query: 122 GSYGEALAKHEERFQHNTD------HMERLQKWKIALTQAANLSG 160
G EA KHEE TD ER+++WK ALT+AANLSG
Sbjct: 129 GDLAEAFLKHEEGIGEGTDGKKREAKQERVKQWKKALTEAANLSG 173
>UniRef100_Q9FPK8 Putative resistance protein [Glycine max]
Length = 522
Score = 166 bits (419), Expect = 4e-39
Identities = 94/180 (52%), Positives = 120/180 (66%), Gaps = 9/180 (5%)
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
M S SSS + +++ VFLSFRG DTR GF GNLYKALT+KG HTF + +L RG+EI
Sbjct: 1 MAGSERSSSRVWHYEFDVFLSFRGEDTRLGFVGNLYKALTEKGFHTFFRE-KLVRGEEIA 59
Query: 61 PS---LDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHV 117
S ++ AI+ SR+F+ VFS NYASS+ CL+EL+ I+ + N R VLPVF+ VDPS V
Sbjct: 60 ASPSVVEKAIQHSRVFVVVFSQNYASSTRCLEELLSILRFSQDNRRPVLPVFYYVDPSDV 119
Query: 118 RHHRGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSG--DHRSPGYEYKLTGKI 175
G YGEALA HE+RF +D ++ KW+ AL +AA LSG GYEY+L KI
Sbjct: 120 GLQTGMYGEALAMHEKRFNSESD---KVMKWRKALCEAAALSGWPFKHGDGYEYELIEKI 176
>UniRef100_Q84ZV7 R 12 protein [Glycine max]
Length = 893
Score = 166 bits (419), Expect = 4e-39
Identities = 92/164 (56%), Positives = 110/164 (66%), Gaps = 11/164 (6%)
Query: 14 FKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIF 73
F Y VFLSF DT GFT LYKAL D+GI+TF D EL R E+TP L AI SR+
Sbjct: 10 FIYDVFLSFIREDTHRGFTFYLYKALNDRGIYTFFYDQELPRETEVTPGLYKAILASRVA 69
Query: 74 IPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEE 133
I V S NYA SSFCLDELV I+H ++ V+PVF VDPS VRH +GSYGEA+AKH++
Sbjct: 70 IIVLSENYAFSSFCLDELVTILHCERE----VIPVFHNVDPSDVRHQKGSYGEAMAKHQK 125
Query: 134 RFQHNTDHMERLQKWKIALTQAANLSGDHRSPG--YEYKLTGKI 175
RF+ ++LQKW++AL Q ANL G H G YEY L G+I
Sbjct: 126 RFK-----AKKLQKWRMALKQVANLCGYHFKDGGSYEYMLIGRI 164
Score = 45.1 bits (105), Expect = 0.011
Identities = 21/34 (61%), Positives = 22/34 (63%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKW 602
AF KM LK LII NG FS G Y P LRVL+W
Sbjct: 550 AFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEW 583
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.355 0.155 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,335,831,635
Number of Sequences: 2790947
Number of extensions: 48058468
Number of successful extensions: 265754
Number of sequences better than 10.0: 336
Number of HSP's better than 10.0 without gapping: 309
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 264742
Number of HSP's gapped (non-prelim): 584
length of query: 984
length of database: 848,049,833
effective HSP length: 137
effective length of query: 847
effective length of database: 465,690,094
effective search space: 394439509618
effective search space used: 394439509618
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 80 (35.4 bits)
Medicago: description of AC123574.2


