

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122727.10 - phase: 1 /pseudo
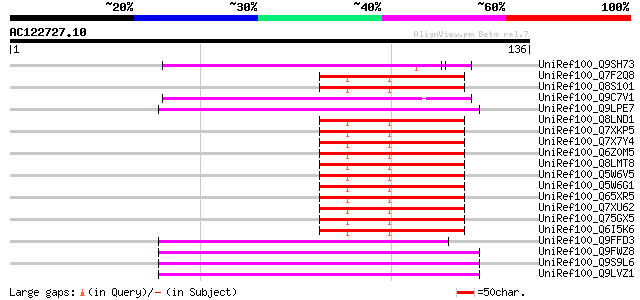
(136 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SH73 F22C12.1 [Arabidopsis thaliana] 50 8e-06
UniRef100_Q7F2Q8 P0478H03.14 protein [Oryza sativa] 48 4e-05
UniRef100_Q8S101 P0445E10.20 protein [Oryza sativa] 48 4e-05
UniRef100_Q9C7V1 Hypothetical protein F15H21.15 [Arabidopsis tha... 47 1e-04
UniRef100_Q9LPE7 T12C22.11 protein [Arabidopsis thaliana] 46 2e-04
UniRef100_Q8LND1 Putative transposable element [Oryza sativa] 46 2e-04
UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa] 46 2e-04
UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa] 46 2e-04
UniRef100_Q6Z0M5 Hypothetical protein OJ1212_C09.22 [Oryza sativa] 46 2e-04
UniRef100_Q8LMT8 Putative retroelement [Oryza sativa] 46 2e-04
UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa] 46 2e-04
UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa] 46 2e-04
UniRef100_Q65XR5 Putative polyprotein [Oryza sativa] 46 2e-04
UniRef100_Q7XU62 OSJNBb0091E11.13 protein [Oryza sativa] 46 2e-04
UniRef100_Q75GX5 Hypothetical protein OSJNBa0034D21.14 [Oryza sa... 46 2e-04
UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sa... 46 2e-04
UniRef100_Q9FFD3 Mutator-like transposase-like protein [Arabidop... 45 3e-04
UniRef100_Q9FWZ8 F11O6.3 protein [Arabidopsis thaliana] 45 5e-04
UniRef100_Q9S9L6 F26C17.4 protein [Arabidopsis thaliana] 45 5e-04
UniRef100_Q9LVZ1 Mutator-like transposase [Arabidopsis thaliana] 45 5e-04
>UniRef100_Q9SH73 F22C12.1 [Arabidopsis thaliana]
Length = 3290
Score = 50.4 bits (119), Expect = 8e-06
Identities = 22/73 (30%), Positives = 37/73 (50%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
F R F + + ++R G + + Y N I++ + + KW+D P+ +W AHD GR
Sbjct: 2326 FSRVFPSFCLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDNGR 2385
Query: 101 RWGHITTNISECF 113
R+G + N F
Sbjct: 2386 RYGIMEINTKALF 2398
Score = 48.5 bits (114), Expect = 3e-05
Identities = 23/73 (31%), Positives = 36/73 (48%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
F F++ +K V G + + Y N+I + + KW+D P+ QW QAHD GR
Sbjct: 1552 FHYVFQDDYLKNLVYEAGSTSEKEEFDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDSGR 1611
Query: 101 RWGHITTNISECF 113
R+ +T + F
Sbjct: 1612 RYRVMTIDAENLF 1624
Score = 46.6 bits (109), Expect = 1e-04
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 3/77 (3%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
F F++ ++ V+ G + + Y I + + KW+D P+ QW AHD GR
Sbjct: 724 FSGIFRDYYLEDLVKRAGSTSQKEEFDSYMKDIEKKNSEARKWLDQFPQNQWALAHDNGR 783
Query: 101 RWGHI---TTNISECFN 114
R+G + TT + E FN
Sbjct: 784 RYGIMEIETTTLFEDFN 800
Score = 46.6 bits (109), Expect = 1e-04
Identities = 24/81 (29%), Positives = 40/81 (48%), Gaps = 1/81 (1%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
FL F++ ++ V G + + Y N I++ + + KW+D P+ +W AHD G
Sbjct: 3049 FLGVFRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGL 3108
Query: 101 RWGHITTNISECFNNVLKGGP 121
R+G I + E V +G P
Sbjct: 3109 RYGIIEID-REALFAVCRGFP 3128
>UniRef100_Q7F2Q8 P0478H03.14 protein [Oryza sativa]
Length = 1171
Score = 48.1 bits (113), Expect = 4e-05
Identities = 22/41 (53%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FNNVL G
Sbjct: 516 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNNVLVG 556
>UniRef100_Q8S101 P0445E10.20 protein [Oryza sativa]
Length = 1130
Score = 48.1 bits (113), Expect = 4e-05
Identities = 22/41 (53%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FNNVL G
Sbjct: 516 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNNVLVG 556
>UniRef100_Q9C7V1 Hypothetical protein F15H21.15 [Arabidopsis thaliana]
Length = 719
Score = 46.6 bits (109), Expect = 1e-04
Identities = 24/81 (29%), Positives = 40/81 (48%), Gaps = 1/81 (1%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
FL F++ ++ V G + + Y N I++ + + KW+D P+ +W AHD G
Sbjct: 478 FLGVFRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGL 537
Query: 101 RWGHITTNISECFNNVLKGGP 121
R+G I + E V +G P
Sbjct: 538 RYGIIEID-REALFAVCRGFP 557
>UniRef100_Q9LPE7 T12C22.11 protein [Arabidopsis thaliana]
Length = 1206
Score = 46.2 bits (108), Expect = 2e-04
Identities = 23/84 (27%), Positives = 41/84 (48%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
N +KN + + V+N GY+ T + QI + + K++ D W + + +G
Sbjct: 925 NIQTKYKNKALTQLVKNAGYAFTGTKFKEFYGQIETTNQNCGKYLHDIGMANWTRHYFRG 984
Query: 100 RRWGHITTNISECFNNVLKGGPQS 123
+R+ +T+NI+E N L G S
Sbjct: 985 QRFNLMTSNIAETLNKALNKGRSS 1008
>UniRef100_Q8LND1 Putative transposable element [Oryza sativa]
Length = 1592
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 682 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 722
>UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa]
Length = 1495
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 607 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 647
>UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa]
Length = 1575
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 639 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 679
>UniRef100_Q6Z0M5 Hypothetical protein OJ1212_C09.22 [Oryza sativa]
Length = 981
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 50 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 90
>UniRef100_Q8LMT8 Putative retroelement [Oryza sativa]
Length = 1460
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 739 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 779
>UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa]
Length = 1525
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 682 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 722
>UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa]
Length = 1567
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 697 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 737
>UniRef100_Q65XR5 Putative polyprotein [Oryza sativa]
Length = 1378
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 492 KWLEDELVDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 532
>UniRef100_Q7XU62 OSJNBb0091E11.13 protein [Oryza sativa]
Length = 1107
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 686 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 726
>UniRef100_Q75GX5 Hypothetical protein OSJNBa0034D21.14 [Oryza sativa]
Length = 1038
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 542 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 582
>UniRef100_Q6I5K6 Hypothetical protein OSJNBb0088F07.13 [Oryza sativa]
Length = 1564
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/41 (51%), Positives = 32/41 (77%), Gaps = 3/41 (7%)
Query: 82 KWVDDN--PKKQWLQAHDQ-GRRWGHITTNISECFNNVLKG 119
KW++D K +W +A+D+ GRRWG++TTN++E FN+VL G
Sbjct: 754 KWLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVG 794
>UniRef100_Q9FFD3 Mutator-like transposase-like protein [Arabidopsis thaliana]
Length = 597
Score = 45.4 bits (106), Expect = 3e-04
Identities = 25/76 (32%), Positives = 38/76 (49%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
NF TFKNT++ + Y++T N++ + S DVV+W + W A+ QG
Sbjct: 317 NFRDTFKNTKLVNIFWSAVYALTPAEFETKSNEMIEISQDVVQWFELYLPHLWAVAYFQG 376
Query: 100 RRWGHITTNISECFNN 115
R+GH I+E N
Sbjct: 377 VRYGHFGLGITEVLYN 392
>UniRef100_Q9FWZ8 F11O6.3 protein [Arabidopsis thaliana]
Length = 884
Score = 44.7 bits (104), Expect = 5e-04
Identities = 25/84 (29%), Positives = 39/84 (45%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
N FKN + + V+N GY T NQI + +K++ D W + + G
Sbjct: 603 NIQARFKNRGLTQLVKNAGYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPG 662
Query: 100 RRWGHITTNISECFNNVLKGGPQS 123
+R+ +T+NI+E N L G S
Sbjct: 663 QRFNLMTSNIAETLNKALFKGRSS 686
>UniRef100_Q9S9L6 F26C17.4 protein [Arabidopsis thaliana]
Length = 872
Score = 44.7 bits (104), Expect = 5e-04
Identities = 25/84 (29%), Positives = 39/84 (45%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
N FKN + + V+N GY T NQI + +K++ D W + + G
Sbjct: 621 NIQARFKNRGLTQLVKNAGYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPG 680
Query: 100 RRWGHITTNISECFNNVLKGGPQS 123
+R+ +T+NI+E N L G S
Sbjct: 681 QRFNLMTSNIAETLNKALFKGRSS 704
>UniRef100_Q9LVZ1 Mutator-like transposase [Arabidopsis thaliana]
Length = 875
Score = 44.7 bits (104), Expect = 5e-04
Identities = 25/84 (29%), Positives = 39/84 (45%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
N FKN + + V+N GY T NQI + +K++ D W + + G
Sbjct: 621 NIQARFKNRGLTQLVKNAGYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPG 680
Query: 100 RRWGHITTNISECFNNVLKGGPQS 123
+R+ +T+NI+E N L G S
Sbjct: 681 QRFNLMTSNIAETLNKALFKGRSS 704
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.329 0.139 0.467
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 235,501,222
Number of Sequences: 2790947
Number of extensions: 8775996
Number of successful extensions: 25815
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 120
Number of HSP's that attempted gapping in prelim test: 25672
Number of HSP's gapped (non-prelim): 179
length of query: 136
length of database: 848,049,833
effective HSP length: 112
effective length of query: 24
effective length of database: 535,463,769
effective search space: 12851130456
effective search space used: 12851130456
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC122727.10


