

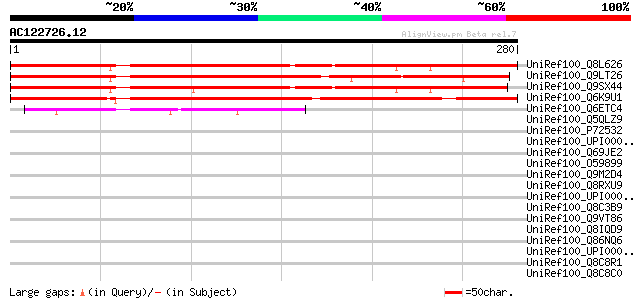
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122726.12 + phase: 0
(280 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L626 Hypothetical protein At1g50260 [Arabidopsis tha... 291 1e-77
UniRef100_Q9LT26 Arabidopsis thaliana genomic DNA, chromosome 3,... 290 3e-77
UniRef100_Q9SX44 F14I3.13 protein [Arabidopsis thaliana] 276 3e-73
UniRef100_Q6K9U1 C2 domain-containing protein-like [Oryza sativa] 270 3e-71
UniRef100_Q6ETC4 Putative CLB1 protein [Oryza sativa] 49 1e-04
UniRef100_Q5QLZ9 Putative synaptotagmin C [Oryza sativa] 43 0.010
UniRef100_P72532 Fibronectin-binding protein II [Streptococcus p... 42 0.014
UniRef100_UPI00003C1595 UPI00003C1595 UniRef100 entry 42 0.023
UniRef100_Q69JE2 Putative CLB1 protein [Oryza sativa] 42 0.023
UniRef100_O59899 Putative Cel1p [Cryptococcus neoformans] 42 0.023
UniRef100_Q9M2D4 Putative anthranilate phosphoribosyltransferase... 40 0.052
UniRef100_Q8RXU9 Putative phosphoribosylanthranilate transferase... 40 0.052
UniRef100_UPI00001CECA6 UPI00001CECA6 UniRef100 entry 40 0.067
UniRef100_Q8C3B9 Mus musculus 15 days embryo head cDNA, RIKEN fu... 40 0.067
UniRef100_Q9VT86 CG6611-PA, isoform A [Drosophila melanogaster] 40 0.067
UniRef100_Q8IQD9 CG6611-PC, isoform C [Drosophila melanogaster] 40 0.067
UniRef100_Q86NQ6 RE01075p [Drosophila melanogaster] 40 0.067
UniRef100_UPI000043027E UPI000043027E UniRef100 entry 40 0.088
UniRef100_Q8C8R1 Mus musculus adult retina cDNA, RIKEN full-leng... 40 0.088
UniRef100_Q8C8C0 Mus musculus 10 days neonate cerebellum cDNA, R... 40 0.088
>UniRef100_Q8L626 Hypothetical protein At1g50260 [Arabidopsis thaliana]
Length = 675
Score = 291 bits (745), Expect = 1e-77
Identities = 171/294 (58%), Positives = 204/294 (69%), Gaps = 24/294 (8%)
Query: 1 MQEGNMDSVGELSVTLVDARKLPY-FFGKTDPYVILSLGDQTIRSKKNSQTTVIG--WRP 57
MQEGN D VGELSVTLVDA+KL Y FFGKTDPY IL LGDQ IRSK+NSQTTVIG +P
Sbjct: 392 MQEGNKDFVGELSVTLVDAQKLRYMFFGKTDPYAILRLGDQVIRSKRNSQTTVIGAPGQP 451
Query: 58 -WNANLESAPQDFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRIV 116
WN QDF LVSNP++Q L I+V D LGFAD+ IGTGEVDL LQDTVPTDRIV
Sbjct: 452 IWN-------QDFQFLVSNPREQVLQIEVNDRLGFADMAIGTGEVDLRFLQDTVPTDRIV 504
Query: 117 ALQGGWGFLRKGLSGEILLRLTYKAYVEDEEDDKTEEDSIDIDVSDDELSDTEEANVTDK 176
L+GGW KG +GEILLRLTYK+YVE+EEDDKT + ID SDDE+SD+EE +
Sbjct: 505 VLRGGWSLFGKGSAGEILLRLTYKSYVEEEEDDKTNVKA--IDTSDDEMSDSEELGSFVR 562
Query: 177 TGVRESAYPTDKESFMDVLAAIIVSEEFQGIVASET--GFTKGLDN-GSNTGSKASKS-- 231
G + S+ D+ESFM+VL+A+IVSEEFQGIV+SE G G D+ G+ SK S
Sbjct: 563 KG-KLSSDDIDQESFMNVLSALIVSEEFQGIVSSEARDGIIDGGDSLGAPVPSKPDTSKG 621
Query: 232 -----PVANAESIPPSADNSEGSSGGSALFWLAVITSIAVLIAVNISGSSIFNP 280
V+N + + ++ G GG AL W +IT I VL+A+N+ GSS FNP
Sbjct: 622 SERNADVSNLDLLVANSGRGAGGDGGLALLWFGIITGILVLVAINMEGSSFFNP 675
>UniRef100_Q9LT26 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MPN9
[Arabidopsis thaliana]
Length = 683
Score = 290 bits (742), Expect = 3e-77
Identities = 169/298 (56%), Positives = 206/298 (68%), Gaps = 34/298 (11%)
Query: 1 MQEGNMDSVGELSVTLVDARKLPYFF-GKTDPYVILSLGDQTIRSKKNSQTTVIG--WRP 57
MQEGN D VGELSVTLV+A+KLPY F G+TDPYVIL +GDQ IRSKKNSQTTVIG +P
Sbjct: 377 MQEGNKDFVGELSVTLVNAQKLPYMFSGRTDPYVILRIGDQVIRSKKNSQTTVIGAPGQP 436
Query: 58 -WNANLESAPQDFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRIV 116
WN QDF LVSNP++Q L I+V D LGFAD+ IG GEVDL SL DTVPTDR V
Sbjct: 437 IWN-------QDFQFLVSNPREQVLQIEVNDCLGFADMAIGIGEVDLESLPDTVPTDRFV 489
Query: 117 ALQGGWGFLRKGLSGEILLRLTYKAYVEDEEDDKTEEDSIDIDVSDDELSDTEEANVTDK 176
+L+GGW KG +GEILLRLTYKAYVEDEEDDK +I D SDDE+SD+EE +
Sbjct: 490 SLRGGWSLFGKGSTGEILLRLTYKAYVEDEEDDKRNAKAIYADASDDEMSDSEEPS---- 545
Query: 177 TGVRESAYPTD---KESFMDVLAAIIVSEEFQGIVASETGFTKGLDNGSNTGSKASKSPV 233
+ V+ P+D +ESFM+VL+A+I+SEEFQGIV+SETG K +D+G ++ S
Sbjct: 546 SFVQNDKIPSDDIGQESFMNVLSALILSEEFQGIVSSETGNNK-VDDGESSVSPVPSMSG 604
Query: 234 ANAESIPPSADNSEGS---------------SGGSALFWLAVITSIAVLIAVNISGSS 276
A++ES P A N + S +GG AL W VITS+ VL+A+N+ GSS
Sbjct: 605 ADSESRPKDAGNGDVSDLEVKNAKSDRGSINNGGLALLWFGVITSVLVLVAINMGGSS 662
>UniRef100_Q9SX44 F14I3.13 protein [Arabidopsis thaliana]
Length = 737
Score = 276 bits (707), Expect = 3e-73
Identities = 167/293 (56%), Positives = 200/293 (67%), Gaps = 28/293 (9%)
Query: 1 MQEGNMDSVGELSVTLVDARKLPY-FFGKTDPYVILSLGDQTIRSKKNSQTTVIG--WRP 57
MQEGN D VGELSVTLVDA+KL Y FFGKTDPY IL LGDQ IRSK+NSQTTVIG +P
Sbjct: 380 MQEGNKDFVGELSVTLVDAQKLRYMFFGKTDPYAILRLGDQVIRSKRNSQTTVIGAPGQP 439
Query: 58 -WNANLESAPQDFHMLVSNPKKQKLSIQVKDALGFADLTIGTGE----VDLGSLQDTVPT 112
WN QDF LVSNP++Q L I+V D LGFAD+ IGTGE VDL LQDTVPT
Sbjct: 440 IWN-------QDFQFLVSNPREQVLQIEVNDRLGFADMAIGTGELYVQVDLRFLQDTVPT 492
Query: 113 DRIVALQGGWGFLRKGLSGEILLRLTYKAYVEDEEDDKTEEDSIDIDVSDDELSDTEEAN 172
DRIV L+GGW KG +GEILLRLTYK+YVE+EEDDKT + ID SDDE+SD+EE
Sbjct: 493 DRIVVLRGGWSLFGKGSAGEILLRLTYKSYVEEEEDDKTNVKA--IDTSDDEMSDSEELG 550
Query: 173 VTDKTGVRESAYPTDKESFMDVLAAIIVSEEFQGIVASET--GFTKGLDN-GSNTGSKAS 229
+ G + S+ D+ESFM+VL+A+IVSEEFQGIV+SE G G D+ G+ SK
Sbjct: 551 SFVRKG-KLSSDDIDQESFMNVLSALIVSEEFQGIVSSEARDGIIDGGDSLGAPVPSKPD 609
Query: 230 KS-------PVANAESIPPSADNSEGSSGGSALFWLAVITSIAVLIAVNISGS 275
S V+N + + ++ G GG AL W +IT I VL+A+N+ GS
Sbjct: 610 TSKGSERNADVSNLDLLVANSGRGAGGDGGLALLWFGIITGILVLVAINMEGS 662
>UniRef100_Q6K9U1 C2 domain-containing protein-like [Oryza sativa]
Length = 658
Score = 270 bits (690), Expect = 3e-71
Identities = 153/286 (53%), Positives = 197/286 (68%), Gaps = 25/286 (8%)
Query: 1 MQEGNMDSVGELSVTLVDARKLPY-FFGKTDPYVILSLGDQTIRSKKNSQTTVIGWRP-- 57
+Q+GN D VGELSVTLVDARKL + FGKTDPYV++ LGDQ I+SKKNSQTTVIG +P
Sbjct: 392 IQDGNKDFVGELSVTLVDARKLSFVLFGKTDPYVVMILGDQEIKSKKNSQTTVIG-QPGE 450
Query: 58 --WNANLESAPQDFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRI 115
WN QDFHMLV+NP+KQKL IQVKD++G D+TIGTGEV+LGSL+DTVPTD+I
Sbjct: 451 PIWN-------QDFHMLVANPRKQKLCIQVKDSVGLTDVTIGTGEVELGSLKDTVPTDKI 503
Query: 116 VALQGGWGFLRKGLSGEILLRLTYKAYVEDEEDDKTEEDSIDIDVSDDELSDTEEANVTD 175
V L GGWG L K GE+LLRLTYKAYVEDEED+ + + VSD+++ D V D
Sbjct: 504 VTLYGGWGLLGKRSKGEVLLRLTYKAYVEDEEDEGVKNEFAAGYVSDEDVLD----YVQD 559
Query: 176 KTGVRESAYPTDKESFMDVLAAIIVSEEFQGIV-ASETGFTKGLDNGSNTGSKASKSPVA 234
T + ++E+FMD+LAA++VSEEFQGIV +SE G + + + + + + A
Sbjct: 560 STSKQSDMDGKERETFMDLLAALLVSEEFQGIVSSSEPGSLRDSEQAAKSRDGENAAAAA 619
Query: 235 NAESIPPSADNSEGSSGGSALFWLAVITSIAVLIAVNISGSSIFNP 280
+ ++ SS +AL WLA ITS+ VL++ N+ GS FNP
Sbjct: 620 DTGTV-------SNSSTDTALVWLAAITSVMVLVSSNLGGSGYFNP 658
>UniRef100_Q6ETC4 Putative CLB1 protein [Oryza sativa]
Length = 538
Score = 48.9 bits (115), Expect = 1e-04
Identities = 48/162 (29%), Positives = 72/162 (43%), Gaps = 15/162 (9%)
Query: 9 VGELSVTLVDARKLPY--FFGKTDPYVILSLGDQTIRSKKNSQTTVIGWRPWNANLESAP 66
VG L V +V A KL F GK+DPYV L L ++ + SKK S WN
Sbjct: 259 VGILHVNIVRAVKLTKKDFLGKSDPYVKLKLTEEKLPSKKTSVKRSNLNPEWN------- 311
Query: 67 QDFHMLVSNPKKQKLSIQVKD--ALGFADLTIGTGEVDLGSLQDTVPTDRIVALQGGWGF 124
+DF ++V +P+ Q L + V D +G D IG + L L + L
Sbjct: 312 EDFKLVVKDPESQALELTVYDWEQVGKHD-KIGMSVIPLKELIPDEAKSLTLDLHKTMDA 370
Query: 125 ---LRKGLSGEILLRLTYKAYVEDEEDDKTEEDSIDIDVSDD 163
G++ + +TYK + E + D T ++S I+ + D
Sbjct: 371 NDPANDKFRGQLTVDVTYKPFKEGDSDVDTSDESGTIEKAPD 412
>UniRef100_Q5QLZ9 Putative synaptotagmin C [Oryza sativa]
Length = 514
Score = 42.7 bits (99), Expect = 0.010
Identities = 41/140 (29%), Positives = 61/140 (43%), Gaps = 16/140 (11%)
Query: 9 VGELSVTLVDARKLPYF--FGKTDPYVILSLGDQTIRSKKNSQTTVIGWRPWNANLESAP 66
VG L V ++ A+ L GK DPYV L + + SKK + WN
Sbjct: 259 VGILLVKVLRAQNLREKGPLGKRDPYVKLKMSGSKLPSKKTAVKHSNLNPEWN------- 311
Query: 67 QDFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQD--TVPTDRIVALQGGWGF 124
Q+F ++ +P+ Q+L I D +G ++ L L V TD ++ G
Sbjct: 312 QEFKFVIRDPETQELDINFGK-----DEKLGMCKISLKKLTPGTEVITDNLIKTMEPNGI 366
Query: 125 LRKGLSGEILLRLTYKAYVE 144
+ +GEI L LTYK + E
Sbjct: 367 QNEKSAGEITLELTYKPFKE 386
>UniRef100_P72532 Fibronectin-binding protein II [Streptococcus pyogenes]
Length = 1025
Score = 42.4 bits (98), Expect = 0.014
Identities = 31/90 (34%), Positives = 48/90 (52%), Gaps = 4/90 (4%)
Query: 140 KAYVEDEEDDKTEEDSIDIDVSDDELSDTEEANVTDKTGVRESAYPTDK-ESFMDVLAAI 198
K V+ EED K ++ +I+ DE D +++ G +E DK E+F D LA++
Sbjct: 564 KRQVKSEEDKKKAKEKENIEKKRDEKFDNYSKQMSE--GGKEFFNDVDKAENFKDTLASV 621
Query: 199 IVSEEFQGIVASETGFTKGLDNGSNTGSKA 228
V+E F V+ E+G K GSN+GS +
Sbjct: 622 TVTETFGNNVSVESGSWK-TSLGSNSGSSS 650
>UniRef100_UPI00003C1595 UPI00003C1595 UniRef100 entry
Length = 736
Score = 41.6 bits (96), Expect = 0.023
Identities = 36/122 (29%), Positives = 54/122 (43%), Gaps = 18/122 (14%)
Query: 145 DEEDDKTEEDSIDIDVSDDELSDTEEANVTDKTGVRESAYPTDKESFMDVLAAIIVSEEF 204
D DD+ +D D D +DE + EE T +G ++ + +A++ S
Sbjct: 375 DGNDDEDCDDG-DEDEDEDENENDEEVTATSTSGSSSNSNAASGSN-----SAVVSSSNS 428
Query: 205 QGIV-ASETGFTKGLDNGSNTGSKASKS-----------PVANAESIPPSADNSEGSSGG 252
G AS TG G + GS GSK++ S +NA + P SA +S SSG
Sbjct: 429 SGASSASNTGSNGGSNKGSTGGSKSASSSGADVSSNSSGSASNAGAGPVSASSSSASSGS 488
Query: 253 SA 254
S+
Sbjct: 489 SS 490
>UniRef100_Q69JE2 Putative CLB1 protein [Oryza sativa]
Length = 539
Score = 41.6 bits (96), Expect = 0.023
Identities = 41/160 (25%), Positives = 73/160 (45%), Gaps = 22/160 (13%)
Query: 9 VGELSVTLVDARKLPY--FFGKTDPYVILSLGDQTIRSKKNSQTTVIGWRPWNANLESAP 66
VG L V ++ A+ L GK+DPYV L + D + SKK + WN
Sbjct: 259 VGILLVKVLRAQNLRKKDLLGKSDPYVKLKMSDDKLPSKKTTVKRSNLNPEWN------- 311
Query: 67 QDFHMLVSNPKKQKLSIQVKD--------ALGFADLTIGTGEVDLGSLQDTVPTDRIVAL 118
+DF +V++P+ Q L I V D +G ++ + +L + + V T ++
Sbjct: 312 EDFKFVVTDPETQALEINVFDWEQVGKHEKMGMNNILL----KELPADETKVMTVNLLKT 367
Query: 119 QGGWGFLRKGLSGEILLRLTYKAYVEDEEDDKTEEDSIDI 158
+ G++ L +TYK + ++E+ +K D+ D+
Sbjct: 368 MDPNDVQNEKSRGQLTLEVTYKPF-KEEDMEKEGIDNADV 406
>UniRef100_O59899 Putative Cel1p [Cryptococcus neoformans]
Length = 365
Score = 41.6 bits (96), Expect = 0.023
Identities = 27/84 (32%), Positives = 46/84 (54%), Gaps = 5/84 (5%)
Query: 193 DVLAAIIVSEEFQGIVASETGFTKGLDNGSNTGSKASKSPVANAESIPPSADNSEGSSGG 252
D ++++ EF GI G+T G+ NG++ +K+ S +++ S+ PS D S+ +S G
Sbjct: 252 DEWQGLLINWEFPGIAVYPGGYTTGVVNGASAAAKSGMSSSSSSSSVAPS-DASDSTSSG 310
Query: 253 SALFWLAVITSIAVLIAVNISGSS 276
+ LAV S + N+S SS
Sbjct: 311 A----LAVSISATSSPSSNVSVSS 330
>UniRef100_Q9M2D4 Putative anthranilate phosphoribosyltransferase [Arabidopsis
thaliana]
Length = 972
Score = 40.4 bits (93), Expect = 0.052
Identities = 39/150 (26%), Positives = 64/150 (42%), Gaps = 18/150 (12%)
Query: 12 LSVTLVDARKLPYF--FGKTDPYVILSLGDQTIRSK--KNSQTTVIGWRPWNANLESAPQ 67
L + +V AR LP G DPY+ + LG+ T ++K + +Q V WN +
Sbjct: 251 LFIKIVKARNLPSMDLTGSLDPYIEVKLGNYTGKTKHFEKNQNPV-----WN-------E 298
Query: 68 DFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRIVALQGGWGFLRK 127
F SN + L + V D D +G DL + V D +A + W +
Sbjct: 299 VFAFSKSNQQSNVLEVIVMDKDMVKDDFVGLIRFDLNQIPTRVAPDSPLAPE--WYRVNN 356
Query: 128 GLSGEILLRLTYKAYVEDEEDDKTEEDSID 157
GEI+L + + ++ D T D+++
Sbjct: 357 EKGGEIMLAVWFGTQADEAFSDATYSDALN 386
>UniRef100_Q8RXU9 Putative phosphoribosylanthranilate transferase [Arabidopsis
thaliana]
Length = 1006
Score = 40.4 bits (93), Expect = 0.052
Identities = 52/185 (28%), Positives = 75/185 (40%), Gaps = 31/185 (16%)
Query: 4 GNMDSVGELS---VTLVDARKLPY--FFGKTDPYVILSLGD---QTIRSKKNSQTTVIGW 55
G D V E+ V +V AR LP G DPYV++ +G+ T KN+
Sbjct: 259 GTYDLVEEMKFLYVRVVKARDLPNKDLTGSLDPYVVVKIGNFKGVTTHFNKNTDPE---- 314
Query: 56 RPWNANLESAPQDFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRI 115
WN Q F N + L + VKD D +G + DL +Q VP D
Sbjct: 315 --WN-------QVFAFAKDNLQSNFLEVMVKDKDILLDDFVGIVKFDLREVQSRVPPDSP 365
Query: 116 VALQGGWGFLR----KGLSGEILLRLTYKAYVEDEEDDKTEEDSIDIDVSDDELSDTEEA 171
+A Q W L + + EI+L + ++ D T DS+ D + S+ A
Sbjct: 366 LAPQ--WYRLENKRGEKKNYEIMLAVWSGTQADEAFGDATFSDSL----VDSDSSNIISA 419
Query: 172 NVTDK 176
N+ K
Sbjct: 420 NLRSK 424
>UniRef100_UPI00001CECA6 UPI00001CECA6 UniRef100 entry
Length = 546
Score = 40.0 bits (92), Expect = 0.067
Identities = 47/154 (30%), Positives = 63/154 (40%), Gaps = 24/154 (15%)
Query: 10 GELSVTLVDARKLPYFF--GKTDPYVILSLGDQTIRSKKNSQTTVIGWRPWNANLESAPQ 67
G +S+TL++ R L G +DPYV LG Q +SK +T WR
Sbjct: 203 GIVSITLIEGRDLKAMDSNGLSDPYVKFRLGHQKYKSKIMPKTLNPQWR--------EQF 254
Query: 68 DFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRIVALQGGWGFLRK 127
DFH+ ++ KDA G D IG +VDL SL + L+ G G L
Sbjct: 255 DFHLYEERGGVMDITAWDKDA-GKRDDFIGRCQVDLSSLSREQTHKLELQLEEGEGHL-- 311
Query: 128 GLSGEILLRLTYKA-------YVEDEEDDKTEED 154
+L+ LT A V ED K E+
Sbjct: 312 ----VLLVTLTASATVSISDLSVNSMEDHKEREE 341
>UniRef100_Q8C3B9 Mus musculus 15 days embryo head cDNA, RIKEN full-length enriched
library, clone:D930024E11 product:hypothetical C2 domain
containing protein, full insert sequence [Mus musculus]
Length = 826
Score = 40.0 bits (92), Expect = 0.067
Identities = 44/155 (28%), Positives = 66/155 (42%), Gaps = 30/155 (19%)
Query: 10 GELSVTLVDARKLPY------FFGKTDPYVILSLGDQTIRSK---KNSQTTVIGWRPWNA 60
G + V L++A+KL GK+DPY +S+G Q RS+ KN T WN
Sbjct: 310 GVIRVHLLEAKKLAQKDNFLGLGGKSDPYAKVSIGLQHCRSRTIYKNLNPT------WN- 362
Query: 61 NLESAPQDFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRIVALQG 120
+ F +V Q L + + D D +G+ ++ LG + D
Sbjct: 363 ------EVFEFMVYEVPGQDLEVDLYDEDTDKDDFLGSLQICLGDVMKNRVVDE------ 410
Query: 121 GWGFLRKGLSGEILLRLTYKAYVEDEEDDKTEEDS 155
W L SG + LRL + + + D+E TE DS
Sbjct: 411 -WFALNDTTSGRLHLRLEWLSLLTDQE-ALTENDS 443
>UniRef100_Q9VT86 CG6611-PA, isoform A [Drosophila melanogaster]
Length = 581
Score = 40.0 bits (92), Expect = 0.067
Identities = 31/116 (26%), Positives = 56/116 (47%), Gaps = 9/116 (7%)
Query: 144 EDEEDDKTEEDSIDIDVSDDELSDTEEANVTDKTGV---RESAYPTDKESFMDVLAAIIV 200
+DEE+D ++D +D + DD L D E+ + D+T ++SA +D +A
Sbjct: 105 DDEEEDDDDDDYLD-RLFDDILGDDEDEDDDDETPAATAQQSAVAAAPAQTLDEVAPAAA 163
Query: 201 SEEFQGI-----VASETGFTKGLDNGSNTGSKASKSPVANAESIPPSADNSEGSSG 251
S GI + +E+G + N ++ + VA A++ S+ N+EG +G
Sbjct: 164 SSVSAGISDGVDLPAESGNAAEVLAAGNAADESVSAAVAPAQNAAASSSNNEGIAG 219
>UniRef100_Q8IQD9 CG6611-PC, isoform C [Drosophila melanogaster]
Length = 283
Score = 40.0 bits (92), Expect = 0.067
Identities = 31/116 (26%), Positives = 56/116 (47%), Gaps = 9/116 (7%)
Query: 144 EDEEDDKTEEDSIDIDVSDDELSDTEEANVTDKTGV---RESAYPTDKESFMDVLAAIIV 200
+DEE+D ++D +D + DD L D E+ + D+T ++SA +D +A
Sbjct: 105 DDEEEDDDDDDYLD-RLFDDILGDDEDEDDDDETPAATAQQSAVAAAPAQTLDEVAPAAA 163
Query: 201 SEEFQGI-----VASETGFTKGLDNGSNTGSKASKSPVANAESIPPSADNSEGSSG 251
S GI + +E+G + N ++ + VA A++ S+ N+EG +G
Sbjct: 164 SSVSAGISDGVDLPAESGNAAEVLAAGNAADESVSAAVAPAQNAAASSSNNEGIAG 219
>UniRef100_Q86NQ6 RE01075p [Drosophila melanogaster]
Length = 280
Score = 40.0 bits (92), Expect = 0.067
Identities = 31/116 (26%), Positives = 56/116 (47%), Gaps = 9/116 (7%)
Query: 144 EDEEDDKTEEDSIDIDVSDDELSDTEEANVTDKTGV---RESAYPTDKESFMDVLAAIIV 200
+DEE+D ++D +D + DD L D E+ + D+T ++SA +D +A
Sbjct: 105 DDEEEDDDDDDYLD-RLFDDILGDDEDEDDDDETPAATAQQSAVAAAPAQTLDEVAPAAA 163
Query: 201 SEEFQGI-----VASETGFTKGLDNGSNTGSKASKSPVANAESIPPSADNSEGSSG 251
S GI + +E+G + N ++ + VA A++ S+ N+EG +G
Sbjct: 164 SSVSAGISDGVDLPAESGNAAEVLAAGNAADESVSAAVAPAQNAAASSSNNEGIAG 219
>UniRef100_UPI000043027E UPI000043027E UniRef100 entry
Length = 365
Score = 39.7 bits (91), Expect = 0.088
Identities = 26/84 (30%), Positives = 45/84 (52%), Gaps = 5/84 (5%)
Query: 193 DVLAAIIVSEEFQGIVASETGFTKGLDNGSNTGSKASKSPVANAESIPPSADNSEGSSGG 252
D ++++ EF GI G+T G+ NG++ +K+ +++ S+ PS D S+ +S G
Sbjct: 252 DEWQGLLINWEFPGIAVYPGGYTTGVVNGASAAAKSGMPSSSSSSSVAPS-DASDSTSSG 310
Query: 253 SALFWLAVITSIAVLIAVNISGSS 276
+ LAV S + N+S SS
Sbjct: 311 A----LAVSISATSSPSSNVSVSS 330
>UniRef100_Q8C8R1 Mus musculus adult retina cDNA, RIKEN full-length enriched library,
clone:A930030A18 product:membrane bound C2 domain
containing protein, full insert sequence [Mus musculus]
Length = 893
Score = 39.7 bits (91), Expect = 0.088
Identities = 29/121 (23%), Positives = 54/121 (43%), Gaps = 18/121 (14%)
Query: 27 GKTDPYVILSLGDQTIRSKKNSQTTVIGWRPWNANLESAPQDFHMLVSNPKKQKLSIQVK 86
GK+DPY ++ +G QT S+ + W + + ++V +Q++ ++V
Sbjct: 344 GKSDPYALVRVGTQTFCSRVIDEELNPHWG----------ETYEVIVHEVPRQEIEVEVF 393
Query: 87 DALGFADLTIGTGEVDLGSLQDTVPTDRIVALQGGWGFLRKGLSGEILLRLTYKAYVEDE 146
D D +G ++D+G + D LQGG G++ LRL + + + D
Sbjct: 394 DKDPDKDDFLGRMKLDVGKVLQAGVLDNWYPLQGG--------QGQVHLRLEWLSLLPDA 445
Query: 147 E 147
E
Sbjct: 446 E 446
>UniRef100_Q8C8C0 Mus musculus 10 days neonate cerebellum cDNA, RIKEN full-length
enriched library, clone:B930088K05 product:hypothetical
C2 domain containing protein, full insert sequence [Mus
musculus]
Length = 694
Score = 39.7 bits (91), Expect = 0.088
Identities = 47/154 (30%), Positives = 63/154 (40%), Gaps = 24/154 (15%)
Query: 10 GELSVTLVDARKLPYFF--GKTDPYVILSLGDQTIRSKKNSQTTVIGWRPWNANLESAPQ 67
G +S+TL++ R L G +DPYV LG Q +SK +T WR
Sbjct: 165 GIVSITLIEGRDLKAMDSNGLSDPYVKFRLGHQKYKSKIMPKTLNPQWR--------EQF 216
Query: 68 DFHMLVSNPKKQKLSIQVKDALGFADLTIGTGEVDLGSLQDTVPTDRIVALQGGWGFLRK 127
DFH+ ++ KDA G D IG +VDL SL + L+ G G L
Sbjct: 217 DFHLYEERGGIMDITAWDKDA-GKRDDFIGRCQVDLSSLSREQTHKLELHLEEGEGHL-- 273
Query: 128 GLSGEILLRLTYKA-------YVEDEEDDKTEED 154
+L+ LT A V ED K E+
Sbjct: 274 ----VLLVTLTASATVCISDLSVNSMEDQKEREE 303
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.131 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 457,705,672
Number of Sequences: 2790947
Number of extensions: 19385796
Number of successful extensions: 97553
Number of sequences better than 10.0: 308
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 257
Number of HSP's that attempted gapping in prelim test: 95760
Number of HSP's gapped (non-prelim): 1133
length of query: 280
length of database: 848,049,833
effective HSP length: 126
effective length of query: 154
effective length of database: 496,390,511
effective search space: 76444138694
effective search space used: 76444138694
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC122726.12


