

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122725.8 - phase: 0 /pseudo
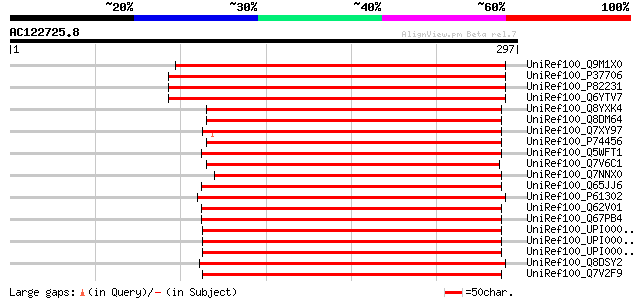
(297 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M1X0 Ribosome recycling factor, chloroplast precurso... 305 1e-81
UniRef100_P37706 Ribosome recycling factor, chloroplast precurso... 303 3e-81
UniRef100_P82231 Ribosome recycling factor, chloroplast precurso... 300 3e-80
UniRef100_Q6YTV7 Putative Ribosome recycling factor, chloroplast... 278 1e-73
UniRef100_Q8YXK4 Ribosome recycling factor [Anabaena sp.] 205 1e-51
UniRef100_Q8DM64 Ribosome recycling factor [Synechococcus elonga... 194 2e-48
UniRef100_Q7XY97 Ribosome recycling factor [Griffithsia japonica] 194 2e-48
UniRef100_P74456 Ribosome recycling factor [Synechocystis sp.] 192 1e-47
UniRef100_Q5WFT1 Ribosome recycling factor [Bacillus clausii] 191 3e-47
UniRef100_Q7V6C1 Ribosome recycling factor [Prochlorococcus mari... 188 2e-46
UniRef100_Q7NNX0 Ribosome recycling factor [Gloeobacter violaceus] 188 2e-46
UniRef100_Q65JJ6 Frr [Bacillus licheniformis] 187 4e-46
UniRef100_P61302 Ribosome recycling factor [Bdellovibrio bacteri... 185 1e-45
UniRef100_Q62V01 Ribosome recycling factor [Bacillus licheniformis] 184 2e-45
UniRef100_Q67PB4 Ribosome recycling factor [Symbiobacterium ther... 182 7e-45
UniRef100_UPI0000346352 UPI0000346352 UniRef100 entry 182 9e-45
UniRef100_UPI00002771CB UPI00002771CB UniRef100 entry 181 2e-44
UniRef100_UPI00002BEA74 UPI00002BEA74 UniRef100 entry 180 5e-44
UniRef100_Q8DSY2 Ribosome recycling factor [Streptococcus mutans] 180 5e-44
UniRef100_Q7V2F9 Ribosome recycling factor [Prochlorococcus mari... 180 5e-44
>UniRef100_Q9M1X0 Ribosome recycling factor, chloroplast precursor [Arabidopsis
thaliana]
Length = 275
Score = 305 bits (781), Expect = 1e-81
Identities = 152/193 (78%), Positives = 181/193 (93%)
Query: 98 VRAATIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPV 157
+R+ATIEEIEAEK++IE DVKS+ME+T++ ++T+F+SIRTGR+ +MLDKIEVEYYGSPV
Sbjct: 80 IRSATIEEIEAEKSAIETDVKSKMEKTIETLRTSFNSIRTGRSNAAMLDKIEVEYYGSPV 139
Query: 158 SLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDR 217
SLK+IAQISTPD SSLL+ PYDKSSLKAIEKAIVNSD+G+TPNNDG+VIRLS+P LTSDR
Sbjct: 140 SLKSIAQISTPDGSSLLLQPYDKSSLKAIEKAIVNSDLGVTPNNDGDVIRLSLPPLTSDR 199
Query: 218 RKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIK 277
RKEL+K+V+KQ EEGKVALRNIRRDALK+Y+KLEK+KKLSEDNVKDLSSDLQKL D Y+K
Sbjct: 200 RKELSKVVAKQSEEGKVALRNIRRDALKSYDKLEKEKKLSEDNVKDLSSDLQKLIDVYMK 259
Query: 278 KVDALYKQKEKVL 290
K++ LYKQKEK L
Sbjct: 260 KIEELYKQKEKEL 272
>UniRef100_P37706 Ribosome recycling factor, chloroplast precursor [Daucus carota]
Length = 227
Score = 303 bits (777), Expect = 3e-81
Identities = 151/197 (76%), Positives = 181/197 (91%)
Query: 94 RKGFVRAATIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYY 153
R G ++ AT+EEIEAEK+ IE VK RME+T++NVK +F+SIRT R+ P MLDKI+VEYY
Sbjct: 28 RTGTLKCATMEEIEAEKSLIEKSVKERMEKTIENVKASFNSIRTXRSNPDMLDKIKVEYY 87
Query: 154 GSPVSLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQL 213
G+P SLK+IAQISTPD+SSLLV+PYDKSSLK IEKAIVNSD+G+TPNNDG+VIRLSIPQL
Sbjct: 88 GTPTSLKSIAQISTPDSSSLLVNPYDKSSLKDIEKAIVNSDLGITPNNDGDVIRLSIPQL 147
Query: 214 TSDRRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTD 273
T+DRRKEL+KIV+KQ EEGKVALRNIRRDA+K+Y+KLEK+KKLSEDNVKDLSSDLQK+ D
Sbjct: 148 TADRRKELSKIVAKQAEEGKVALRNIRRDAIKSYDKLEKEKKLSEDNVKDLSSDLQKVID 207
Query: 274 DYIKKVDALYKQKEKVL 290
+YIKKVD+++KQKEK L
Sbjct: 208 EYIKKVDSIFKQKEKEL 224
>UniRef100_P82231 Ribosome recycling factor, chloroplast precursor [Spinacia
oleracea]
Length = 271
Score = 300 bits (768), Expect = 3e-80
Identities = 147/197 (74%), Positives = 180/197 (90%)
Query: 94 RKGFVRAATIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYY 153
R G R AT+EE+EAEK+ IE + K RME+T++ +++NF+S+RT RA+P+MLD+IEVEYY
Sbjct: 72 RAGTFRCATMEEVEAEKSLIETNTKQRMEKTIETIRSNFNSVRTNRASPTMLDRIEVEYY 131
Query: 154 GSPVSLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQL 213
G+PVSLK+IAQISTPDASSLL+SPYDKSSLKAIEKAIV S +G++PNNDGEVIRLS+P L
Sbjct: 132 GTPVSLKSIAQISTPDASSLLISPYDKSSLKAIEKAIVTSQLGVSPNNDGEVIRLSLPPL 191
Query: 214 TSDRRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTD 273
TSDRRKEL K+VSK EEGKVA+RNIRRDALK+YEKLEK+KKLSEDNVKDLS+DLQKLTD
Sbjct: 192 TSDRRKELAKVVSKLAEEGKVAVRNIRRDALKSYEKLEKEKKLSEDNVKDLSADLQKLTD 251
Query: 274 DYIKKVDALYKQKEKVL 290
+Y+KKV+++YKQKE+ L
Sbjct: 252 EYMKKVESIYKQKEQEL 268
>UniRef100_Q6YTV7 Putative Ribosome recycling factor, chloroplast [Oryza sativa]
Length = 266
Score = 278 bits (711), Expect = 1e-73
Identities = 133/197 (67%), Positives = 175/197 (88%)
Query: 94 RKGFVRAATIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYY 153
++ +R ATIEEIEAEK+ IE+ + RME+ ++ V+ NF+++RTGRA P+MLD+IEVEYY
Sbjct: 67 KRAVLRHATIEEIEAEKSVIEDQARERMEKAIETVQNNFNTVRTGRANPAMLDRIEVEYY 126
Query: 154 GSPVSLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQL 213
G+PV+LK+IAQI+TPDA+SLL+ PYDKSSLK IEK IV +++G+TP+NDGEVIR+++P L
Sbjct: 127 GTPVNLKSIAQINTPDATSLLIQPYDKSSLKLIEKTIVAANLGVTPSNDGEVIRVTVPPL 186
Query: 214 TSDRRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTD 273
TSDRRKEL K V+K EEGKVA+RNIRRDA+KAY+KLEK+KKLSEDNVKDLS+DLQK+TD
Sbjct: 187 TSDRRKELAKTVAKLAEEGKVAIRNIRRDAIKAYDKLEKEKKLSEDNVKDLSADLQKVTD 246
Query: 274 DYIKKVDALYKQKEKVL 290
+Y+KK++A+ KQKE+ L
Sbjct: 247 EYMKKIEAIQKQKEQEL 263
>UniRef100_Q8YXK4 Ribosome recycling factor [Anabaena sp.]
Length = 182
Score = 205 bits (522), Expect = 1e-51
Identities = 100/173 (57%), Positives = 134/173 (76%)
Query: 116 DVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLV 175
+ +S+M+ T++ + F++IRTGRA S+LDK+ V+YYGSP LK++A ISTPDA+++L+
Sbjct: 5 EAESKMQHTVEATQRAFNTIRTGRANASLLDKVLVDYYGSPTPLKSLANISTPDATTILI 64
Query: 176 SPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVA 235
PYDKSSL +EKAI SDVG+TP+NDG VIRL+IP LTSDRRKEL KI +K EEG+VA
Sbjct: 65 QPYDKSSLNIVEKAISLSDVGLTPSNDGAVIRLNIPPLTSDRRKELVKIAAKYAEEGRVA 124
Query: 236 LRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
+RNIRRDA+ + KLEK+ ++SED KD LQKLT+ Y ++D L +KEK
Sbjct: 125 IRNIRRDAVDSIRKLEKNAEVSEDEAKDQQDKLQKLTNKYTARIDELLVEKEK 177
>UniRef100_Q8DM64 Ribosome recycling factor [Synechococcus elongatus]
Length = 182
Score = 194 bits (494), Expect = 2e-48
Identities = 93/173 (53%), Positives = 134/173 (76%)
Query: 116 DVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLV 175
DV+ RM+++++ + +F+SIRTGRA ++LD++ V+YYG+ L+++A +STPDA+++L+
Sbjct: 5 DVEERMQKSVEATQHDFNSIRTGRANAALLDRVMVDYYGTETPLRSLANVSTPDATTILI 64
Query: 176 SPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVA 235
PYD+S+L +IEKAI SD+G+TPNNDG +RL+IP LT++RRKEL K +K E GKVA
Sbjct: 65 QPYDRSTLASIEKAIQLSDLGLTPNNDGSSVRLNIPPLTTERRKELVKTAAKIAEGGKVA 124
Query: 236 LRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
+RNIRRDA+ +K K +L+ED VKDL +QKLTD YI K+DAL +KEK
Sbjct: 125 IRNIRRDAIDHIKKAGKAGELTEDEVKDLQEKVQKLTDKYIGKIDALLAEKEK 177
>UniRef100_Q7XY97 Ribosome recycling factor [Griffithsia japonica]
Length = 230
Score = 194 bits (493), Expect = 2e-48
Identities = 91/179 (50%), Positives = 135/179 (74%), Gaps = 4/179 (2%)
Query: 114 ENDV----KSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPD 169
END ++RM++T+D+VK+ F+S+RTGRA+PS+LD+I+V YYG+ L ++A +S
Sbjct: 47 ENDTAEKSEARMQKTIDSVKSAFNSVRTGRASPSLLDRIQVNYYGADTPLNSLATVSVSG 106
Query: 170 ASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQV 229
S+L+V PYDKS + IE++++ SD+G+TPN+DG IRLSIPQLT +RR +L K V
Sbjct: 107 TSTLVVEPYDKSCISDIERSLMESDIGLTPNSDGSTIRLSIPQLTKERRSQLVKQVKSMA 166
Query: 230 EEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
E+G+VA+RNIRRDA+ + +KLEK +L +D K L +QKLTD Y+K ++++YKQKE+
Sbjct: 167 EDGRVAIRNIRRDAVDSIKKLEKKSELGKDESKTLQDRVQKLTDKYVKTIESVYKQKEE 225
>UniRef100_P74456 Ribosome recycling factor [Synechocystis sp.]
Length = 182
Score = 192 bits (487), Expect = 1e-47
Identities = 91/173 (52%), Positives = 129/173 (73%)
Query: 116 DVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLV 175
++K M+++++ + +F++IRTGRA S+LD+I VEYYG+ LK++A I TPDAS++++
Sbjct: 5 ELKDHMQKSVEATQRSFNTIRTGRANASLLDRITVEYYGAETPLKSLATIGTPDASTIVI 64
Query: 176 SPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVA 235
P+D S+ IEKAI SD+G+TPNNDG+VIRL+IP LT++RRKEL K+ K EEGKVA
Sbjct: 65 QPFDMGSIGTIEKAISLSDLGLTPNNDGKVIRLNIPPLTAERRKELVKVAGKLAEEGKVA 124
Query: 236 LRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
+RNIRRDA+ K EK+ +SED +DL ++QKLTD K++D L KEK
Sbjct: 125 IRNIRRDAVDEVRKQEKNSDISEDEARDLQEEIQKLTDQSTKRIDELLAAKEK 177
>UniRef100_Q5WFT1 Ribosome recycling factor [Bacillus clausii]
Length = 185
Score = 191 bits (484), Expect = 3e-47
Identities = 86/176 (48%), Positives = 133/176 (74%)
Query: 113 IENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASS 172
I+ D K+RM++ +D + + +R GRA P++LD++ VEYYG+ L +A ++ P+A
Sbjct: 5 IKADAKTRMDKAIDVLSRELAKLRAGRANPALLDRVTVEYYGAQTPLNQLASVTVPEARL 64
Query: 173 LLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEG 232
LL++PYDK+++ +IEKAI+ SD+G+TP+NDG+VIR++IP LT +RRK+L ++VSK EE
Sbjct: 65 LLITPYDKTAVASIEKAILKSDLGLTPSNDGQVIRIAIPPLTEERRKDLVRLVSKTAEES 124
Query: 233 KVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
KVA+RNIRRDA +KL+KD +++ED ++ + D+QKLTD Y+ K+D KQKE+
Sbjct: 125 KVAVRNIRRDANDELKKLQKDGEMTEDELRSATDDIQKLTDTYVAKIDEKAKQKEQ 180
>UniRef100_Q7V6C1 Ribosome recycling factor [Prochlorococcus marinus]
Length = 182
Score = 188 bits (477), Expect = 2e-46
Identities = 86/172 (50%), Positives = 127/172 (73%)
Query: 116 DVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLV 175
++K M ++++ + NF++IRTGRA PS+LD+I VEYYG+ LK++A ISTPD+ ++ +
Sbjct: 5 ELKDNMHKSVEATQRNFATIRTGRANPSLLDRINVEYYGTETPLKSLATISTPDSQTIAI 64
Query: 176 SPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVA 235
P+D S+ IEKAI SD+G+TPNNDG++IR+++P LT +RRKE K+ ++ EEGKVA
Sbjct: 65 QPFDNGSMGLIEKAIATSDLGLTPNNDGKIIRINVPPLTEERRKEFCKLAARYAEEGKVA 124
Query: 236 LRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKE 287
LRNIRRDA+ +KLEKD +LS+D D +QKLTD +I +++ +KE
Sbjct: 125 LRNIRRDAIDKVKKLEKDAELSKDQSHDEQDGIQKLTDTFITEIEKYLAEKE 176
>UniRef100_Q7NNX0 Ribosome recycling factor [Gloeobacter violaceus]
Length = 182
Score = 188 bits (477), Expect = 2e-46
Identities = 89/168 (52%), Positives = 127/168 (74%)
Query: 121 MERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLVSPYDK 180
M + ++ NF++IRTGRA+ S+LD+I VEYYG P LKT+A I+TPDAS++L+ PYD
Sbjct: 10 MRKAIEATAGNFATIRTGRASTSLLDRINVEYYGQPTPLKTLATITTPDASTVLIQPYDP 69
Query: 181 SSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVALRNIR 240
SSL+ IEK I+ SD+G+ P+NDG+ IRL+IP LT++RRK+L K++ EEG+VA+RNIR
Sbjct: 70 SSLRLIEKTILESDLGLPPSNDGKTIRLNIPPLTAERRKDLVKVLRNLAEEGRVAVRNIR 129
Query: 241 RDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
R A+ K EKD K+SED + L ++QKLTD I++++ L++ KEK
Sbjct: 130 RHAIDEVRKEEKDAKVSEDEARRLQDEVQKLTDKSIQQIEKLFEAKEK 177
>UniRef100_Q65JJ6 Frr [Bacillus licheniformis]
Length = 185
Score = 187 bits (474), Expect = 4e-46
Identities = 86/176 (48%), Positives = 129/176 (72%)
Query: 113 IENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASS 172
+ N+ K RM++ + + +++R GRA PS+LDK+ VEYYG+ L IA I+ P+A
Sbjct: 5 VMNETKERMQKAISAYQRELATVRAGRANPSLLDKVAVEYYGAQTPLNQIASITVPEARM 64
Query: 173 LLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEG 232
L+++PYDK++L IEKAI +D+G+TP+NDG +IR++IP LT +RRKEL K+V K E+
Sbjct: 65 LVITPYDKTALGDIEKAIQKADLGVTPSNDGNIIRITIPPLTEERRKELAKLVKKYSEDA 124
Query: 233 KVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
KVA+RNIRRDA +KLEK+ +++ED ++ + D+QKLTD+Y+ K+D + K KEK
Sbjct: 125 KVAVRNIRRDANDDLKKLEKNGEMTEDELRSSTEDVQKLTDEYVSKIDEITKDKEK 180
>UniRef100_P61302 Ribosome recycling factor [Bdellovibrio bacteriovorus]
Length = 186
Score = 185 bits (470), Expect = 1e-45
Identities = 90/180 (50%), Positives = 129/180 (71%)
Query: 111 ASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDA 170
A ++ + +++ME+TL+++ +RTGRA SMLD I V YYG+P L +A ISTPDA
Sbjct: 4 ADVKKNAQAKMEKTLNSLSEELKKVRTGRAQVSMLDGIRVNYYGTPSPLSQVASISTPDA 63
Query: 171 SSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVE 230
S L++P++ + LK IE+AI+ S++GM P NDG+VIRL +P LT +RRK+L K V K E
Sbjct: 64 KSFLIAPWEVAILKDIEQAIIKSELGMAPMNDGKVIRLKVPDLTEERRKDLAKQVKKIAE 123
Query: 231 EGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEKVL 290
E +VA+R RRDA +KL+KDK +SED K +D+QK+TDD+IKKVD + ++KEK +
Sbjct: 124 EARVAVRMARRDANDEVKKLQKDKAVSEDEGKKAEADIQKVTDDFIKKVDQVAEEKEKAI 183
>UniRef100_Q62V01 Ribosome recycling factor [Bacillus licheniformis]
Length = 185
Score = 184 bits (467), Expect = 2e-45
Identities = 85/176 (48%), Positives = 128/176 (72%)
Query: 113 IENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASS 172
+ N+ K RM++ + + +++R GRA PS+LDK+ VEYYG+ L IA I+ P+A
Sbjct: 5 VMNETKERMQKAISAYQRELATVRAGRANPSLLDKVAVEYYGAQTPLNQIASITVPEARM 64
Query: 173 LLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEG 232
L+++PYDK++L I KAI +D+G+TP+NDG +IR++IP LT +RRKEL K+V K E+
Sbjct: 65 LVITPYDKTALGDIGKAIQKADLGVTPSNDGNIIRITIPPLTEERRKELAKLVKKYSEDA 124
Query: 233 KVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
KVA+RNIRRDA +KLEK+ +++ED ++ + D+QKLTD+Y+ K+D + K KEK
Sbjct: 125 KVAVRNIRRDANDDLKKLEKNGEMTEDELRSSTEDVQKLTDEYVSKIDEITKDKEK 180
>UniRef100_Q67PB4 Ribosome recycling factor [Symbiobacterium thermophilum]
Length = 185
Score = 182 bits (463), Expect = 7e-45
Identities = 89/176 (50%), Positives = 126/176 (71%)
Query: 113 IENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASS 172
I D + RM++ ++ + + +R RATP++LDK+ VE YGS V + +A I PD +
Sbjct: 5 IIKDAEERMKKAVEVFRQELAGMRANRATPALLDKVRVEAYGSEVPVNNVATIEVPDPRT 64
Query: 173 LLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEG 232
L++ P+D+S +KAIE+AI SD+G+ P NDG+VIRLSIP +T +RR+EL K+V+K+ EE
Sbjct: 65 LVIKPWDRSLIKAIERAINASDLGLNPTNDGQVIRLSIPPMTEERRRELVKVVAKRTEEQ 124
Query: 233 KVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
+VA+RNIRRDA + +KLEKDK +SED K ++QKLTD YIK+VD L KEK
Sbjct: 125 RVAIRNIRRDANEQIKKLEKDKAVSEDESKRAQDEVQKLTDKYIKEVDQLMAAKEK 180
>UniRef100_UPI0000346352 UPI0000346352 UniRef100 entry
Length = 182
Score = 182 bits (462), Expect = 9e-45
Identities = 86/175 (49%), Positives = 129/175 (73%)
Query: 114 ENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSL 173
E +++S M +T++ + NF++IRTGRA S+LD+I VEYYG+ +K++A I+T D+ ++
Sbjct: 3 EQEIQSNMNKTVEATQRNFNTIRTGRANASLLDRISVEYYGAETPIKSLASIATVDSQTI 62
Query: 174 LVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGK 233
+ P+D S L+AIEKAI SD+GMTPNNDG+VIR+++P LT +RRKE K+ SK EEGK
Sbjct: 63 SIQPFDISCLQAIEKAISVSDLGMTPNNDGKVIRINVPPLTEERRKEFCKLASKYAEEGK 122
Query: 234 VALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
+ALRNIRRD++ ++ EK+ +S D +D S++QK+TD YI ++ +KEK
Sbjct: 123 IALRNIRRDSVDKEKRDEKEGLISIDESRDNQSEIQKITDKYISLIETKLSEKEK 177
>UniRef100_UPI00002771CB UPI00002771CB UniRef100 entry
Length = 182
Score = 181 bits (460), Expect = 2e-44
Identities = 86/175 (49%), Positives = 128/175 (73%)
Query: 114 ENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSL 173
E +++ M ++++ + NF++IRTGRA S+LD++ VEYYG+ +K++A IST D+ ++
Sbjct: 3 EKEIQENMNKSIEATQRNFNTIRTGRANASLLDRVSVEYYGAETPIKSLATISTVDSQTI 62
Query: 174 LVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGK 233
+ P+D S L+AIEK+I SD+G+TPNNDG+VIR+++P LT +RRKE K+ SK EEGK
Sbjct: 63 SIQPFDISCLQAIEKSISMSDLGITPNNDGKVIRINVPPLTEERRKEFCKLASKYAEEGK 122
Query: 234 VALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
VALRNIRRDA+ +K EKD +S D +D S++QK+TD YI ++ +KEK
Sbjct: 123 VALRNIRRDAVDKEKKDEKDGLISIDESRDNQSEIQKITDKYIALIETKLSEKEK 177
>UniRef100_UPI00002BEA74 UPI00002BEA74 UniRef100 entry
Length = 182
Score = 180 bits (456), Expect = 5e-44
Identities = 85/175 (48%), Positives = 128/175 (72%)
Query: 114 ENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSL 173
E +++ M ++++ + NF++IRTGRA S+LD++ VEYYG+ +K++A IST D+ ++
Sbjct: 3 EQEIQENMNKSIEATQRNFNTIRTGRANASLLDRVSVEYYGAETPIKSLATISTVDSQTI 62
Query: 174 LVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGK 233
+ P+D S L+AIEK+I SD+G+TPNNDG+VIR+++P LT +RRKE K+ SK EEGK
Sbjct: 63 SIQPFDISCLQAIEKSISMSDLGITPNNDGKVIRINVPPLTEERRKEFCKLASKYAEEGK 122
Query: 234 VALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
VALRNIRRDA+ +K EK+ +S D +D S++QK+TD YI ++ +KEK
Sbjct: 123 VALRNIRRDAIDKEKKDEKEGLISIDESRDNQSEIQKITDKYIALIEIKLSEKEK 177
>UniRef100_Q8DSY2 Ribosome recycling factor [Streptococcus mutans]
Length = 185
Score = 180 bits (456), Expect = 5e-44
Identities = 94/179 (52%), Positives = 122/179 (67%)
Query: 112 SIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDAS 171
+I K R E++ ++ F SIR GRA S+LD+IEVEYYG P L +A I+ P+A
Sbjct: 4 AIVEKAKERFEQSHQSLAREFGSIRAGRANASLLDRIEVEYYGVPTPLNQLASITVPEAR 63
Query: 172 SLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEE 231
LLVSP+DKSSLK IE AI SD+G+ P NDG VIRL IP LT + RKEL K V K E
Sbjct: 64 VLLVSPFDKSSLKDIEHAINASDIGINPANDGSVIRLVIPALTEETRKELAKEVKKVGEN 123
Query: 232 GKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEKVL 290
KVA+RNIRRDA+ +K EK K+++ED +K L D+QK+TD+ +K +D++ KEK L
Sbjct: 124 AKVAIRNIRRDAMDEAKKQEKIKEITEDELKSLEKDIQKVTDEAVKHIDSMTANKEKEL 182
>UniRef100_Q7V2F9 Ribosome recycling factor [Prochlorococcus marinus subsp. pastoris]
Length = 182
Score = 180 bits (456), Expect = 5e-44
Identities = 87/175 (49%), Positives = 128/175 (72%)
Query: 114 ENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSL 173
E +++S M ++++ + NF++IRTGRA S+LD+I VEYYG+ +K++A IST D+ ++
Sbjct: 3 EQEIQSSMNKSVEATQRNFNTIRTGRANASLLDRISVEYYGAETPIKSLASISTIDSQTI 62
Query: 174 LVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGK 233
+ P+D SSL+ IEKAI SD+G+TPNNDG+VIR+++P LT +RRKE K+ SK EEGK
Sbjct: 63 SIQPFDISSLQTIEKAISVSDLGITPNNDGKVIRINVPPLTEERRKEFCKLASKYAEEGK 122
Query: 234 VALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
VALRNIRRDA+ +K EK+ +S+D +D ++QK TD YI ++ +KEK
Sbjct: 123 VALRNIRRDAVDKEKKDEKEGLISKDVSRDNQLEIQKFTDKYISLIETKLSEKEK 177
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.133 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 434,135,918
Number of Sequences: 2790947
Number of extensions: 16519289
Number of successful extensions: 89548
Number of sequences better than 10.0: 1062
Number of HSP's better than 10.0 without gapping: 628
Number of HSP's successfully gapped in prelim test: 447
Number of HSP's that attempted gapping in prelim test: 88034
Number of HSP's gapped (non-prelim): 1749
length of query: 297
length of database: 848,049,833
effective HSP length: 126
effective length of query: 171
effective length of database: 496,390,511
effective search space: 84882777381
effective search space used: 84882777381
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC122725.8


