

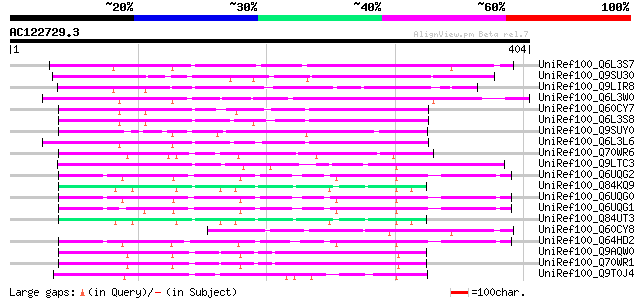
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122729.3 + phase: 0
(404 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum] 138 2e-31
UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis tha... 123 8e-27
UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana] 118 3e-25
UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum] 104 5e-21
UniRef100_Q60CY7 Putative F-box protein [Solanum demissum] 100 1e-19
UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum] 99 3e-19
UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis tha... 98 4e-19
UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum] 95 4e-18
UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hi... 92 3e-17
UniRef100_Q9LTC3 Gb|AAC62811.1 [Arabidopsis thaliana] 90 1e-16
UniRef100_Q6UQG2 S1 self-incompatibility locus-linked putative F... 88 5e-16
UniRef100_Q84KQ9 F-box [Prunus mume] 87 6e-16
UniRef100_Q6UQG0 S3 self-incompatibility locus-linked putative F... 87 8e-16
UniRef100_Q6UQG1 S2 self-incompatibility locus-linked putative F... 87 1e-15
UniRef100_Q84UT3 F-box [Prunus mume] 86 1e-15
UniRef100_Q60CY8 Putative F-box protein [Solanum demissum] 86 2e-15
UniRef100_Q64HD2 S3 putative F-box protein SLF-S3B [Petunia hybr... 82 3e-14
UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein)... 80 1e-13
UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum his... 80 1e-13
UniRef100_Q9T0J4 Hypothetical protein AT4g38870 [Arabidopsis tha... 80 1e-13
>UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum]
Length = 372
Score = 138 bits (348), Expect = 2e-31
Identities = 111/376 (29%), Positives = 182/376 (47%), Gaps = 28/376 (7%)
Query: 32 LFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHL----SQTRPYHL 87
+ ++L ++ +EIL ++P KSLL CVSK+ LIS KF+K HL L + + +
Sbjct: 5 IISVLPHEIIIEILLKVPPKSLLKFMCVSKTWLELISSAKFIKTHLELIANDKEYSHHRI 64
Query: 88 LIRNSELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPI---MIDSCDGIICFENRNDNHV 144
+ + S LPS+ T +P +N I ++ S +G+IC ++ +
Sbjct: 65 IFQESACNFKVCCLPSMLNKERSTELFDIGSPMENPTIYTWIVGSVNGLICLYSKIE--- 121
Query: 145 DLVVWNPCTGKFKILPPL-ENIPNGKTHTL-YSIGYDRFVDNYKVVAFSCHRQINKSYKY 202
+ V+WNP K K LP L + NG ++ L Y GYD D+YKVV C I +
Sbjct: 122 ETVLWNPAVKKSKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYKVVVIQC---IYEDSGS 178
Query: 203 CNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIE-NQKNYDSWVILSL 261
C+S V +++L + WR I F N + GKFV+G I WA+ + ++ I+SL
Sbjct: 179 CDSVVNIYSLKADSWRTINKFQGNFL---VNSPGKFVNGKIYWALSADVDTFNMCNIISL 235
Query: 262 DLGNESYQEISRPD-FGLDD-PVHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNKNSWT 319
DL +E+++ + PD +G P+ + +G LC+ T +W+ KD G + SWT
Sbjct: 236 DLADETWRRLELPDSYGKGSYPLALGVVGSHLSVLCLNCIEGTNSDVWIRKDCGVEVSWT 295
Query: 320 KLFAVPYAKVGYHGFGFVDLHYI---SEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQGQP 376
K+F V + K F + + D++ L + YN V+ +D +
Sbjct: 296 KIFTVDHPKDLGEFIFFTSIFSVPCYQSNKDEILLLLPPVILTYNGSTRQVEVVDRFEEC 355
Query: 377 SILYNSSRVYFESLYD 392
+ ++ +Y ESL D
Sbjct: 356 A----AAEIYVESLVD 367
>UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis thaliana]
Length = 408
Score = 123 bits (309), Expect = 8e-27
Identities = 102/368 (27%), Positives = 184/368 (49%), Gaps = 37/368 (10%)
Query: 34 ALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLH-LSQTRPYHLLIRNS 92
A + +D+ +I RLP K+L+ + +SK LI+DP F++ HLH + QT + +++
Sbjct: 2 ATIPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRG 61
Query: 93 ELLLVDSRLPSVTAIIPDTTHNF-RLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNP 151
L L L S+ + + D H R P++ + S +G+I N + DL V+NP
Sbjct: 62 ALRLYSVDLDSLDS-VSDVEHPMKRGGPTE----VFGSSNGLIGLSN---SPTDLAVFNP 113
Query: 152 CTGKFKILPPLE-NIPNGKT---HTLYSIGYDRFVDNYKVV-----AFSCHRQINKSYKY 202
T + LPP ++P+G + + Y +GYD D+YKVV ++ S+ Y
Sbjct: 114 STRQIHRLPPSSIDLPDGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKIDSEDELGCSFPY 173
Query: 203 CNSQVRVHTLGTNFWRRIPNFPSNIMGL---------PNGYVGKFVSGTINWAIENQKNY 253
+V+V +L N W+RI + S+I L GY G +++W + +
Sbjct: 174 ---EVKVFSLKKNSWKRIESVASSIQLLFYFYYHLLYRRGY-GVLAGNSLHWVLPRRPGL 229
Query: 254 DSW-VILSLDLGNESYQEISRPDFGLDDPVHI-FTLGVSKDCLCVLV-YTETLLGIWVMK 310
++ +I+ DL E ++ + P+ + V I +GV CLC++ Y ++ + +W+MK
Sbjct: 230 IAFNLIVRFDLALEEFEIVRFPEAVANGNVDIQMDIGVLDGCLCLMCNYDQSYVDVWMMK 289
Query: 311 DYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHF-CSKVYVYNYKNSTVKT 369
+Y ++SWTK+F V K F ++ S++ +V L +K+ ++ ++ + T
Sbjct: 290 EYNVRDSWTKVFTVQKPK-SVKSFSYMRPLVYSKDKKKVLLELNNTKLVWFDLESKKMST 348
Query: 370 LDIQGQPS 377
L I+ PS
Sbjct: 349 LRIKDCPS 356
>UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana]
Length = 364
Score = 118 bits (296), Expect = 3e-25
Identities = 99/338 (29%), Positives = 159/338 (46%), Gaps = 30/338 (8%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKF-VKHHLHL------SQTRPYHLLIR 90
L++ EIL RLPVKSL KCV S SLIS+ F +KH L L + T+ + +I
Sbjct: 16 LEMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTSTKSPYGVIT 75
Query: 91 NSELLLVDSRLPSV--TAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVV 148
S L + S+ + + + H+ L D + + + +C G++CF D L +
Sbjct: 76 TSRYHLKSCCIHSLYNASTVYVSEHDGELLGRDYYQV-VGTCHGLVCFHVDYDK--SLYL 132
Query: 149 WNPCTGKFKILPPLE-NIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQV 207
WNP + L + + + Y GYD D+YKVVA R K +
Sbjct: 133 WNPTIKLQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVALLQQRHQVK------IET 186
Query: 208 RVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNES 267
++++ WR +FPS ++ G +++GT+NWA + + SW I+S D+ +
Sbjct: 187 KIYSTRQKLWRSNTSFPSGVVVADKSRSGIYINGTLNWAATSSSS--SWTIISYDMSRDE 244
Query: 268 YQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTE-TLLGIWVMKDYGNKNSWTKLFAVPY 326
++E+ P TLG + CL ++ Y + +WVMK++G SW+KL ++P
Sbjct: 245 FKELPGP-VCCGRGCFTMTLGDLRGCLSMVCYCKGANADVWVMKEFGEVYSWSKLLSIPG 303
Query: 327 AKVGYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKN 364
FV +IS + V L F S + +YN N
Sbjct: 304 LT------DFVRPLWIS-DGLVVLLEFRSGLALYNCSN 334
>UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum]
Length = 427
Score = 104 bits (259), Expect = 5e-21
Identities = 102/391 (26%), Positives = 162/391 (41%), Gaps = 37/391 (9%)
Query: 26 SSDPFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRP- 84
+SD LL +L EIL +LPVKSL CVSKS LIS P FVK+H+ L+
Sbjct: 43 ASDSIIPLLLLPDELITEILLKLPVKSLSKFMCVSKSWLQLISSPTFVKNHIKLTADDKG 102
Query: 85 ---YHLLIRNSELLLVDSRLPSVTAIIPDTTHNFRL-NPSDNHPI---MIDSCDGIICFE 137
+ L+ RN + LP + T F + +P + + ++ S +G+IC
Sbjct: 103 YIHHRLIFRNIDGNFKFCSLPPLFTKQQHTEELFHIDSPIERSTLSTHIVGSVNGLICVV 162
Query: 138 NRNDNHVDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQIN 197
+ + +WNP K K LP + + Y GYD D+YKVV F + +
Sbjct: 163 H---GQKEAYIWNPTITKSKELPKFTS-NMCSSSIKYGFGYDESRDDYKVV-FIHYPYNH 217
Query: 198 KSYKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKN-YDSW 256
S + V +++L N W F + + G+FV+G + W N Y
Sbjct: 218 SSSSNMTTVVHIYSLRNNSW---TTFRDQLQCFLVNHYGRFVNGKLYWTSSTCINKYKVC 274
Query: 257 VILSLDLGNESYQEISRPDFGLDD-PVHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNK 315
I S DL + ++ + P G D+ +++ +G L +W+MK
Sbjct: 275 NITSFDLADGTWGSLDLPICGKDNFDINLGVVGSDLSLLYTCQRGAATSDVWIMKHSAVN 334
Query: 316 NSWTKLFAVPYAK--VGYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQ 373
SWTKLF + Y + + F V I ++ L S + +Y+ +K
Sbjct: 335 VSWTKLFTIKYPQNIKTHRCFAPVFTFSIHFRHSEILLLLRSAIMIYDGSTRQLK----- 389
Query: 374 GQPSILYNSSRVYFESLYDSSGVYVESLISP 404
+ + +YVESL++P
Sbjct: 390 ------------HTSDVMQCEEIYVESLVNP 408
>UniRef100_Q60CY7 Putative F-box protein [Solanum demissum]
Length = 383
Score = 99.8 bits (247), Expect = 1e-19
Identities = 90/304 (29%), Positives = 132/304 (42%), Gaps = 32/304 (10%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRP---YH-LLIRNSEL 94
+L EIL RLP+KSL CVSKS LIS P FVK H+ L+ YH L+ RN+
Sbjct: 14 ELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKKHIKLTANDKGYIYHRLIFRNTNN 73
Query: 95 LLVDSRLPSV---TAIIPDTTH-NFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWN 150
LP + +I + H + + + ++ S +G+IC + + +WN
Sbjct: 74 DFKFCPLPPLFTNQQLIEEILHIDSPIERTTLSTHIVGSVNGLICVAHVRQR--EAYIWN 131
Query: 151 PCTGKFKILPPLENIPNGKTHTLYS------IGYDRFVDNYKVVAFSCHRQINKSYKYCN 204
P K K LP T L S GYD D+YKVV + N
Sbjct: 132 PAITKSKELPK-------STSNLCSDGIKCGFGYDESRDDYKVVFIDYPIRHNH-----R 179
Query: 205 SQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIEN-QKNYDSWVILSLDL 263
+ V +++L TN W + + I L G+FV+G + W NY I S DL
Sbjct: 180 TVVNIYSLRTNSWTTLHDQLQGIFLL--NLHGRFVNGKLYWTSSTCINNYKVCNITSFDL 237
Query: 264 GNESYQEISRPDFGLDDP-VHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNKNSWTKLF 322
+ ++ + P G D+ +++ +G L +W+MK G SWTKLF
Sbjct: 238 ADGTWGSLELPSCGKDNSYINVGVVGSDLSLLYTCQLGAATSDVWIMKHSGVNVSWTKLF 297
Query: 323 AVPY 326
+ Y
Sbjct: 298 TIKY 301
>UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum]
Length = 327
Score = 98.6 bits (244), Expect = 3e-19
Identities = 87/301 (28%), Positives = 137/301 (44%), Gaps = 25/301 (8%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRP---YH-LLIRNSEL 94
+L EIL +LP+KSLL CVSKS LIS P FVK+H+ L+ YH L+ RN+
Sbjct: 15 ELITEILLKLPIKSLLKFMCVSKSWLQLISSPAFVKNHIKLTADDKGYIYHRLIFRNTND 74
Query: 95 LLVDSRLPSV---TAIIPDTTH-NFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWN 150
LP + +I + H + + + ++ S +G+IC + + +WN
Sbjct: 75 DFKFCPLPPLFTQQQLIKELYHIDSPIERTTLSTHIVGSVNGLICAAHVRQR--EAYIWN 132
Query: 151 PCTGKFKILP-PLENIPNGKTHTLYSIGYDRFVDNYKVV--AFSCHRQINKSYKYCNSQV 207
P K K LP N+ + GYD D+YKVV + HR +++ V
Sbjct: 133 PTITKSKELPKSRSNLCSDGIKC--GFGYDESRDDYKVVFIDYPIHRHNHRTV------V 184
Query: 208 RVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIEN-QKNYDSWVILSLDLGNE 266
+++L T W + + L G+FV+G + W + NY I S DL +
Sbjct: 185 NIYSLRTKSWTTLHDQLQGFFLL--NLHGRFVNGKLYWTSSSCINNYKVCNITSFDLADG 242
Query: 267 SYQEISRPDFGLDDP-VHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNKNSWTKLFAVP 325
+++ + P G D+ +++ +G L +W+MK G SWTKLF +
Sbjct: 243 TWERLELPSCGKDNSYINVGVVGSDLSLLYTCQRGAATSDVWIMKHSGVNVSWTKLFTIK 302
Query: 326 Y 326
Y
Sbjct: 303 Y 303
>UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis thaliana]
Length = 394
Score = 98.2 bits (243), Expect = 4e-19
Identities = 91/312 (29%), Positives = 147/312 (46%), Gaps = 43/312 (13%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
DL E+ RL +L+ + +SK SLI P+FV HL HL+I LL
Sbjct: 7 DLINEMFLRLRATTLVKCRVLSKPCFSLIDSPEFVSSHLRRRLETGEHLMI-----LLRG 61
Query: 99 SRLPSVTAIIPDTTHNFRLNPSDNHPI-------MIDSCDGIICFENRNDNHVDLVVWNP 151
RL + + D+ N P HP+ + S +G+I N + VDL ++NP
Sbjct: 62 PRL--LRTVELDSPENVSDIP---HPLQAGGFTEVFGSFNGVIGLCN---SPVDLAIFNP 113
Query: 152 CTGKFKILP------PLENIPNGKTHTLYSIGYDRFVDNYKVVAF-SCHRQINKSYKYCN 204
T K LP P +I + + Y +GYD D++KVV C + K C
Sbjct: 114 STRKIHRLPIEPIDFPERDIT--REYVFYGLGYDSVGDDFKVVRIVQCKLKEGKKKFPCP 171
Query: 205 SQVRVHTLGTNFWRRI-PNFPSNIMG-------LPNGYVGKFVSGTINWAIENQKNYDSW 256
+V+V +L N W+R+ F I+ LP G V+ ++W + ++ ++
Sbjct: 172 VEVKVFSLKKNSWKRVCLMFEFQILWISYYYHLLPRRGYGVVVNNHLHWILPRRQGVIAF 231
Query: 257 -VILSLDLGNESYQEISRP-DFGLDDPVHIFTLGVSKDCLCVLVYTE-TLLGIWVMKDYG 313
I+ DL ++ +S P + ++D + I GV C+C++ Y E + + +WV+K+Y
Sbjct: 232 NAIIKYDLASDDIGVLSFPQELYIEDNMDI---GVLDGCVCLMCYDEYSHVDVWVLKEYE 288
Query: 314 NKNSWTKLFAVP 325
+ SWTKL+ VP
Sbjct: 289 DYKSWTKLYRVP 300
>UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum]
Length = 384
Score = 94.7 bits (234), Expect = 4e-18
Identities = 92/312 (29%), Positives = 137/312 (43%), Gaps = 22/312 (7%)
Query: 26 SSDPFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRP- 84
+SD LL +L EIL RLP+KSL CVSKS LIS P FVK+H+ L+
Sbjct: 2 ASDSIIPLLLLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKNHIKLTANGKG 61
Query: 85 --YH-LLIRNSELLLVDSRLPSVTAIIPDTTHNFRL-NPSDNHPI---MIDSCDGIICFE 137
YH L+ RN+ LPS+ F + +P + + ++ S +G+IC
Sbjct: 62 YIYHRLIFRNTNDDFKFCPLPSLFTKQQLIEELFDIVSPIERTTLSTHIVGSVNGLICAA 121
Query: 138 NRNDNHVDLVVWNPCTGKFKILP-PLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQI 196
+ + +WNP K K LP N+ + GYD D+YKVV +
Sbjct: 122 HVRQR--EAYIWNPTITKSKELPKSRSNLCSDGIKC--GFGYDESHDDYKVVFIN----- 172
Query: 197 NKSYKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIEN-QKNYDS 255
S+ S V +++L TN W + + I L +FV + W NY
Sbjct: 173 YPSHHNHRSVVNIYSLRTNSWTTLHDQLQGIFLL--NLHCRFVKEKLYWTSSTCINNYKV 230
Query: 256 WVILSLDLGNESYQEISRPDFGLDDP-VHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGN 314
I S DL + +++ + P G D+ +++ +G L +W+MK G
Sbjct: 231 CNITSFDLADGTWESLELPSCGKDNSYINVGVVGSDLSLLYTCQRGAANSDVWIMKHSGV 290
Query: 315 KNSWTKLFAVPY 326
SWTKLF + Y
Sbjct: 291 NVSWTKLFTIKY 302
>UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hispanicum]
Length = 374
Score = 91.7 bits (226), Expect = 3e-17
Identities = 83/316 (26%), Positives = 136/316 (42%), Gaps = 34/316 (10%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIR-------- 90
D+ EIL LPVKSL+ LKC SK L+ LI F+ H+ + LL+R
Sbjct: 10 DILKEILVWLPVKSLIRLKCASKHLDMLIKSQAFITSHMIKQRRNDGMLLVRRILPPSTY 69
Query: 91 NSELLLVDSRLPSVTAIIPDTTHNFRLNPSDN--HPIMID---SCDGIICFENRNDNHVD 145
N D P + ++P NP + +P ++D C+GI+C + D
Sbjct: 70 NDVFSFHDVNSPELEEVLPKLPITLLSNPDEASFNPNIVDVLGPCNGIVCITGQE----D 125
Query: 146 LVVWNPCTGKFKILPPLENIPNGKTHTLYSI----GY-DRFVDNYKVVAFSCHRQINKSY 200
+++ NP +F+ LP + P YSI G+ +N+KV+ +
Sbjct: 126 IILCNPALREFRKLP---SAPISCRPPCYSIRTGGGFGSTCTNNFKVILMNTLYTARVDG 182
Query: 201 KYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGK--FVSGTINWAIENQKNYDSWVI 258
+ ++ ++ + WR I +F + +M + Y F G +W VI
Sbjct: 183 RDAQHRIHLYNSNNDSWREINDF-AIVMPVVFSYQCSELFFKGACHWNGRTSGETTPDVI 241
Query: 259 LSLDLGNESYQEISRPD-FGLDDPV-HIFTLGVSKDCLCVL--VYTETLLGIWVMKDYGN 314
L+ D+ E + + P F L + H F + +C + L+ +WVMK+YG
Sbjct: 242 LTFDVSTEVFGQFEHPSGFKLCTGLQHNFM--ILNECFASVRSEVVRCLIEVWVMKEYGI 299
Query: 315 KNSWTKLFAVPYAKVG 330
K SWTK F + ++G
Sbjct: 300 KQSWTKKFVIGPHEIG 315
>UniRef100_Q9LTC3 Gb|AAC62811.1 [Arabidopsis thaliana]
Length = 362
Score = 89.7 bits (221), Expect = 1e-16
Identities = 91/364 (25%), Positives = 146/364 (40%), Gaps = 37/364 (10%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLV 97
++L EIL R+P K L L+ SK N+L F K H + P +++++S + L
Sbjct: 8 VELQEEILSRVPAKYLARLRSTSKQWNALSKTGSFAKKHSANATKEPLIIMLKDSRVYLA 67
Query: 98 DSRLPSVTAIIPDTTH-NFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCTGKF 156
L V + + RL D H + CDG++ + +N L VWNPC+G+
Sbjct: 68 SVNLHGVHNNVAQSFELGSRLYLKDPHISNVFHCDGLLLLCSIKEN--TLEVWNPCSGEA 125
Query: 157 KILPPLENIPNGKTHTLYSIGYDR--FVDNYKVVAFSCHRQINKSYK-------YCNSQV 207
K++ P + K Y++GYD YKV+ + +K + N
Sbjct: 126 KLIKPRHSY--YKESDFYALGYDNKSSCKKYKVLRVISQVHVQGDFKIEYEIYDFTNDSW 183
Query: 208 RVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNES 267
RVH T R + S V G+ W + N + + LS D E
Sbjct: 184 RVHGATTELSIRQKHPVS-------------VKGSTYWVVRN-RYFPYKYFLSFDFSTER 229
Query: 268 YQEISRPDFGLDDPVHIFTLG-VSKDCLCVLVY-----TETLLGIWVMKDYGNKNSWTKL 321
+Q +S P P + L V ++ LC+ Y T L +WV G+ SW+K
Sbjct: 230 FQSLSLPQ---PFPYLVTDLSVVREEQLCLFGYYNWSTTSEDLNVWVTTSLGSVVSWSKF 286
Query: 322 FAVPYAKVGYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQGQPSILYN 381
+ K F + + E++ + KV + ++ LD G+ S L +
Sbjct: 287 LTIQIIKPRVDMFDYGMSFLVDEQNKSLVCWISQKVLHIVGEIYHIQDLDDHGRDSALRS 346
Query: 382 SSRV 385
S V
Sbjct: 347 SCSV 350
>UniRef100_Q6UQG2 S1 self-incompatibility locus-linked putative F-box protein S1-
A134 [Petunia integrifolia subsp. inflata]
Length = 379
Score = 87.8 bits (216), Expect = 5e-16
Identities = 95/386 (24%), Positives = 166/386 (42%), Gaps = 65/386 (16%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYH----LLIRNSEL 94
D+ + I LPVKSLL KC K+ +I F+ +LHL+ T ++ LL R+ E
Sbjct: 7 DVVIYIFVMLPVKSLLRFKCTCKTFYHIIKSSTFI--NLHLNHTTNFNDELVLLKRSFET 64
Query: 95 LLVDSRLPSVTAIIPDTTHNFR-LNPSDNHPIM-----------IDSCDGIICFENRNDN 142
+ ++ + ++F+ ++P P + I C+G+I +
Sbjct: 65 DEYNFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACICHRLIGPCNGLIVLTDS--- 121
Query: 143 HVDLVVWNPCTGKFKILPPLE-NIPNGKTHTLYSIGY--DRFVDNYKVVAFSCHRQINKS 199
+ +V+NP T K++++PP IP G ++ IG+ D ++YKVV S ++ K
Sbjct: 122 -LTTIVFNPATLKYRLIPPCPFGIPRGFRRSISGIGFGFDSDANDYKVVRLS---EVYKE 177
Query: 200 YKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNY------ 253
+V ++ + WR + +G +V F I + ++N+
Sbjct: 178 PCDKEMKVDIYDFSVDSWREL-------LGQDVPFVFWFPCAEILY----KRNFHWFAFA 226
Query: 254 DSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVY--------TETLLG 305
D VIL D+ E + + PD D + L + C+ ++ Y TE L
Sbjct: 227 DDVVILCFDMNTEKFHNMGMPDACHFDDGKSYGLVILFKCMTLICYPDPMPSSPTEKLTD 286
Query: 306 IWVMKDYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHF-CSKVYVYNYKN 364
IW+MK+YG K SW K ++ + +D+ + LH + Y+ +
Sbjct: 287 IWIMKEYGEKESWIKRCSIRL---------LPESPLAVWKDEILLLHSKMGHLIAYDLNS 337
Query: 365 STVKTLDIQGQPSILYNSSRVYFESL 390
+ V+ LD+ G P L +Y ESL
Sbjct: 338 NEVQELDLHGYPESL--RIIIYRESL 361
>UniRef100_Q84KQ9 F-box [Prunus mume]
Length = 428
Score = 87.4 bits (215), Expect = 6e-16
Identities = 91/352 (25%), Positives = 141/352 (39%), Gaps = 76/352 (21%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQ----TRPYHLLIRNSEL 94
D+ IL RLP KSL+ +CV KS + LI P FV HHL +S T ++L++++ L
Sbjct: 14 DMMGNILSRLPPKSLMRFRCVLKSWHDLIDKPSFVDHHLSISMDNKVTSSTYVLLKHNVL 73
Query: 95 ----LLVDSRLPSVTAIIPDTTHNFRLNPSDNHPIMID---------------------- 128
+ D + T PD+ L S N ++D
Sbjct: 74 TDPSIKDDEKAVRATLFNPDSNQRDILLSSLNLGSLVDDGLEIENHVVPPPMRGYALSLE 133
Query: 129 ---SCDGIICFENRNDNHVDLVVWNPCTGKFKILPPL---------ENIPNGKTHTLY-- 174
SCDG+IC N D+V+ NP ++++LP + Y
Sbjct: 134 ISGSCDGLICLNTFNSE--DIVLCNPALEEYRVLPKSCILLPPRVPRQFEENEDDDYYEE 191
Query: 175 -------------SIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTNFWRRIP 221
GYD +YKVV + + ++ + S+V V++L + WR +P
Sbjct: 192 DDDDEIESNPKCVGFGYDPNSKDYKVVRAA--QFVSGVFTQHPSKVEVYSLAADTWREVP 249
Query: 222 NFPSNIMGLPNGYVGKFVSGTINWAI--ENQKNYDSWVILSLDLGNESYQEISRPDFGLD 279
G N + G W ++N VILS D+ E + +I+ P+ G
Sbjct: 250 -VDIQPHGSLNPSYQMYFKGFFYWIAYWTEERN----VILSFDMSEEVFHDIALPESG-P 303
Query: 280 DPVHIFTLGVSKDCLCVLVY-----TETLLGIWVMKD--YGNKNSWTKLFAV 324
D ++ V KD L +L Y L +WV+ + G K WTK A+
Sbjct: 304 DAYEYTSIAVWKDSLALLTYPVENEAPKTLDLWVLDEDLKGAKGLWTKHLAI 355
>UniRef100_Q6UQG0 S3 self-incompatibility locus-linked putative F-box protein S3-
A134 [Petunia integrifolia subsp. inflata]
Length = 379
Score = 87.0 bits (214), Expect = 8e-16
Identities = 95/386 (24%), Positives = 166/386 (42%), Gaps = 65/386 (16%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYH----LLIRNSEL 94
D+ + I LPVKSLL KC K+ +I F+ +LHL+ T ++ LL R+ E
Sbjct: 7 DVVIYIFVMLPVKSLLRSKCTCKTFCHIIKSSTFI--NLHLNHTTNFNDELVLLKRSFET 64
Query: 95 LLVDSRLPSVTAIIPDTTHNFR-LNPSDNHPIM-----------IDSCDGIICFENRNDN 142
+ ++ + ++F+ ++P P + I C+G+I +
Sbjct: 65 DEYNFYKSILSFLFAKKDYDFKPISPDVKIPHLTTTAACICHRLIGPCNGLIVLTDS--- 121
Query: 143 HVDLVVWNPCTGKFKILPPLE-NIPNGKTHTLYSIGY--DRFVDNYKVVAFSCHRQINKS 199
+ +V+NP T K++++PP IP G ++ IG+ D ++YKVV S ++ K
Sbjct: 122 -LTTIVFNPATLKYRLIPPCPFGIPRGFRRSISGIGFGFDSDANDYKVVRLS---EVYKG 177
Query: 200 YKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNY------ 253
+V ++ + WR + +G +V F I + ++N+
Sbjct: 178 TCDKKMKVDIYDFSVDSWREL-------LGQDVPFVFWFPCAEILY----KRNFHWFAFA 226
Query: 254 DSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVY--------TETLLG 305
D VIL D+ E + + PD D + L + C+ ++ Y TE L
Sbjct: 227 DDVVILCFDMNTEKFHNMGMPDACHFDDGKSYGLVILFKCMTLICYPDPMPSSPTEKLTD 286
Query: 306 IWVMKDYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHF-CSKVYVYNYKN 364
IW+MK+YG K SW K ++ + +D+ + LH + Y+ +
Sbjct: 287 IWIMKEYGEKESWIKRCSIRL---------LPESPLAVWKDEILLLHSKMGHLIAYDLNS 337
Query: 365 STVKTLDIQGQPSILYNSSRVYFESL 390
+ V+ LD+ G P L +Y ESL
Sbjct: 338 NEVQELDLHGYPESL--RIIIYRESL 361
>UniRef100_Q6UQG1 S2 self-incompatibility locus-linked putative F-box protein S2-
A134 [Petunia integrifolia subsp. inflata]
Length = 379
Score = 86.7 bits (213), Expect = 1e-15
Identities = 95/391 (24%), Positives = 166/391 (42%), Gaps = 75/391 (19%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
D+ + I LPVKSLL KC K+ +I F+ +LHL+ T + N EL+L+
Sbjct: 7 DVVIYIFVMLPVKSLLRFKCTCKTFCHIIKSSTFI--NLHLNHTTNF-----NDELVLLK 59
Query: 99 SRLPS---------VTAIIPDTTHNFR-LNPSDNHPIM-----------IDSCDGIICFE 137
+ ++ + ++F+ ++P P + I C+G+I
Sbjct: 60 RSFETDEYKFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACVCHRLIGPCNGLIVLT 119
Query: 138 NRNDNHVDLVVWNPCTGKFKILPPLE-NIPNGKTHTLYSIGY--DRFVDNYKVVAFSCHR 194
+ + +V+NP T K++++PP IP G ++ IG+ D ++YKVV S
Sbjct: 120 DS----LTTIVFNPATLKYRLIPPCPFGIPRGFRRSISGIGFGFDSDANDYKVVRLS--- 172
Query: 195 QINKSYKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNY- 253
++ K +V ++ + WR + +G +V F I + ++N+
Sbjct: 173 EVYKEPCDKEMKVDIYDFSVDSWREL-------LGQDVPFVFWFPCAEILY----KRNFH 221
Query: 254 -----DSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVY--------T 300
D VIL D+ E + + PD D + L + C+ ++ Y T
Sbjct: 222 WFAFADVVVILCFDMNTEKFHNMGMPDACHFDDGKCYGLVILFKCMTLICYPDPKPSSPT 281
Query: 301 ETLLGIWVMKDYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHF-CSKVYV 359
E L IW+MK+YG K SW K ++ + +D+ + LH +
Sbjct: 282 EKLTDIWIMKEYGEKESWMKRCSIRL---------LPESPLAVWKDEILLLHSKMGHLMA 332
Query: 360 YNYKNSTVKTLDIQGQPSILYNSSRVYFESL 390
Y+ ++ V+ LD+ G P L +Y ESL
Sbjct: 333 YDLNSNEVQELDLHGYPESL--RIIIYRESL 361
>UniRef100_Q84UT3 F-box [Prunus mume]
Length = 428
Score = 86.3 bits (212), Expect = 1e-15
Identities = 91/352 (25%), Positives = 140/352 (38%), Gaps = 76/352 (21%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQ----TRPYHLLIRNSEL 94
D+ IL RLP KSL+ +CV KS + LI P FV HHL +S T ++L++++ L
Sbjct: 14 DMMGNILSRLPPKSLMRFRCVLKSWHDLIDKPSFVDHHLSISMDNKVTSSTYVLLKHNVL 73
Query: 95 ----LLVDSRLPSVTAIIPDTTHNFRLNPSDNHPIMID---------------------- 128
+ D + T PD+ L S N ++D
Sbjct: 74 TDPSIKDDEKAVRATLFNPDSNQRDILLSSLNLGSLVDDGLEIENHIVPPPMRGYALSLE 133
Query: 129 ---SCDGIICFENRNDNHVDLVVWNPCTGKFKILPPL---------ENIPNGKTHTLY-- 174
SCDG+IC N D+V+ NP ++++LP + Y
Sbjct: 134 ISGSCDGLICLNTFNSE--DIVLCNPALEEYRVLPKSCILLPPRVPRQFEENEDDDYYEE 191
Query: 175 -------------SIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTNFWRRIP 221
GYD +YKVV + + ++ + S+V V++L + WR +P
Sbjct: 192 DDDDEIESNPKCVGFGYDPNSKDYKVVRAA--QFVSGVFTQHPSKVEVYSLAADTWREVP 249
Query: 222 NFPSNIMGLPNGYVGKFVSGTINWAI--ENQKNYDSWVILSLDLGNESYQEISRPDFGLD 279
G N + G W ++N VILS D+ E + +I+ P+ G
Sbjct: 250 -VDIQPHGSLNPSYQMYFKGFFYWIAYWTEERN----VILSFDMSEEVFHDIALPESG-P 303
Query: 280 DPVHIFTLGVSKDCLCVLVY-----TETLLGIWVMKD--YGNKNSWTKLFAV 324
D ++ V KD L +L Y L +WV + G K WTK A+
Sbjct: 304 DAYEYTSIAVWKDSLVLLTYPVENEAPKTLDLWVFDEDLKGAKGLWTKHLAI 355
>UniRef100_Q60CY8 Putative F-box protein [Solanum demissum]
Length = 261
Score = 85.9 bits (211), Expect = 2e-15
Identities = 71/247 (28%), Positives = 117/247 (46%), Gaps = 20/247 (8%)
Query: 155 KFKILPPL-ENIPNGKTHTL-YSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTL 212
K K LP L + NG ++ L Y GYD D+YKVV C + + S C+S V +++L
Sbjct: 21 KSKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYKVVVIQCIYEDSGS---CDSVVNIYSL 77
Query: 213 GTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIE-NQKNYDSWVILSLDLGNESYQEI 271
+ WR I F N + GKFV+G + WA+ + ++ I+SLDL +E+++ +
Sbjct: 78 KADSWRTINKFQGNFLV---NSPGKFVNGKLYWALSADVDTFNMCNIISLDLADETWRRL 134
Query: 272 SRPDFGLDDPVHIFTLGVSKDCL---CVLVYTETLLGIWVMKDYGNKNSWTKLFAVPYAK 328
PD + LGV + L C+ T +W+ KD G + SWTK+F V + K
Sbjct: 135 ELPD-SYGKGSYPLALGVVESHLSVFCLNCIEGTNSDVWIRKDCGVEVSWTKIFTVDHPK 193
Query: 329 VGYHGFGFVDLHYI---SEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQGQPSILYNSSRV 385
F + + + D++ L + YN V+ +D + + ++ +
Sbjct: 194 DLGEFIFFTSIFSVPCYQTKKDEILLLLPPVIMTYNGSTRQVEVVDRFEECA----AAEI 249
Query: 386 YFESLYD 392
Y E+L D
Sbjct: 250 YAENLVD 256
>UniRef100_Q64HD2 S3 putative F-box protein SLF-S3B [Petunia hybrida]
Length = 379
Score = 82.0 bits (201), Expect = 3e-14
Identities = 92/386 (23%), Positives = 165/386 (41%), Gaps = 65/386 (16%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYH----LLIRNSEL 94
D+ + I LPVKSLL KC K+ +I F+ +LHL+ T ++ LL R+ E
Sbjct: 7 DVVIYIFVMLPVKSLLRFKCTCKTFCHIIKSSTFI--NLHLNHTTNFNDELVLLKRSFET 64
Query: 95 LLVDSRLPSVTAIIPDTTHNFR-LNPSDNHP-----------IMIDSCDGIICFENRNDN 142
+ ++ + ++F+ ++P P +I C+G+I +
Sbjct: 65 DEYNFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACVCHRLIGPCNGLIVL----TD 120
Query: 143 HVDLVVWNPCTGKFKILPPLE-NIPNGKTHTLYSI--GYDRFVDNYKVVAFSCHRQINKS 199
+ +V+NP T K++++PP IP G ++ I G+D ++YKVV S ++ K
Sbjct: 121 SLTTIVFNPATLKYRLIPPCPFGIPRGFRRSISGIGFGFDSDANDYKVVRLS---EVYKE 177
Query: 200 YKYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNY------ 253
+V ++ + WR ++G +V F I + ++N+
Sbjct: 178 PCDKEMKVDIYDFSVDSWR-------ELLGQDVPFVFWFPCAEILY----KRNFHWFAFA 226
Query: 254 DSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVY--------TETLLG 305
D VIL ++ E + + PD + L + C+ ++ Y TE
Sbjct: 227 DVVVILCFEMNTEKFHNMGMPDACHFADGKCYGLVILFKCMTLICYPDPMPSSPTEXWTD 286
Query: 306 IWVMKDYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHF-CSKVYVYNYKN 364
IW+MK+YG K SW K ++ + +D+ + LH + Y++ +
Sbjct: 287 IWIMKEYGEKESWIKRCSIRL---------LPESPLAVWKDEILLLHSKTGHLIAYDFNS 337
Query: 365 STVKTLDIQGQPSILYNSSRVYFESL 390
+ V+ LD+ G P L +Y ESL
Sbjct: 338 NEVQELDLHGYPESL--RIIIYRESL 361
>UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein) [Antirrhinum
hispanicum]
Length = 376
Score = 80.1 bits (196), Expect = 1e-13
Identities = 78/310 (25%), Positives = 132/310 (42%), Gaps = 34/310 (10%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIR-------N 91
D+ EIL VKSLL +CVSKS SLI F+ +HL QT ++++
Sbjct: 10 DVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 92 SELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPI--------MIDSCDGIICFENRNDNH 143
+ P + ++PD + + N ++ ++ C+G+IC +
Sbjct: 70 DMFSFYNINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLICLAYGD--- 126
Query: 144 VDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGY---DRFVDNYKVVAFSCHRQINKSY 200
+++ NP + K LPP N + H IGY + D YKVV +
Sbjct: 127 -CVLLSNPALREIKRLPPTP-FANPEGHCTDIIGYGFGNTCNDCYKVVLI---ESVGPED 181
Query: 201 KYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGK-FVSGTINWAIENQKNYDSWVIL 259
+ N + V+ TN W+ I + + I + + + F G +W + + + IL
Sbjct: 182 HHIN--IYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHWNANSTDIFYADFIL 239
Query: 260 SLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTE-----TLLGIWVMKDYGN 314
+ D+ E ++E++ P +L +CL ++ Y E L IWVM YG
Sbjct: 240 TFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMVRYKEWMEDPELFDIWVMNQYGV 299
Query: 315 KNSWTKLFAV 324
+ SWTK + +
Sbjct: 300 RESWTKQYVI 309
>UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum hispanicum]
Length = 376
Score = 80.1 bits (196), Expect = 1e-13
Identities = 78/310 (25%), Positives = 132/310 (42%), Gaps = 34/310 (10%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIR-------N 91
D+ EIL VKSLL +CVSKS SLI F+ +HL QT ++++
Sbjct: 10 DVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 92 SELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPI--------MIDSCDGIICFENRNDNH 143
+ P + ++PD + + N ++ ++ C+G+IC +
Sbjct: 70 DMFSFYNINSPKLEELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLICLAYGD--- 126
Query: 144 VDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGY---DRFVDNYKVVAFSCHRQINKSY 200
+++ NP + K LPP N + H IGY + D YKVV +
Sbjct: 127 -CVLLSNPALREIKRLPPTP-FANPEGHCTDIIGYGFGNTCNDCYKVVLI---ESVGPED 181
Query: 201 KYCNSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGK-FVSGTINWAIENQKNYDSWVIL 259
+ N + V+ TN W+ I + + I + + + F G +W + + + IL
Sbjct: 182 HHIN--IYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHWNANSTDIFYADFIL 239
Query: 260 SLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTE-----TLLGIWVMKDYGN 314
+ D+ E ++E++ P +L +CL ++ Y E L IWVM YG
Sbjct: 240 TFDIITEVFKEMAYPHCLAQFSNSFLSLMSLNECLAMVRYKEWMEEPELFDIWVMNQYGV 299
Query: 315 KNSWTKLFAV 324
+ SWTK + +
Sbjct: 300 RESWTKQYVI 309
>UniRef100_Q9T0J4 Hypothetical protein AT4g38870 [Arabidopsis thaliana]
Length = 426
Score = 79.7 bits (195), Expect = 1e-13
Identities = 88/315 (27%), Positives = 141/315 (43%), Gaps = 46/315 (14%)
Query: 35 LLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLL------ 88
LL +DL +EIL +L +K L+ CVSK S+I DP F+K L+ S RP L+
Sbjct: 52 LLPVDLIMEILKKLSLKPLIRFLCVSKLWASIIRDPYFMKLFLNESLKRPKSLVFVFRAQ 111
Query: 89 ----IRNSELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPIMID-SCDGIICFENRNDNH 143
I +S L + S ++ ++ + + + I S G+IC+ +
Sbjct: 112 SLGSIFSSVHLKSTREISSSSSSSSASSITYHVTCYTQQRMTISPSVHGLICYGPPS--- 168
Query: 144 VDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYC 203
LV++NPCT + LP I G+ IGYD NYKVV + + ++ +
Sbjct: 169 -SLVIYNPCTRRSITLP---KIKAGRRAINQYIGYDPLDGNYKVVCITRGMPMLRNRRGL 224
Query: 204 NSQVRVHTLGT--NFWRRI-----PNFPSNIMGLPNG--YVGKFVSGTINWAIENQKNYD 254
+++V TLGT + WR I P+ P + NG Y F+ +N +
Sbjct: 225 AEEIQVLTLGTRDSSWRMIHDIIPPHSPVSEELCINGVLYYRAFIGTKLNES-------- 276
Query: 255 SWVILSLDLGNESYQEISRP-DFGLDDPVHIFTLGVSKDCLCVLVY---TETLLGIWVMK 310
I+S D+ +E + I P +F L + L V+ Y T ++G+W+++
Sbjct: 277 --AIMSFDVRSEKFDLIKVPCNFR-----SFSKLAKYEGKLAVIFYEKKTSGIIGLWILE 329
Query: 311 DYGNKNSWTKLFAVP 325
D N K FA+P
Sbjct: 330 DASNGEWSKKTFALP 344
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 749,569,248
Number of Sequences: 2790947
Number of extensions: 34166676
Number of successful extensions: 64847
Number of sequences better than 10.0: 358
Number of HSP's better than 10.0 without gapping: 180
Number of HSP's successfully gapped in prelim test: 178
Number of HSP's that attempted gapping in prelim test: 64307
Number of HSP's gapped (non-prelim): 448
length of query: 404
length of database: 848,049,833
effective HSP length: 130
effective length of query: 274
effective length of database: 485,226,723
effective search space: 132952122102
effective search space used: 132952122102
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC122729.3


