

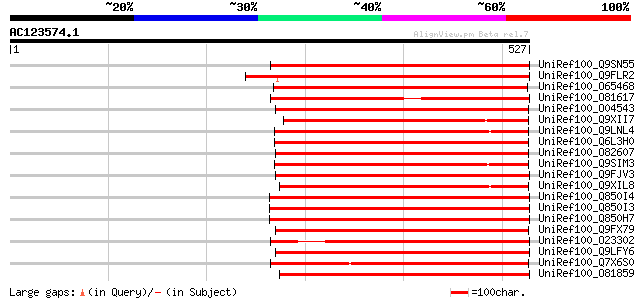
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.1 + phase: 0 /pseudo
(527 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SN55 Putative retrotransposon polyprotein [Arabidops... 298 3e-79
UniRef100_Q9FLR2 Polyprotein-like [Arabidopsis thaliana] 295 3e-78
UniRef100_O65468 Hypothetical protein F21P8.50 [Arabidopsis thal... 278 3e-73
UniRef100_O81617 F8M12.17 protein [Arabidopsis thaliana] 276 1e-72
UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana] 273 7e-72
UniRef100_Q9XII7 Putative retroelement pol polyprotein [Arabidop... 273 9e-72
UniRef100_Q9LNL4 F12K21.14 [Arabidopsis thaliana] 272 2e-71
UniRef100_Q6L3H0 Putative receptor kinase [Solanum demissum] 272 2e-71
UniRef100_O82607 T2L5.9 protein [Arabidopsis thaliana] 269 2e-70
UniRef100_Q9SIM3 Putative retroelement pol polyprotein [Arabidop... 265 2e-69
UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis ... 263 7e-69
UniRef100_Q9XIL8 Putative retroelement pol polyprotein [Arabidop... 263 9e-69
UniRef100_Q850I4 Gag-pol polyprotein [Vitis vinifera] 262 2e-68
UniRef100_Q850I3 Gag-pol polyprotein [Vitis vinifera] 261 3e-68
UniRef100_Q850H7 Gag-pol polyprotein [Vitis vinifera] 261 5e-68
UniRef100_Q9FX79 Putative retroelement polyprotein [Arabidopsis ... 259 2e-67
UniRef100_O23302 Retrovirus-related like polyprotein [Arabidopsi... 258 3e-67
UniRef100_Q9LFY6 T7N9.5 [Arabidopsis thaliana] 258 3e-67
UniRef100_Q7X6S0 OSJNBb0011N17.2 protein [Oryza sativa] 258 3e-67
UniRef100_O81859 Putative LTR retrotransposon [Arabidopsis thali... 257 7e-67
>UniRef100_Q9SN55 Putative retrotransposon polyprotein [Arabidopsis thaliana]
Length = 1203
Score = 298 bits (762), Expect = 3e-79
Identities = 143/262 (54%), Positives = 192/262 (72%)
Query: 266 SEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPI 325
S N+K +LR+ FKIKDLG ++FLGL++A S +GIS+CQRKY +LL D GL+ KP
Sbjct: 734 SAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVCQRKYAQNLLEDVGLSGCKPS 793
Query: 326 STPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHH 385
S P DP++ L ++ + ++YR LVGRLLYL TRPDITF LSQFL+ T +H
Sbjct: 794 SIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDITFAVHTLSQFLSAPTDIHM 853
Query: 386 SAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISW 445
AA +VL+YLK + G+GL + S L + GFSDADW C DSRRS++G C +LG SLI+W
Sbjct: 854 QAAHKVLRYLKGNPGQGLMYSASSELCLNGFSDADWGTCKDSRRSVTGFCIYLGTSLITW 913
Query: 446 RTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAAN 505
++KKQ +VSRSS++ EYR+LA ATCE+ WL LL+DL + + L+CDN+SALH+A N
Sbjct: 914 KSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATN 973
Query: 506 PIFHERTKHLEIDCHLVQDKLQ 527
P+FHERTKH+EIDCH V+D+++
Sbjct: 974 PVFHERTKHIEIDCHTVRDQIK 995
>UniRef100_Q9FLR2 Polyprotein-like [Arabidopsis thaliana]
Length = 509
Score = 295 bits (754), Expect = 3e-78
Identities = 153/294 (52%), Positives = 199/294 (67%), Gaps = 6/294 (2%)
Query: 240 YLPNLQLLHSPSFWSM*MILF*QEIHSEFNN------IKSVLRASFKIKDLGQLKYFLGL 293
Y+ N + S + M+++ +I NN +KSVL A +KIKDLG K+FLGL
Sbjct: 163 YVDNTLFVKITSTAIVAMLIYVDDILIVSNNDEVVCAVKSVLAARYKIKDLGPAKFFLGL 222
Query: 294 QVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVKLHRDSSPPYTDISAYRRLV 353
++A + GIS+CQRKYCLDLLA+SGL KP S P DP V L +D D YR L+
Sbjct: 223 EIARNSDGISICQRKYCLDLLANSGLLGCKPKSVPMDPKVVLTKDLGTLLEDGRPYRELI 282
Query: 354 GRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQI 413
GRLLYL TRPDITF LSQFL+ T VH AA +VLKYLK + G+GLF + L +
Sbjct: 283 GRLLYLCVTRPDITFAVHNLSQFLSCPTNVHLHAAHQVLKYLKNNPGQGLFSSAGTELYL 342
Query: 414 LGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAATCELQ 473
GF+DADW CLDSRRS+SG C FLG SLI+W++KKQ + S SS++ EYR++A AT EL
Sbjct: 343 NGFADADWGTCLDSRRSVSGVCVFLGTSLITWKSKKQEVASGSSTEAEYRSMAVATKELL 402
Query: 474 WLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEIDCHLVQDKLQ 527
WL +L+DL ++ L+CDN+SA+HIA N +FHERTKH+EIDCH +D+++
Sbjct: 403 WLAQMLKDLHVEMEFQVKLFCDNKSAMHIANNSVFHERTKHVEIDCHTTRDRVK 456
>UniRef100_O65468 Hypothetical protein F21P8.50 [Arabidopsis thaliana]
Length = 1240
Score = 278 bits (711), Expect = 3e-73
Identities = 132/257 (51%), Positives = 181/257 (70%)
Query: 269 NNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTP 328
+ +KS L++ FK++DLG LKYFLGL++A S GI++CQRKY LDLL ++GL KP S P
Sbjct: 298 DELKSQLKSCFKLRDLGPLKYFLGLEIARSAAGINICQRKYALDLLDETGLLGCKPSSVP 357
Query: 329 SDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAA 388
DPSV S + D AYRRL+GRL+YL TR DI+F +LSQF H A
Sbjct: 358 MDPSVTFSAHSGGDFVDAKAYRRLIGRLMYLQITRLDISFAVNKLSQFSEAPRLAHQQAV 417
Query: 389 MRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTK 448
M++L Y+K ++G+GLF+ + +Q+ FSDA + C D+RRS +G C FLG SLISW++K
Sbjct: 418 MKILHYIKGTVGQGLFYSSQAEMQLQVFSDASFQSCKDTRRSTNGYCMFLGTSLISWKSK 477
Query: 449 KQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIF 508
KQ +VS+SS++ EYRAL+ AT E+ WL ++LQL SK +L+CDN +A+HIA N +F
Sbjct: 478 KQQVVSKSSAEAEYRALSFATDEMMWLAQFFRELQLPLSKPTLLFCDNTAAIHIATNAVF 537
Query: 509 HERTKHLEIDCHLVQDK 525
HERTKH+E DCH V+++
Sbjct: 538 HERTKHIESDCHSVRER 554
>UniRef100_O81617 F8M12.17 protein [Arabidopsis thaliana]
Length = 1633
Score = 276 bits (705), Expect = 1e-72
Identities = 136/262 (51%), Positives = 183/262 (68%), Gaps = 16/262 (6%)
Query: 266 SEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPI 325
S N+K +LR+ FKIKDLG ++FLGL++A S +GIS+CQRKY +LL D GL+ KP
Sbjct: 1148 SAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVCQRKYAQNLLEDVGLSGCKPS 1207
Query: 326 STPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHH 385
S P DP++ L ++ + ++YR LVGRLLYL TRPDITF LSQFL+ T +H
Sbjct: 1208 SIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDITFAVHTLSQFLSAPTDIHM 1267
Query: 386 SAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISW 445
AA +VL+YLK + G+ DADW C DSRRS++G C +LG SLI+W
Sbjct: 1268 QAAHKVLRYLKGNPGQ----------------DADWGTCKDSRRSVTGFCIYLGTSLITW 1311
Query: 446 RTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAAN 505
++KKQ +VSRSS++ EYR+LA ATCE+ WL LL+DL + + L+CDN+SALH+A N
Sbjct: 1312 KSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATN 1371
Query: 506 PIFHERTKHLEIDCHLVQDKLQ 527
P+FHERTKH+EIDCH V+D+++
Sbjct: 1372 PVFHERTKHIEIDCHTVRDQIK 1393
>UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana]
Length = 1315
Score = 273 bits (699), Expect = 7e-72
Identities = 128/256 (50%), Positives = 185/256 (72%)
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSD 330
+KS +++ FK++DLG+LKYFLGL++ S +GI + QRKY LDLL ++G KP S P D
Sbjct: 1015 LKSQMKSFFKLRDLGELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMD 1074
Query: 331 PSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMR 390
PS+ DS + ++ YRRL+GRL+YLN TRPDITF +L+QF + H A +
Sbjct: 1075 PSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYK 1134
Query: 391 VLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQ 450
+L+Y+K ++G+GLF+ S LQ+ +++AD+ C DSRRS SG C FLG SLI W+++KQ
Sbjct: 1135 ILQYIKGTIGQGLFYSATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQ 1194
Query: 451 LIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHE 510
+VS+SS++ EYR+L+ AT EL WL L++LQ+ SK +L+CDN++A+HIA N +FHE
Sbjct: 1195 DVVSKSSAEAEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHE 1254
Query: 511 RTKHLEIDCHLVQDKL 526
RTKH+E DCH V+++L
Sbjct: 1255 RTKHIESDCHSVRERL 1270
>UniRef100_Q9XII7 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1454
Score = 273 bits (698), Expect = 9e-72
Identities = 131/248 (52%), Positives = 179/248 (71%), Gaps = 1/248 (0%)
Query: 279 FKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVKLHRD 338
FK++DLG LKYFLGL+VA + GIS+CQRKY L+LL +G+ KP+S P P++K+ +D
Sbjct: 1162 FKLRDLGDLKYFLGLEVARTTAGISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKD 1221
Query: 339 SSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTS 398
DI YRR+VG+L+YL TRPDITF +L QF + H +AA RVL+Y+K +
Sbjct: 1222 DGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGT 1281
Query: 399 LGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSS 458
+G+GLF+ S L + GF+D+DWA C DSRRS + F+G SLISWR+KKQ VSRSS+
Sbjct: 1282 VGQGLFYSASSDLTLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSA 1341
Query: 459 KEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKHLEID 518
+ EYRALA ATCE+ WL LL LQ + + +LY D+ +A++IA NP+FHERTKH+++D
Sbjct: 1342 EAEYRALALATCEMVWLFTLLVSLQ-ASPPVPILYSDSTAAIYIATNPVFHERTKHIKLD 1400
Query: 519 CHLVQDKL 526
CH V+++L
Sbjct: 1401 CHTVRERL 1408
>UniRef100_Q9LNL4 F12K21.14 [Arabidopsis thaliana]
Length = 427
Score = 272 bits (696), Expect = 2e-71
Identities = 134/257 (52%), Positives = 183/257 (71%), Gaps = 1/257 (0%)
Query: 270 NIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPS 329
++ L+ SFK+++LG LKYFLGL+VA + +GISLCQRKY L+LL +G+ KP + P
Sbjct: 127 SLTEALKESFKLRELGPLKYFLGLEVARTTEGISLCQRKYALELLTSAGMLDCKPSTIPM 186
Query: 330 DPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAM 389
P+++L + D YR LVGRL+YL TRPDITF +L QF + H +A
Sbjct: 187 TPNLRLSKADGVLLEDAEMYRSLVGRLMYLTITRPDITFAVNKLCQFSSAPRTSHLTAVY 246
Query: 390 RVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKK 449
+VL+Y+K ++G+GLF+ DS L + G++DADW C DSRRS +G F+G SLISWR+KK
Sbjct: 247 KVLQYIKGTVGQGLFYSADSDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKK 306
Query: 450 QLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFH 509
Q VSRSS++ EYRALA A+CE+ WL LL L++ TS + +LY D+ +A++IA NP+FH
Sbjct: 307 QPTVSRSSAEAEYRALALASCEMAWLFTLLIALRVATS-VPILYSDSTAAVYIAINPVFH 365
Query: 510 ERTKHLEIDCHLVQDKL 526
ERTKH+EIDCH V+DKL
Sbjct: 366 ERTKHIEIDCHTVRDKL 382
>UniRef100_Q6L3H0 Putative receptor kinase [Solanum demissum]
Length = 1358
Score = 272 bits (696), Expect = 2e-71
Identities = 129/257 (50%), Positives = 179/257 (69%)
Query: 270 NIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPS 329
++K L F+ KDLG+LKYFLG++VA S+ GI + QRKY LD+L ++G+ +P+ TP
Sbjct: 1057 DLKQHLFKHFQTKDLGRLKYFLGIEVAQSRSGIVISQRKYALDILEETGMMGCRPVDTPM 1116
Query: 330 DPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAM 389
DP+VKL P ++ YRRLVG+L YL TRPDI+F +SQF+ H A +
Sbjct: 1117 DPNVKLLPGQGEPLSNPERYRRLVGKLNYLTVTRPDISFPVSVVSQFMTSPCDSHWEAVV 1176
Query: 390 RVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKK 449
R+L+Y+K++ GKGL F I+G++DADWAG RRS SG C +G +L+SW++KK
Sbjct: 1177 RILRYIKSAPGKGLLFEDQGHEHIIGYTDADWAGSPSDRRSTSGYCVLVGGNLVSWKSKK 1236
Query: 450 QLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFH 509
Q +V+RSS++ EYRA+A ATCEL W+ LL +L+ L CDNQ+ALHIA+NP+FH
Sbjct: 1237 QNVVARSSAESEYRAMATATCELVWIKQLLGELKFGKVDKMELVCDNQAALHIASNPVFH 1296
Query: 510 ERTKHLEIDCHLVQDKL 526
ERTKH+EIDCH V++K+
Sbjct: 1297 ERTKHIEIDCHFVREKI 1313
>UniRef100_O82607 T2L5.9 protein [Arabidopsis thaliana]
Length = 1244
Score = 269 bits (687), Expect = 2e-70
Identities = 128/256 (50%), Positives = 182/256 (71%)
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSD 330
+ + L+ SFK++DLG LKYFLGL++A + GIS+CQRKY L+LLA +G+ KP + P
Sbjct: 944 LTNALKESFKLRDLGPLKYFLGLEIARTAAGISICQRKYALELLAFTGMLDCKPSTIPMV 1003
Query: 331 PSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMR 390
P++KL + D YR LVG+L+YL TRPDITF +L Q+ A H + +
Sbjct: 1004 PNLKLSKADGELLEDREFYRSLVGKLMYLTITRPDITFAVNKLCQYSAAPRTSHLTTVYK 1063
Query: 391 VLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQ 450
VL+Y+K ++G+GLF+ D L + GF+D+DW C D+RRS +G FLG SLI+WR+KKQ
Sbjct: 1064 VLQYIKGTVGQGLFYSSDPDLTLKGFADSDWGTCPDTRRSTTGLTMFLGSSLITWRSKKQ 1123
Query: 451 LIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHE 510
VSRSS++ EYRALA A+CE+ WL LL DL++ T + +++ D+ +A++IA NP+FHE
Sbjct: 1124 PTVSRSSAEAEYRALALASCEMVWLASLLLDLKIITGSVPIVFSDSTAAIYIATNPVFHE 1183
Query: 511 RTKHLEIDCHLVQDKL 526
RTKH+EIDCHLV+++L
Sbjct: 1184 RTKHIEIDCHLVRERL 1199
>UniRef100_Q9SIM3 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1461
Score = 265 bits (677), Expect = 2e-69
Identities = 130/257 (50%), Positives = 182/257 (70%), Gaps = 1/257 (0%)
Query: 270 NIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPS 329
++ L+ASFK+++LG LKYFLGL+VA + +GISL QRKY L+LL + + KP S P
Sbjct: 1160 SLTEALKASFKLRELGPLKYFLGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIPM 1219
Query: 330 DPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAM 389
P+++L ++ D YRRLVG+L+YL TRPDITF +L QF + H +A
Sbjct: 1220 TPNIRLSKNDGLLLEDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVY 1279
Query: 390 RVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKK 449
+VL+Y+K ++G+GLF+ + L + G++DADW C DSRRS +G F+G SLISWR+KK
Sbjct: 1280 KVLQYIKGTVGQGLFYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKK 1339
Query: 450 QLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFH 509
Q VSRSS++ EYRALA A+CE+ WL LL L++ S + +LY D+ +A++IA NP+FH
Sbjct: 1340 QPTVSRSSAEAEYRALALASCEMAWLSTLLLALRVH-SGVPILYSDSTAAVYIATNPVFH 1398
Query: 510 ERTKHLEIDCHLVQDKL 526
ERTKH+EIDCH V++KL
Sbjct: 1399 ERTKHIEIDCHTVREKL 1415
>UniRef100_Q9FJV3 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1475
Score = 263 bits (673), Expect = 7e-69
Identities = 125/258 (48%), Positives = 182/258 (70%), Gaps = 1/258 (0%)
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSD 330
+K L +FK++DLG +KYFLGL++A +K+GIS+CQRKY ++LL D+GL +P + P +
Sbjct: 1174 LKKDLAKAFKLRDLGPMKYFLGLEIARTKEGISVCQRKYTMELLEDTGLLGCRPSTIPME 1233
Query: 331 PSVKLHRDSSPPYTDI-SAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAM 389
PS+KL + + D YRRLVG+L+YL TRPDIT+ +L QF + H AA
Sbjct: 1234 PSLKLSQHNDEHVIDNPEVYRRLVGKLMYLTITRPDITYAINRLCQFSSSPKNSHLKAAQ 1293
Query: 390 RVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKK 449
+V+ YLK ++G GLF+ S L + ++DADW C+DSRRS SG C FLG SLISW++KK
Sbjct: 1294 KVVHYLKGTIGLGLFYSSKSDLCLKAYTDADWGSCVDSRRSTSGICMFLGDSLISWKSKK 1353
Query: 450 QLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFH 509
Q + S SS++ EYRA+A + E+ WL+ LL + Q++ +K L+CD+ +A+HIA N +FH
Sbjct: 1354 QNMASSSSAESEYRAMAMGSREIAWLVKLLAEFQVKQTKPVPLFCDSTAAIHIANNAVFH 1413
Query: 510 ERTKHLEIDCHLVQDKLQ 527
ERTKH+E DCH+ +D+++
Sbjct: 1414 ERTKHIENDCHITRDRIE 1431
>UniRef100_Q9XIL8 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1264
Score = 263 bits (672), Expect = 9e-69
Identities = 132/252 (52%), Positives = 178/252 (70%), Gaps = 1/252 (0%)
Query: 275 LRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVK 334
L+ FK++DLG LKYFLGL++ ++ GISLCQRKY L+LLA +G+ + K +S P P++K
Sbjct: 968 LQNLFKLRDLGDLKYFLGLEITRTEAGISLCQRKYALELLASTGMINCKLVSVPMVPNLK 1027
Query: 335 LHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKY 394
L + D YRR+VG L+YL TRPDITF +L QF T H AA RVL+Y
Sbjct: 1028 LMKVDGELLEDREQYRRIVGTLMYLTITRPDITFAVNKLCQFSYAPTTAHLQAAHRVLQY 1087
Query: 395 LKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVS 454
+K ++G+GLF+ S L + GF+D+DWA C DSR S +G F+G SLIS R+KKQ +VS
Sbjct: 1088 IKGTVGQGLFYSASSDLTLKGFADSDWASCPDSRHSTTGFTMFVGDSLISLRSKKQHVVS 1147
Query: 455 RSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKH 514
RSS++ EYRALA ATCEL WL LL L T+ + +L+ D+ +A++IA NP+FHERTKH
Sbjct: 1148 RSSAEAEYRALALATCELVWLHTLLASLTAATT-IPILFSDSTAAIYIAINPVFHERTKH 1206
Query: 515 LEIDCHLVQDKL 526
+EIDCH V++K+
Sbjct: 1207 IEIDCHTVREKI 1218
>UniRef100_Q850I4 Gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 262 bits (669), Expect = 2e-68
Identities = 122/263 (46%), Positives = 183/263 (69%)
Query: 265 HSEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKP 324
H+ + +K+ + + F KDLG+LKYFLG++V+ SK+G+ L QRKY LDLL ++G +KP
Sbjct: 20 HAGISELKTFMHSKFHTKDLGELKYFLGIEVSRSKKGMFLSQRKYVLDLLKETGKIEAKP 79
Query: 325 ISTPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVH 384
+TP P+V+L D P+ + YRR+VG+L YL TRPDI + +SQF + T H
Sbjct: 80 CTTPMVPNVQLMPDDGDPFYNPERYRRVVGKLNYLTVTRPDIAYAVSVVSQFTSAPTIKH 139
Query: 385 HSAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLIS 444
+A ++L YLK + G G+ + +I FSDADWAG RRS +G C F G +L++
Sbjct: 140 WAALEQILCYLKKAPGLGILYSSQGHTRIECFSDADWAGSKFDRRSTTGYCVFFGGNLVA 199
Query: 445 WRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAA 504
W++KKQ +VSRSS++ EYRA++ ATCE+ W+ LL ++ ++ + L+CDNQ+ALHIAA
Sbjct: 200 WKSKKQSVVSRSSAESEYRAMSRATCEIIWIHQLLCEVGMKCTMPAKLWCDNQAALHIAA 259
Query: 505 NPIFHERTKHLEIDCHLVQDKLQ 527
NP++HERTKH+E+DCH +++K++
Sbjct: 260 NPVYHERTKHIEVDCHFIREKIE 282
>UniRef100_Q850I3 Gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 261 bits (668), Expect = 3e-68
Identities = 122/263 (46%), Positives = 184/263 (69%)
Query: 265 HSEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKP 324
H+ +++K+ + + F KDLG+LKYFLG++V+ SK+G+ L QRKY LDLL ++G +KP
Sbjct: 20 HAGTSDLKTFMHSKFHTKDLGELKYFLGIEVSRSKKGMFLSQRKYVLDLLKETGKIEAKP 79
Query: 325 ISTPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVH 384
+TP P+V+L D P+ + YRR+VG+L YL TRPDI + +SQF + T H
Sbjct: 80 CTTPMVPNVQLMPDDGDPFYNPERYRRVVGKLNYLTVTRPDIAYAVSVVSQFTSAPTIKH 139
Query: 385 HSAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLIS 444
+A ++L YLK + G G+ + +I FSDADWAG RRS +G C F G +L++
Sbjct: 140 WAALEQILCYLKKAPGLGILYSSQGHTRIECFSDADWAGSKFDRRSTTGYCVFFGGNLVA 199
Query: 445 WRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAA 504
W++KKQ +VSRSS++ EYRA++ ATCE+ W+ LL ++ ++ + L+CDNQ+ALHIAA
Sbjct: 200 WKSKKQSVVSRSSAESEYRAMSQATCEIIWIHQLLCEVGMKCTMPAKLWCDNQAALHIAA 259
Query: 505 NPIFHERTKHLEIDCHLVQDKLQ 527
NP++HERTKH+E+DCH +++K++
Sbjct: 260 NPVYHERTKHIEVDCHFIREKIE 282
>UniRef100_Q850H7 Gag-pol polyprotein [Vitis vinifera]
Length = 326
Score = 261 bits (666), Expect = 5e-68
Identities = 122/263 (46%), Positives = 183/263 (69%)
Query: 265 HSEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKP 324
H+ +++K+ + + F KDLG+LKYFLG++V+ SK+G+ L QRKY LDLL ++G +KP
Sbjct: 20 HAGISDLKTFMHSKFHTKDLGELKYFLGIEVSRSKKGMFLSQRKYVLDLLKETGKIEAKP 79
Query: 325 ISTPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVH 384
+TP P+V+L D P+ + YRR+VG+L YL TRPDI + +SQF + T H
Sbjct: 80 CTTPMVPNVQLMPDDGDPFYNPERYRRVVGKLNYLTVTRPDIAYAVSVVSQFTSAPTIKH 139
Query: 385 HSAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLIS 444
+A ++L YLK + G+ + +I FSDADWAG RRS +G C F G +L++
Sbjct: 140 WAALEQILCYLKKAPSLGILYRSQGHTRIECFSDADWAGSKFDRRSTTGYCVFFGGNLVA 199
Query: 445 WRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAA 504
W++KKQ +VSRSS++ EYRA+A ATCE+ W+ LL ++ ++ + L+CDNQ+ALHIAA
Sbjct: 200 WKSKKQSVVSRSSAESEYRAMAQATCEIIWIHQLLCEVGMKCTMPAKLWCDNQAALHIAA 259
Query: 505 NPIFHERTKHLEIDCHLVQDKLQ 527
NP++HERTKH+E+DCH +++K++
Sbjct: 260 NPVYHERTKHIEVDCHFIREKIE 282
>UniRef100_Q9FX79 Putative retroelement polyprotein [Arabidopsis thaliana]
Length = 1413
Score = 259 bits (661), Expect = 2e-67
Identities = 118/256 (46%), Positives = 181/256 (70%)
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSD 330
++ L+ S K++DLG L+YFLGL++A + GIS+CQRKY L+LLA++GL K S P +
Sbjct: 1129 LRDALQRSSKLRDLGTLRYFLGLEIARNTDGISICQRKYTLELLAETGLLGCKSSSVPME 1188
Query: 331 PSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMR 390
P+ KL ++ D YR+LVG+L+YL TRPDIT+ +L QF + H A +
Sbjct: 1189 PNQKLSQEDGELIDDAEHYRKLVGKLMYLTFTRPDITYAVHRLCQFTSAPRVPHLKAVYK 1248
Query: 391 VLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQ 450
++ YLK ++G+GLF+ + L++ GF+D+D++ C DSR+ +G C FLG SL++W++KKQ
Sbjct: 1249 IIYYLKGTVGQGLFYSANVDLKLSGFADSDFSSCSDSRKLTTGYCMFLGTSLVAWKSKKQ 1308
Query: 451 LIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHE 510
++S SS++ EY+A++ A E+ WL +LL+DL + S+ VLYCDN +A+HIA NP+FHE
Sbjct: 1309 EVISMSSAEAEYKAMSMAVREMMWLRFLLEDLWIDVSEASVLYCDNTAAIHIANNPVFHE 1368
Query: 511 RTKHLEIDCHLVQDKL 526
RTKH+E D H +++K+
Sbjct: 1369 RTKHIERDYHHIREKI 1384
>UniRef100_O23302 Retrovirus-related like polyprotein [Arabidopsis thaliana]
Length = 1489
Score = 258 bits (659), Expect = 3e-67
Identities = 127/262 (48%), Positives = 175/262 (66%), Gaps = 26/262 (9%)
Query: 266 SEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPI 325
+E N+K++LR+ FKIKDLG ++FLGL KP
Sbjct: 1205 AEVENLKALLRSEFKIKDLGPARFFLGL--------------------------LGCKPS 1238
Query: 326 STPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHH 385
S P DP++ L RD P + +AYR+L+GRLLYL TRPDIT+ QLSQF++ + +H
Sbjct: 1239 SIPMDPTLHLVRDMGTPLPNPTAYRKLIGRLLYLTITRPDITYAVHQLSQFISAPSDIHL 1298
Query: 386 SAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISW 445
AA +VL+Y+K + G+GL + D + + GFSDADWA C D+RRSISG C +LG SLISW
Sbjct: 1299 QAAHKVLRYIKANPGQGLMYSADYEICLNGFSDADWAACKDTRRSISGFCIYLGTSLISW 1358
Query: 446 RTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAAN 505
++KKQ + SRSS++ EYR++A ATCE+ WL LL+DL + + L+CDN+SALH + N
Sbjct: 1359 KSKKQAVASRSSTESEYRSMAQATCEIIWLQQLLKDLHIPLTCPAKLFCDNKSALHSSLN 1418
Query: 506 PIFHERTKHLEIDCHLVQDKLQ 527
P+FHERTKH+EIDCH V+D+++
Sbjct: 1419 PVFHERTKHIEIDCHTVRDQIK 1440
>UniRef100_Q9LFY6 T7N9.5 [Arabidopsis thaliana]
Length = 1436
Score = 258 bits (659), Expect = 3e-67
Identities = 122/257 (47%), Positives = 179/257 (69%)
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSD 330
+ S L + F+++DLG+ K+FLG+++A + GISLCQRKY LDLLA S + KP S P +
Sbjct: 1135 LTSQLSSFFQLRDLGEPKFFLGIEIARNADGISLCQRKYVLDLLASSDFSDCKPSSIPME 1194
Query: 331 PSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMR 390
P+ KL +D+ D YRR++G+L YL TRPDI F +L+Q+ + T +H A +
Sbjct: 1195 PNQKLSKDTGTLLEDGKQYRRILGKLQYLCLTRPDINFAVSKLAQYSSAPTDIHLQALHK 1254
Query: 391 VLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQ 450
+L+YLK ++G+GLF+ D+ + GFSD+DW C D+RR ++G F+G SL+SWR+KKQ
Sbjct: 1255 ILRYLKGTIGQGLFYGADTNFDLRGFSDSDWQTCPDTRRCVTGFAIFVGNSLVSWRSKKQ 1314
Query: 451 LIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHE 510
+VS SS++ EYRA++ AT EL WL Y+L ++ + LYCDN++ALHIA N +FHE
Sbjct: 1315 DVVSMSSAEAEYRAMSVATKELIWLGYILTAFKIPFTHPAYLYCDNEAALHIANNSVFHE 1374
Query: 511 RTKHLEIDCHLVQDKLQ 527
RTKH+E DCH V++ ++
Sbjct: 1375 RTKHIENDCHKVRECIE 1391
>UniRef100_Q7X6S0 OSJNBb0011N17.2 protein [Oryza sativa]
Length = 1262
Score = 258 bits (659), Expect = 3e-67
Identities = 128/261 (49%), Positives = 178/261 (68%), Gaps = 1/261 (0%)
Query: 266 SEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPI 325
SE +K L F++KDLGQLKYFLG+++A S +GI L QRKY LDLL+D+G+ +P
Sbjct: 958 SEITRLKQNLSKEFEVKDLGQLKYFLGIEIARSPRGIVLSQRKYALDLLSDTGMLGCRPA 1017
Query: 326 STPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHH 385
STP D + KL +S P Y+RLVGRL+YL TRPDIT+ +S+++ H
Sbjct: 1018 STPVDQNHKLCAESGNPVNK-ERYQRLVGRLIYLCHTRPDITYAVSMVSRYMHDPRSGHM 1076
Query: 386 SAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISW 445
A R+L+YLK S GKGL+F ++ L++ G+ DADWA C D RRS SG C F+G +L+SW
Sbjct: 1077 DAVYRILRYLKGSPGKGLWFKKNGHLEVEGYCDADWASCPDDRRSTSGYCVFVGGNLVSW 1136
Query: 446 RTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAAN 505
R+KKQ +VSRS+++ EYRA++ + EL WL LL +L L L+CDN+SA+ IA N
Sbjct: 1137 RSKKQPVVSRSTAEAEYRAMSVSLSELLWLRNLLSELMLPVDTPMKLWCDNKSAISIANN 1196
Query: 506 PIFHERTKHLEIDCHLVQDKL 526
P+ H+RTKH+E+D +++KL
Sbjct: 1197 PVQHDRTKHVELDRFFIKEKL 1217
>UniRef100_O81859 Putative LTR retrotransposon [Arabidopsis thaliana]
Length = 306
Score = 257 bits (656), Expect = 7e-67
Identities = 119/253 (47%), Positives = 177/253 (69%)
Query: 275 LRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVK 334
L++ FK++DLG KYFLG+++A S++GIS+CQRKY L+LL+ +G SKP S P DPSVK
Sbjct: 8 LKSYFKLRDLGAAKYFLGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPLDPSVK 67
Query: 335 LHRDSSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKY 394
L+++ P TD ++YR+LVG+L+YL TRPDI + L QF T VH +A +VL+Y
Sbjct: 68 LNKEDGVPLTDSTSYRKLVGKLMYLQITRPDIAYAVNTLCQFSHAPTSVHLNAVHKVLRY 127
Query: 395 LKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVS 454
LK ++G+GLF+P D + G++D+D+ C DSRR ++ C F+G SL+SW++KKQ VS
Sbjct: 128 LKGTVGQGLFYPADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDSLVSWKSKKQDTVS 187
Query: 455 RSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHIAANPIFHERTKH 514
S+++ E+RA++ T E+ WL LL D ++ LY DN +ALHI N +FHERTK
Sbjct: 188 MSTAEAEFRAMSQGTKEMIWLSRLLDDFKVPFIPPAYLYSDNTAALHIVNNSVFHERTKF 247
Query: 515 LEIDCHLVQDKLQ 527
+E+DC+ ++ ++
Sbjct: 248 VELDCYKTREAVE 260
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.355 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 756,695,192
Number of Sequences: 2790947
Number of extensions: 28360484
Number of successful extensions: 150716
Number of sequences better than 10.0: 753
Number of HSP's better than 10.0 without gapping: 671
Number of HSP's successfully gapped in prelim test: 82
Number of HSP's that attempted gapping in prelim test: 148864
Number of HSP's gapped (non-prelim): 1085
length of query: 527
length of database: 848,049,833
effective HSP length: 132
effective length of query: 395
effective length of database: 479,644,829
effective search space: 189459707455
effective search space used: 189459707455
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC123574.1


