

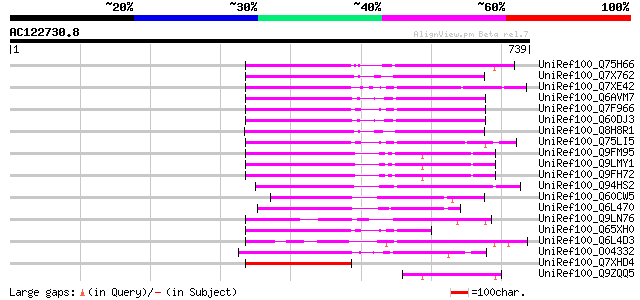
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122730.8 + phase: 0 /pseudo
(739 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sa... 209 3e-52
UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa] 206 3e-51
UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa] 196 2e-48
UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa] 191 7e-47
UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa] 189 2e-46
UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa] 189 3e-46
UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa] 180 1e-43
UniRef100_Q75LI5 Putative transposon protein [Oryza sativa] 173 1e-41
UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabido... 170 1e-40
UniRef100_Q9LMY1 F21F23.10 protein [Arabidopsis thaliana] 170 2e-40
UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5,... 169 3e-40
UniRef100_Q94HS2 Putative transposon protein [Oryza sativa] 168 5e-40
UniRef100_Q60CW5 Putative mutator transposable element-related p... 165 4e-39
UniRef100_Q6L470 Putative mutator transposable element [Solanum ... 163 2e-38
UniRef100_Q9LN76 T12C24.24 [Arabidopsis thaliana] 140 1e-31
UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa] 138 5e-31
UniRef100_Q6L4D3 Hypothetical protein OSJNBa0088M05.7 [Oryza sat... 134 1e-29
UniRef100_O04332 Putative Mutator-like transposase [Arabidopsis ... 122 4e-26
UniRef100_Q7XHD4 Putative mutator-like transposase [Oryza sativa] 117 2e-24
UniRef100_Q9ZQQ5 Mutator-like transposase [Arabidopsis thaliana] 102 4e-20
>UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sativa]
Length = 981
Score = 209 bits (531), Expect = 3e-52
Identities = 119/388 (30%), Positives = 197/388 (50%), Gaps = 43/388 (11%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+PA+ +L PD+++RFCVRHLY NF++ + G+ LK +W A+++ ++ + E+K
Sbjct: 547 KGLIPAVQQLFPDSEHRFCVRHLYQNFQQSFKGEILKNQLWACARSSSVQEWNTKFEEMK 606
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
++E+AYN L Q W ++ FS+ P CD L+NN E FN I+ R+ PI+TMLE I+
Sbjct: 607 ALNEDAYNWLEQMAPNTWVRAFFSDFPKCDILLNNSCEVFNKYILEAREMPILTMLEKIK 666
Query: 456 VYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLF 515
LM R+ +++ + I P IRKK+ K +Q +C
Sbjct: 667 GQLMTRFFNKQKEAQKWQGPICPKIRKKLLK---IAEQ--ANIC---------------- 705
Query: 516 VLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHA 575
V A + +++ R G Y ++++ K C CR+W LTG+PCCHA
Sbjct: 706 --------------YVLPAGKGVFQVEER---GTKYIVDVVTKHCDCRRWDLTGIPCCHA 748
Query: 576 ISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPGR 635
I+C++ + + ++ C+ ++ Y+ I P N ++ W + + PP +++ GR
Sbjct: 749 IACIREDHLSEEDFLPHCYSINAFKAVYAENIIPCNDKANWEKMNGPQILPPVYEKKVGR 808
Query: 636 PKKNRRKDVDEKRDEQQLKKAKYG--MKCSRCKAEGHNKSTCKLPPPPPQPEPGTQ---P 690
PKK+RRK E + K K+G + CS C HNK C+L +P+ +
Sbjct: 809 PKKSRRKQPQEVQGRNGPKLTKHGVTIHCSYCHEANHNKKGCELRKKGIRPKNKIRRNVV 868
Query: 691 PATQFPTSQPTSDHAPTTQVSATQASAS 718
AT+ P+ P P S+ A +
Sbjct: 869 EATEEPSVMPQELMGPQAGGSSLLAEVN 896
>UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa]
Length = 939
Score = 206 bits (523), Expect = 3e-51
Identities = 116/342 (33%), Positives = 175/342 (50%), Gaps = 39/342 (11%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+PA+ DA+ RFCVRHLY NF+ + G+ LK +W A+++ ++ M ++K
Sbjct: 517 KGLVPAVRREFSDAEQRFCVRHLYQNFQVLHKGETLKNQLWAIARSSTVPEWNANMEKMK 576
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+S EAY +L + P W ++ FS+ P CD L+NN +E FN I+ R+ PI++MLE IR
Sbjct: 577 ALSSEAYKYLEEIPPNQWCRAFFSDFPKCDILLNNNSEVFNKYILDAREMPILSMLERIR 636
Query: 456 VYLMER-WEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
+M R + K ++ N+ I P I++K+ K C
Sbjct: 637 NQIMNRLYTKQKELERNWPCGICPKIKRKVEKNTEM-----ANTC--------------- 676
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCH 574
Y + A+ V S G Y + L K C CR+W LTG+PC H
Sbjct: 677 ---YVFPAGMGAFQV---------------SDIGSQYIVELNVKRCDCRRWQLTGIPCNH 718
Query: 575 AISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPG 634
AISC++ + I + VS C+ YE YS+ I P+ W + + +++PP +++ G
Sbjct: 719 AISCLRHERIKPEDVVSFCYSTRCYEQAYSYNIMPLRDSIHWEKMQGIEVKPPVYEKKVG 778
Query: 635 RPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK 676
RPKK RRK E ++ K M CS CK GHNK++CK
Sbjct: 779 RPKKTRRKQPQELEGGTKISKHGVEMHCSYCKNGGHNKTSCK 820
>UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa]
Length = 1005
Score = 196 bits (498), Expect = 2e-48
Identities = 127/403 (31%), Positives = 196/403 (48%), Gaps = 44/403 (10%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GLL ++ L P A++R C RH+Y N+RKK+ ++ ++ W+ AKA F+ ++
Sbjct: 552 KGLLSIVSTLFPFAEHRMCARHIYANWRKKHRLQEYQKRFWKIAKAPNEQLFNHYKRKLA 611
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+ + L +T HWS++ F G C+++ NNM+E+FNS I+ R KPI+TMLEDIR
Sbjct: 612 AKTPRGWQDLEKTNPIHWSRAWFRLGSNCESVDNNMSESFNSWIIESRFKPIITMLEDIR 671
Query: 456 VYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLF 515
+ + R ++NR + + PNI +K K R + F
Sbjct: 672 IQVTRRIQENRSNSERWTMTVCPNIIRKFNK------------IRHRT--------QFCH 711
Query: 516 VLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHA 575
VL WN + +E+R + ++++L K CSCR W ++G+PC HA
Sbjct: 712 VL---------WNG------DAGFEVRDKKWR---FTVDLTSKTCSCRYWQVSGIPCQHA 753
Query: 576 ISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPGR 635
+ + + + + +CF ERY+ Y H++ PV +S W + PP +K+ PGR
Sbjct: 754 CAALFKMAQEPNNCIHECFSLERYKKTYQHVLQPVEHESAWPVSPNPKPLPPRVKKMPGR 813
Query: 636 PKKNRRKDVDEKRDEQQLKKAKYGMK--CSRCKAEGHNKSTCKLPPPPPQPEPGTQPPAT 693
PKKNRRKD K + K +K G K C RC GHN TC + E PPAT
Sbjct: 814 PKKNRRKD-PSKPVKSGTKSSKVGTKIRCRRCGNYGHNSRTCCVKMQQEHAE-SYNPPAT 871
Query: 694 QFPTSQPTSDHAPTTQVSATQASASQVSTTQFVAAKVSSGTQS 736
S P Q T+ + S+++ + SS QS
Sbjct: 872 NASHSSPL--QIEPAQSKQTKGKSIATSSSKSKMPEGSSFIQS 912
>UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa]
Length = 1006
Score = 191 bits (485), Expect = 7e-47
Identities = 107/345 (31%), Positives = 177/345 (51%), Gaps = 41/345 (11%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+PA+ ++ P++++RFCVRHLY+NF+ ++ G+ K +W A+++ +++ M+ ++
Sbjct: 558 KGLIPAVQQVFPESEHRFCVRHLYSNFQLQFKGEVPKNQLWACARSSSVQEWNKNMDVMR 617
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+++ AY L + P W ++ FS P CD L+NN E FN I+ R+ PI+TMLE I+
Sbjct: 618 NLNKSAYEWLEKLPPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPILTMLEKIK 677
Query: 456 VYLMER-WEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
LM R + K ++ + I P IRKK+ K C
Sbjct: 678 GQLMTRHFNKQKELADQFQGLICPKIRKKVLKNA-----DAANTC--------------- 717
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCH 574
Y L A + +++ R Y +++ C CR+W LTG+PC H
Sbjct: 718 ---YAL------------PAGQGIFQVHEREY---QYIVDINAMHCDCRRWDLTGIPCNH 759
Query: 575 AISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPG 634
AISC++ + I+ + + +C+ + + Y I+P N +S W +++PP +++ G
Sbjct: 760 AISCLRHERINAESILPNCYTTDAFSKAYGFNIWPCNDKSKWENINGPEIKPPVYEKKAG 819
Query: 635 RPKKNRRKDVDEKRDEQQLKKAKYG--MKCSRCKAEGHNKSTCKL 677
RPKK+RRK E + K K+G M C C E HN CKL
Sbjct: 820 RPKKSRRKAPYEVIGKNGPKLTKHGVMMHCKYCGEENHNSGGCKL 864
>UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa]
Length = 879
Score = 189 bits (481), Expect = 2e-46
Identities = 107/342 (31%), Positives = 168/342 (48%), Gaps = 38/342 (11%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+ A+ E LP ++R C RH+Y N+RKKY + ++ W+ AKA+ F+ ++
Sbjct: 513 KGLVSAVEEFLPQIEHRMCTRHIYANWRKKYRDQAFQKPFWKCAKASCRPFFNFCRAKLA 572
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+++ T HWS++ F G CD++ NNM E+FN+ I+ +R PI++M E IR
Sbjct: 573 QLTPAGAKXXXSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMFEGIR 632
Query: 456 VYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLF 515
+ R ++NR K + I PNI KK+ K Y L G C
Sbjct: 633 TKVYVRIQQNRSKAKGWLGRICPNILKKLNK----YIDLSGN-CEA-------------- 673
Query: 516 VLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHA 575
WN + +E+ + Y+++L ++ CSCR W L G+PC HA
Sbjct: 674 ----------IWNG------KDGFEVTDKDKR---YTVDLEKRTCSCRYWQLAGIPCAHA 714
Query: 576 ISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPGR 635
I+ + + Y++DC+ E Y Y H + P+ G W T + PP + PGR
Sbjct: 715 ITALFVSSKQPEDYIADCYSVEVYNKIYDHCMMPMEGMMQWPITGHPKPGPPGYVKMPGR 774
Query: 636 PKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKL 677
P+K RR+D E + ++L K +CS+CK HN TC +
Sbjct: 775 PRKERRRDPLEAKKGKKLSKTGTKGRCSQCKQTTHNIRTCPM 816
>UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa]
Length = 1030
Score = 189 bits (480), Expect = 3e-46
Identities = 107/345 (31%), Positives = 177/345 (51%), Gaps = 41/345 (11%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+PA+ ++ P++++RFCVRHLY+NF+ ++ G+ LK +W A+++ +++ M+ ++
Sbjct: 582 KGLIPAVQQVFPESEHRFCVRHLYSNFQLQFKGEVLKNQLWACARSSSVQEWNKNMDVMR 641
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+++ AY L + P W ++ FS P CD L+NN E FN I+ R+ PI+TMLE I+
Sbjct: 642 NLNKSAYEWLEKLPPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPILTMLEKIK 701
Query: 456 VYLMER-WEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
LM R + K ++ + I P IRKK+ K C
Sbjct: 702 GQLMTRHFNKQKELADQFQGLICPKIRKKVLKNA-----DAANTC--------------- 741
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCH 574
Y L A + +++ R Y +++ C CR+W LTG+PC H
Sbjct: 742 ---YAL------------PAGQGIFQVHEREY---QYIVDINAMYCDCRRWDLTGIPCNH 783
Query: 575 AISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPG 634
AISC++ + I+ + + +C+ + + Y I+P N +S W +++PP +++ G
Sbjct: 784 AISCLRHERINAESILPNCYTTDAFSKAYGFNIWPCNDKSKWENVNGPEIKPPVYEKKAG 843
Query: 635 RPKKNRRKDVDEKRDEQQLKKAKYG--MKCSRCKAEGHNKSTCKL 677
RPKK+RRK E + K K+G M C E HN CKL
Sbjct: 844 RPKKSRRKAPYEVIGKNGPKLTKHGVMMHYKYCGEENHNSGGCKL 888
>UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa]
Length = 746
Score = 180 bits (457), Expect = 1e-43
Identities = 101/342 (29%), Positives = 167/342 (48%), Gaps = 38/342 (11%)
Query: 335 MQGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEI 394
++GL+PA+ + PD+++RFCVRHLY NF Y G+ LK +W A++T ++ ++
Sbjct: 392 VEGLIPAIKDEFPDSEHRFCVRHLYQNFAVLYKGEALKNQLWAIARSTTVPEWNVNTEKM 451
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDI 454
K ++++AY +L + P W ++ F + CD L+NN E I+ R+ I++MLE I
Sbjct: 452 KAVNKDAYGYLEEIPPNQWCRAFFRDFSKCDILLNNNLECHVRYILEARELTILSMLEKI 511
Query: 455 RVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
R LM R +++ + +I P I++K+ K + C
Sbjct: 512 RSKLMNRIYTKQEECKKWVFDICPKIKQKVEKNIEM-----SNTC--------------- 551
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCH 574
Y L + + V R + +++ K+C CR+W L G+PC H
Sbjct: 552 ---YALPSRMGVFQVTDR---------------DKQFVVDIKNKQCDCRRWQLIGIPCNH 593
Query: 575 AISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPG 634
AISC++ + I + VS C+ + ++ Y I PV ++ W + + PP +++ G
Sbjct: 594 AISCLRHERIKPEDEVSFCYTIQSFKQAYMFNIMPVRDKTHWEKMNGVPVNPPVYEKKVG 653
Query: 635 RPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK 676
RPK RRK E ++ K + CS CK GHNK CK
Sbjct: 654 RPKTTRRKQPQELDAGTKISKHGVQIHCSYCKNVGHNKKGCK 695
>UniRef100_Q75LI5 Putative transposon protein [Oryza sativa]
Length = 839
Score = 173 bits (439), Expect = 1e-41
Identities = 111/397 (27%), Positives = 184/397 (45%), Gaps = 56/397 (14%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GLL + L P A++R C RH+Y N+RK++ ++ ++ W+ A+++ F+ +++
Sbjct: 437 KGLLSVIEHLFPKAEHRMCARHIYANWRKRHRLQEYQKRFWKIARSSNAVLFNHYKSKLA 496
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+ + L +T HW ++ F G CD++ NN+ E+FN+ I+ R KPI+TMLEDIR
Sbjct: 497 NKTPMGWEDLEKTNPIHWCRAWFKLGSNCDSVENNICESFNNWIIEARFKPIITMLEDIR 556
Query: 456 VYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLF 515
+ + R ++N+ + I PNI KKI K + Q C
Sbjct: 557 MKVTRRIQENKTNSERWTMGICPNILKKINK-----IRHATQFCH--------------- 596
Query: 516 VLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHA 575
WN +E+R + ++++L CSCR W ++G+PC HA
Sbjct: 597 ---------VLWNG------SSGFEVREKKWR---FTVDLSANTCSCRYWQISGIPCQHA 638
Query: 576 ISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPGR 635
+ + + +V+ CF ++Y Y ++ PV +S+W + PP +K+ PG
Sbjct: 639 CAAYFKMAEEPNNHVNMCFSIDQYRNTYQDVLQPVEHESVWPLSTNPRPLPPRVKKMPGS 698
Query: 636 PKKNRRKDVDEKRDEQQLKKAKYG--MKCSRCKAEGHNKSTCK---------LPPPPPQP 684
PK+ RRKD E K +K G +KC C +GHN CK P P Q
Sbjct: 699 PKRARRKDPTEAAG-SSTKSSKRGGSVKCGFCHEKGHNSRGCKKKMEHSSSHYPSHPDQA 757
Query: 685 EPGTQPPATQFPTSQPTSDHAPTTQVSATQASASQVS 721
P + + T +S +Q SA Q++
Sbjct: 758 APSGE------ANDSSVAHRRGKTLLSQSQGSAVQLA 788
>UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabidopsis thaliana]
Length = 733
Score = 170 bits (431), Expect = 1e-40
Identities = 105/363 (28%), Positives = 186/363 (50%), Gaps = 46/363 (12%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGK-KLKELMWRAAKATYTTAFDREMNEI 394
+GL+ A+ +LP A++R C RH++ N +K+Y L ++ W+ A+A T F +++ ++
Sbjct: 365 KGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKM 424
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDI 454
K I EAY+ + ++ +WS++ FS+ + NN++E++N+V+ R+KP+V +LEDI
Sbjct: 425 KTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDI 484
Query: 455 RVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
R ++M +++ N I P + KR K
Sbjct: 485 RRHIMASNLVKLKEMQNVTGLITPKAIAIMEKRKKSLK---------------------- 522
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRK-ECSCRKWLLTGLPCC 573
+ + R I YE+ H + Y +++ K C+CR++ ++G+PCC
Sbjct: 523 ----------WCYPFSNGRGI---YEVDHGK---NKYVVHVRDKTSCTCREYDVSGIPCC 566
Query: 574 HAISCMKSQDIDV---DQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIK 630
H +S M ++ + + + D + E++++CY+ +++PVNG LW + PPP +
Sbjct: 567 HIMSAMWAEYKETKLPETAILDWYSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDR 626
Query: 631 RQPGRPKKNRR-KDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLPPPPPQPEPGTQ 689
PGRPKKN R KD E+ + +KA + CS C GHNK TC++ P P P +
Sbjct: 627 IMPGRPKKNDRIKDPSEEASSENSQKAL--VTCSNCGQTGHNKRTCQIELVPKPPNPSLE 684
Query: 690 PPA 692
P+
Sbjct: 685 FPS 687
>UniRef100_Q9LMY1 F21F23.10 protein [Arabidopsis thaliana]
Length = 753
Score = 170 bits (430), Expect = 2e-40
Identities = 105/363 (28%), Positives = 186/363 (50%), Gaps = 46/363 (12%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGK-KLKELMWRAAKATYTTAFDREMNEI 394
+GL+ A+ +LP A++R C RH++ N +K+Y L ++ W+ A+A T F +++ ++
Sbjct: 385 KGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKM 444
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDI 454
K I EAY+ + ++ +WS++ FS+ + NN++E++N+V+ R+KP+V +LEDI
Sbjct: 445 KTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDI 504
Query: 455 RVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
R ++M +++ N I P + KR K
Sbjct: 505 RRHIMASNLVKIKEMQNVTGLITPKAIAIMEKRKKSLK---------------------- 542
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRK-ECSCRKWLLTGLPCC 573
+ + R I YE+ H + Y +++ K C+CR++ ++G+PCC
Sbjct: 543 ----------WCYPFSNGRGI---YEVDHGK---NKYVVHVRDKTSCTCREYDVSGIPCC 586
Query: 574 HAISCMKSQDIDV---DQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIK 630
H +S M ++ + + + D + E++++CY+ +++PVNG LW + PPP +
Sbjct: 587 HIMSAMWAEYKETKLPETAILDWYSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDR 646
Query: 631 RQPGRPKKNRR-KDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLPPPPPQPEPGTQ 689
PGRPKKN R KD E+ + +KA + CS C GHNK TC++ P P P +
Sbjct: 647 IMPGRPKKNDRIKDPSEEASSENSQKAL--VTCSNCGQTGHNKRTCQIELVPKPPNPSLE 704
Query: 690 PPA 692
P+
Sbjct: 705 FPS 707
>UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MRD20
[Arabidopsis thaliana]
Length = 733
Score = 169 bits (428), Expect = 3e-40
Identities = 105/363 (28%), Positives = 186/363 (50%), Gaps = 46/363 (12%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGK-KLKELMWRAAKATYTTAFDREMNEI 394
+GL+ A+ +LP A++R C RH++ N +K+Y L ++ W+ A+A T F +++ ++
Sbjct: 365 KGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKM 424
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDI 454
K I EAY+ + ++ +WS++ FS+ + NN++E++N+V+ R+KP+V +LEDI
Sbjct: 425 KTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDI 484
Query: 455 RVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
R ++M +++ N I P + KR K
Sbjct: 485 RRHIMASNLVKIKEMQNVTGLITPKAIAIMEKRKKSLK---------------------- 522
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRK-ECSCRKWLLTGLPCC 573
+ + R I YE+ H + Y +++ K C+CR++ ++G+PCC
Sbjct: 523 ----------WCYPFSNGRGI---YEVDHGK---NKYVVHVRDKTSCTCREYDVSGIPCC 566
Query: 574 HAISCMKSQDIDV---DQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIK 630
H +S M ++ + + + D + E++++CY+ +++PVNG LW + PPP +
Sbjct: 567 HIMSAMWAEYKETKLPETAILDWYSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDR 626
Query: 631 RQPGRPKKNRR-KDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLPPPPPQPEPGTQ 689
PGRPKKN R KD E+ + +KA + CS C GHNK TC++ P P P +
Sbjct: 627 IMPGRPKKNDRIKDPSEEASSENSQKAL--VTCSNCGQIGHNKRTCQIELVPKPPNPSLE 684
Query: 690 PPA 692
P+
Sbjct: 685 FPS 687
>UniRef100_Q94HS2 Putative transposon protein [Oryza sativa]
Length = 841
Score = 168 bits (426), Expect = 5e-40
Identities = 107/381 (28%), Positives = 180/381 (47%), Gaps = 45/381 (11%)
Query: 350 DNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKKISEEAYNHLLQTP 409
DN + + + + + G+ LK +W A+++ ++ M E+K ++++AY L + P
Sbjct: 415 DNTYPWTIMTDKQKVHFKGENLKNQLWACARSSSEVEWNANMEEMKSLNQDAYEWLQKMP 474
Query: 410 VKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRVYLMER-WEKNRQK 468
K W K+ FS P CD L+NN E FN I+ R+ PI++M E I+ L+ R + K ++
Sbjct: 475 PKTWVKAYFSEFPKCDILLNNNCEVFNKYILEARELPILSMFEKIKSQLISRHYSKQKEV 534
Query: 469 VANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFVLYPLV*HLYAWN 528
+ I P IRKK+ K A
Sbjct: 535 AEQWHGPICPKIRKKVLKNADM-----------------------------------ANT 559
Query: 529 VLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAISCMKSQDIDVDQ 588
V A + +++ R+ Y ++L K C CR+W LTG+PC HAISC++S+ I +
Sbjct: 560 CYVLPAGKGIFQVEDRNFK---YIVDLSAKHCDCRRWDLTGIPCNHAISCLRSERISAES 616
Query: 589 YVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPGRPKKNRRKDVDEKR 648
+ C+ E + Y+ I+P N + W + +++PP +++ GRP K+RRK E +
Sbjct: 617 ILPPCYSLEAFSRAYAFNIWPYNDMTKWVQVNGPEVKPPIYEKKVGRPPKSRRKAPHEVQ 676
Query: 649 DEQQLKKAKYG--MKCSRCKAEGHNKSTCKLPPPPPQPEPGTQPPATQFPTSQPTSDHAP 706
+ K +K+G M CS CK HNK C L +P+ T ++ P + T +
Sbjct: 677 GKNGPKMSKHGVEMHCSFCKEPRHNKKGCPLRKARLRPKLKT----SRIPAASSTKEWHE 732
Query: 707 TTQVSATQASASQVSTTQFVA 727
+ Q A + SQ + Q ++
Sbjct: 733 SKQEPAEVLTQSQSAYDQLLS 753
>UniRef100_Q60CW5 Putative mutator transposable element-related protein [Solanum
tuberosum]
Length = 493
Score = 165 bits (418), Expect = 4e-39
Identities = 99/311 (31%), Positives = 154/311 (48%), Gaps = 44/311 (14%)
Query: 372 KELMWRAAKATYTTAFDREMNEIKKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNM 431
K W AAKAT FD M I+++ A + L WS+S S+ CD L+NN+
Sbjct: 163 KTAFWAAAKATTVKEFDACMVRIRELDPNAVDWLNDKEPSQWSRSHLSSDAKCDILLNNI 222
Query: 432 TEAFNSVIVVLRKKPIVTMLEDIRVYLMERWEKNRQKVANYADN-ILPNIRKKI*KRV*F 490
E FNS+I R KPIVT+LE +R LM R NR+K ++ N + P I+ + K
Sbjct: 223 CEVFNSMIFDARDKPIVTLLEKLRYLLMARMLANREKAHKWSSNDVCPKIKDILHKN--- 279
Query: 491 YKQLGGQVCRQQL*TLN*Y*IYFLFVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDS 550
A + R++ + YEI +++ DS
Sbjct: 280 --------------------------------QTAAGEYIPRKSNQRKYEIIGATIH-DS 306
Query: 551 YSINLLRKECSCRKWLLTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPV 610
++++L + CSC KW + G+PC HAI+ ++++ ++ YV DC++ E Y Y H I +
Sbjct: 307 WAVDLENRICSCTKWSIMGIPCKHAIAAIRAKKDNILDYVDDCYKVETYRRIYEHAILSI 366
Query: 611 NGQSLWRRTEYTDLQPPPI----KRQPGRPKKNRRKDVDE-KRDEQQLKKAKYGMKCSRC 665
NG +W ++ T + P P+ ++ GR +K RRK+ DE ++K+ + + CS C
Sbjct: 367 NGPQMWPKS--TKVPPLPLTIVGNKKTGRKQKARRKEADEVGASRTKIKRKQQSLDCSTC 424
Query: 666 KAEGHNKSTCK 676
GHNK TCK
Sbjct: 425 NKPGHNKKTCK 435
>UniRef100_Q6L470 Putative mutator transposable element [Solanum demissum]
Length = 707
Score = 163 bits (412), Expect = 2e-38
Identities = 100/292 (34%), Positives = 149/292 (50%), Gaps = 42/292 (14%)
Query: 353 FCVRHLYNNFRKK-YPGKKLKELMWRAAKATYTTAFDREMNEIKKISEEAYNHLLQTPVK 411
FCVRHL+NNF++ Y G LK +W+AA AT FD M ++ ++ ++AY L
Sbjct: 448 FCVRHLHNNFKRAGYSGMALKNALWKAASATTIDRFDACMTDLFELDKDAYAWLSAKVPS 507
Query: 412 HWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRVYLMERWEKNRQKVAN 471
WS+S FS P CD L+NN E FN I+ R KPIV +LE IR LM R R+K
Sbjct: 508 EWSRSHFSPLPKCDILLNNQCEVFNKFILDARDKPIVKLLETIRHLLMTRINSIREKAEK 567
Query: 472 Y-ADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFVLYPLV*HLYAWNVL 530
+ ++I P I+KK+ K + A N +
Sbjct: 568 WNLNDICPTIKKKLAKTM-----------------------------------KKAANYI 592
Query: 531 VRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAISCMKSQDIDVDQYV 590
+R+ ++YE+ + GD+++++L + CSCR+W L+G+PC HAIS + ++ +V YV
Sbjct: 593 PQRSNMWNYEV-IGPVEGDNWAVDLYNRTCSCRQWELSGVPCKHAISSIWLKNDEVLNYV 651
Query: 591 SDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQPGRPKKNRRK 642
DC++ + Y Y I P+NG LW ++ L PPP KK +K
Sbjct: 652 DDCYKVDTYRKIYEASILPMNGLDLWPKS----LNPPPFPPSYLNNKKKGKK 699
>UniRef100_Q9LN76 T12C24.24 [Arabidopsis thaliana]
Length = 839
Score = 140 bits (353), Expect = 1e-31
Identities = 99/360 (27%), Positives = 151/360 (41%), Gaps = 72/360 (20%)
Query: 337 GLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKK 396
GL+ A+ +LP A++R C +H+ +N+++ +L+ L W+ +++ F+ M +K
Sbjct: 444 GLVKAIHNVLPQAEHRQCSKHIMDNWKRDSHDMELQRLFWKISRSYTIEEFNTHMANLKS 503
Query: 397 ISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRV 456
+ +AY L T W+ I R+KP++ MLEDIR
Sbjct: 504 YNPQAYASLQLTSPMTWT------------------------IRQARRKPLLDMLEDIRR 539
Query: 457 YLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFV 516
M R K P +I K + G C + L N + IY
Sbjct: 540 QCMVRTAKRFIIAERLKSRFTPRAHAEIEKMI-----AGSAGCERHLARNNLHEIY---- 590
Query: 517 LYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAI 576
+N Y +++ +K C CRKW + G+PC H
Sbjct: 591 -----------------------------VNDVGYFVDMDKKTCGCRKWEMVGIPCVHTP 621
Query: 577 SCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKR-QPGR 635
+ + V+ YVSD + K R+ Y I PV G LW R + PPP +R PGR
Sbjct: 622 CVIIGRKEKVEDYVSDYYTKVRWRETYRDGIRPVQGMPLWPRMSRLPVLPPPWRRGNPGR 681
Query: 636 ----PKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK-----LPPPPPQPEP 686
+K R + ++ ++ +A M CS CK EGHNKS+CK LPPP P+ P
Sbjct: 682 QSNYARKKGRYETASSSNKNKMSRANRIMTCSNCKQEGHNKSSCKNATVLLPPPRPRGRP 741
>UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa]
Length = 792
Score = 138 bits (348), Expect = 5e-31
Identities = 80/265 (30%), Positives = 131/265 (49%), Gaps = 38/265 (14%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+ A+ E LP ++R C RH+Y N+RKKY + ++ W+ AKA+ F+ ++
Sbjct: 513 KGLVSAVEEFLPQIEHRMCTRHIYANWRKKYRDQAFQKPFWKCAKASCRPFFNFCRAKLA 572
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+++ ++ T HWS++ F G CD++ NNM E+FN+ I+ +R PI++M E IR
Sbjct: 573 QLTPAGAKDMMSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMFEGIR 632
Query: 456 VYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLF 515
+ R ++NR K + I PNI KK+ K Y L G C
Sbjct: 633 TKVYVRIQQNRSKAKGWLGRICPNILKKLNK----YIDLSGN-CEA-------------- 673
Query: 516 VLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHA 575
WN + +E+ + Y+++L ++ CSCR W L G+PC HA
Sbjct: 674 ----------IWNG------KDGFEVTDKDKR---YTVDLEKRTCSCRYWQLAGIPCAHA 714
Query: 576 ISCMKSQDIDVDQYVSDCFRKERYE 600
I+ + + Y++DC+ E E
Sbjct: 715 ITALFVSSKQPEDYIADCYSVEVQE 739
>UniRef100_Q6L4D3 Hypothetical protein OSJNBa0088M05.7 [Oryza sativa]
Length = 1092
Score = 134 bits (337), Expect = 1e-29
Identities = 100/411 (24%), Positives = 171/411 (41%), Gaps = 83/411 (20%)
Query: 337 GLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKK 396
G+L A+ + +A++R RH+Y N+++ + K+ ++ WR ++ +
Sbjct: 680 GILNAVEKWAHEAEHRNYARHIYTNWKRHFHEKQFQKKFWRV--------------KLAQ 725
Query: 397 ISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRV 456
+++ +L T +HWS++ F I+ R PI+TMLE I
Sbjct: 726 VTQAGAQAILNTHPEHWSRAWFR------------------WILESRFHPIITMLETIWR 767
Query: 457 YLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFV 516
+M R + A + + P I KK+
Sbjct: 768 KVMVRINDQKAAGAKWTTVVCPGILKKL-------------------------------- 795
Query: 517 LYPLV*HLYAWNVLVRRAI---EYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCC 573
++Y AI +E++H ++++L +KECSCR W L GLPC
Sbjct: 796 ------NVYITESAFCHAICNGGDSFEVKHHDHR---FTVHLDKKECSCRYWQLLGLPCP 846
Query: 574 HAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKRQP 633
HAISC+ + +D Y++ C+ + + Y H + P+ G S W + + L P + P
Sbjct: 847 HAISCIFYRTNKLDDYIAPCYYVDAFRSTYVHCLQPLEGMSAWPQDDREPLNAPGYIKMP 906
Query: 634 GRPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLPPPPPQPEPGTQ---- 689
GRPK RR++ E ++ K ++C+RCK GHNKS+C G+Q
Sbjct: 907 GRPKTERRREKHEPPKPTKMPKYGTVIRCTRCKQVGHNKSSCSKHQSSGSGTVGSQQQNP 966
Query: 690 PPATQFPTSQPTSDHAPTTQ---VSATQASASQVSTTQFVAAKVSSGTQSV 737
P+ Q S A + + VS T + S T+ +K +Q +
Sbjct: 967 SPSQQMVLSNTLGSSAHSKKRKFVSLTTTDTTSQSRTKHYKSKAPMESQEM 1017
>UniRef100_O04332 Putative Mutator-like transposase [Arabidopsis thaliana]
Length = 754
Score = 122 bits (306), Expect = 4e-26
Identities = 95/362 (26%), Positives = 155/362 (42%), Gaps = 56/362 (15%)
Query: 327 KLMTYPS*MQGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTA 386
KL S Q ++ + P A + CV L + R ++ L L+W AAK
Sbjct: 438 KLTILSSRDQSIVDGVDTNFPTAFHGLCVHCLTESVRTQFNNSILVNLVWEAAKCLTDFE 497
Query: 387 FDREMNEIKKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKP 446
F+ +M EI +IS EA + + W+ F G L N++E+ NS + P
Sbjct: 498 FEGKMGEIAQISPEAASWIRNIQHSQWATYCFE-GTRFGHLTANVSESLNSWVQDASGLP 556
Query: 447 IVTMLEDIRVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TL 506
I+ MLE IR LM + + R+ ++ ++P+ + + + + + CR
Sbjct: 557 IIQMLESIRRQLMTLFNERRETSMQWSGMLVPSAERHVLEAI--------EECR------ 602
Query: 507 N*Y*IYFLFVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWL 566
LYP V +A E +E+ + + +++ + C CR W
Sbjct: 603 ----------LYP-----------VHKANEAQFEVM---TSEGKWIVDIRCRTCYCRGWE 638
Query: 567 LTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTD--- 623
L GLPC HA++ + + +V ++ F Y Y+ I+PV ++ W+ TE
Sbjct: 639 LYGLPCSHAVAALLACRQNVYRFTESYFTVANYRRTYAETIHPVPDKTEWKTTEPAGESG 698
Query: 624 -------LQPPPIKRQPGRPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK 676
++PP RQP RPKK R + D R K ++CSRC GH ++TC
Sbjct: 699 EDGEEIVIRPPRDLRQPPRPKKRRSQGEDRGRQ-------KRVVRCSRCNQAGHFRTTCT 751
Query: 677 LP 678
P
Sbjct: 752 AP 753
>UniRef100_Q7XHD4 Putative mutator-like transposase [Oryza sativa]
Length = 812
Score = 117 bits (292), Expect = 2e-24
Identities = 57/152 (37%), Positives = 93/152 (60%), Gaps = 1/152 (0%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+PA+ A+ RFCVRHLY NF+ + G+ LK +W A+++ ++ M ++K
Sbjct: 519 KGLVPAVRREFSHAEQRFCVRHLYQNFQVLHKGETLKNQLWAIARSSTVPEWNANMEKMK 578
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+S EAY +L + P W ++ FS+ P CD L+NN +E FN I+ R+ PI++MLE IR
Sbjct: 579 ALSSEAYKYLEEIPPNQWCRAFFSDFPKCDILLNNNSEVFNKYILDAREMPILSMLERIR 638
Query: 456 VYLMER-WEKNRQKVANYADNILPNIRKKI*K 486
+M R + K ++ N+ + P I++K+ K
Sbjct: 639 NQIMNRLYTKQKELERNWPCGLCPKIKRKVEK 670
>UniRef100_Q9ZQQ5 Mutator-like transposase [Arabidopsis thaliana]
Length = 241
Score = 102 bits (254), Expect = 4e-20
Identities = 56/154 (36%), Positives = 85/154 (54%), Gaps = 13/154 (8%)
Query: 560 CSCRKWLLTGLPCCHAISCMKSQDIDV---DQYVSDCFRKERYEICYSHIIYPVNGQSLW 616
C+CR + ++G+PCCH +S M + + + VSD + E++++CYS + +PVNG LW
Sbjct: 81 CTCRAYDVSGIPCCHIMSAMWEEYKETKLPETVVSDWYSIEKWKLCYSSLFFPVNGMELW 140
Query: 617 RRTEYTDLQPPPIKRQPGRPKKNRR-KDVDEKRDEQQL--KKAKYGMKCSRCKAEGHNKS 673
+ + PPP + PGRPK N R +D E R Q++ + K M CS C+ GHNK
Sbjct: 141 KTHIDVVVMPPPDRIMPGRPKNNDRIRDPTEDRPPQKVPSTREKLQMTCSNCQQIGHNKR 200
Query: 674 TCKLPP-PPPQPEPGTQP------PATQFPTSQP 700
+CK P P +P +P A + P +QP
Sbjct: 201 SCKRKAVPKPSKKPQGRPRKKQKTMAEENPITQP 234
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.342 0.148 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,157,702,442
Number of Sequences: 2790947
Number of extensions: 46568735
Number of successful extensions: 410677
Number of sequences better than 10.0: 742
Number of HSP's better than 10.0 without gapping: 351
Number of HSP's successfully gapped in prelim test: 427
Number of HSP's that attempted gapping in prelim test: 401677
Number of HSP's gapped (non-prelim): 4518
length of query: 739
length of database: 848,049,833
effective HSP length: 135
effective length of query: 604
effective length of database: 471,271,988
effective search space: 284648280752
effective search space used: 284648280752
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 79 (35.0 bits)
Medicago: description of AC122730.8


