

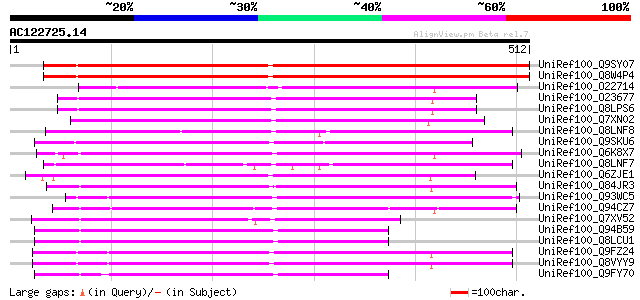
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122725.14 + phase: 0
(512 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SY07 Hypothetical protein T5J8.14 [Arabidopsis thali... 584 e-165
UniRef100_Q8W4P4 Hypothetical protein At4g02820; T5J8.14 [Arabid... 583 e-165
UniRef100_O22714 F8A5.28 protein [Arabidopsis thaliana] 273 9e-72
UniRef100_O23677 T7I23.8 protein [Arabidopsis thaliana] 256 1e-66
UniRef100_Q8LPS6 At1g02150/T7I23.8 [Arabidopsis thaliana] 255 2e-66
UniRef100_Q7XN02 OSJNBb0038F03.9 protein [Oryza sativa] 247 5e-64
UniRef100_Q8LNF8 Putative leaf protein [Oryza sativa] 241 4e-62
UniRef100_Q9SKU6 Hypothetical protein At2g20710 [Arabidopsis tha... 237 7e-61
UniRef100_Q6K8X7 Pentatricopeptide (PPR) repeat-containing prote... 236 9e-61
UniRef100_Q8LNF7 Putative leaf protein [Oryza sativa] 236 1e-60
UniRef100_Q6ZJE1 Putative DNA-binding protein [Oryza sativa] 233 1e-59
UniRef100_Q84JR3 Hypothetical protein At4g21705 [Arabidopsis tha... 221 4e-56
UniRef100_Q93WC5 AT4g01990/T7B11_26 [Arabidopsis thaliana] 219 1e-55
UniRef100_Q94CZ7 P0682B08.20 protein [Oryza sativa] 219 1e-55
UniRef100_Q7XV52 OSJNBa0086B14.3 protein [Oryza sativa] 211 4e-53
UniRef100_Q94B59 Hypothetical protein T5E8_250 [Arabidopsis thal... 209 1e-52
UniRef100_Q8LCU1 Hypothetical protein [Arabidopsis thaliana] 209 2e-52
UniRef100_Q9FZ24 T6A9.6 protein [Arabidopsis thaliana] 206 1e-51
UniRef100_Q8VYY9 Hypothetical protein At1g02370 [Arabidopsis tha... 206 1e-51
UniRef100_Q9FY70 Hypothetical protein T5E8_250 [Arabidopsis thal... 195 3e-48
>UniRef100_Q9SY07 Hypothetical protein T5J8.14 [Arabidopsis thaliana]
Length = 532
Score = 584 bits (1506), Expect = e-165
Identities = 289/479 (60%), Positives = 376/479 (78%), Gaps = 5/479 (1%)
Query: 34 GGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCE 93
G DTLG RLLSLVY KRSAV+ I KWKEEGH++ RKY+LNR++RELRK KRYKHALE+CE
Sbjct: 59 GRDTLGGRLLSLVYTKRSAVVTIRKWKEEGHSV-RKYELNRIVRELRKIKRYKHALEICE 117
Query: 94 WMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNL 153
WM +Q DIKL GDYAV LDLI+K+RGLNSAEKFFED+PD+MRG CT+LLH+YVQN L
Sbjct: 118 WMVVQEDIKLQAGDYAVHLDLISKIRGLNSAEKFFEDMPDQMRGHAACTSLLHSYVQNKL 177
Query: 154 TNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNTSPDVVTFNLL 213
++KAEAL KM ECGFL+S +PYN M+S+YIS G+ EKVP L +ELK+ TSPD+VT+NL
Sbjct: 178 SDKAEALFEKMGECGFLKSCLPYNHMLSMYISRGQFEKVPVLIKELKIRTSPDIVTYNLW 237
Query: 214 LTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEME 273
LTA AS NDVE AE+V L+ K+ K++PDWVTYS LTNLY + D +EKA LKEME
Sbjct: 238 LTAFASGNDVEGAEKVYLKAKEEKLNPDWVTYSVLTNLYAKT----DNVEKARLALKEME 293
Query: 274 KRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFA 333
K S++ RVAY+SL+SLHAN+G+ D VN W K+K+ F KM+D EY+ MIS++VKLG+F
Sbjct: 294 KLVSKKNRVAYASLISLHANLGDKDGVNLTWKKVKSSFKKMNDAEYLSMISAVVKLGEFE 353
Query: 334 GVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLT 393
+ LY EWESVSGT D R+ NL+L Y+++ ++ + E F ++VEKG+ SYS+WE+LT
Sbjct: 354 QAKGLYDEWESVSGTGDARIPNLILAEYMNRDEVLLGEKFYERIVEKGINPSYSTWEILT 413
Query: 394 RGYLKKKDVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAG 453
YLK+KD++K L FGKAI SVK+W + RLV+ A ++EQ +++GAE+L+ +L+ AG
Sbjct: 414 WAYLKRKDMEKVLDCFGKAIDSVKKWTVNVRLVKGACKELEEQGNVKGAEKLMTLLQKAG 473
Query: 454 HVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLLDLTSKMCVSDVSGILS 512
+VNT +YN L+TYA AG+M L+V ERM KDNV+LD+ET L+ LTS+M V+++S +S
Sbjct: 474 YVNTQLYNSLLRTYAKAGEMALIVEERMAKDNVELDEETKELIRLTSQMRVTEISSTIS 532
>UniRef100_Q8W4P4 Hypothetical protein At4g02820; T5J8.14 [Arabidopsis thaliana]
Length = 532
Score = 583 bits (1503), Expect = e-165
Identities = 288/479 (60%), Positives = 376/479 (78%), Gaps = 5/479 (1%)
Query: 34 GGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCE 93
G DTLG RLLSLVY KRSAV+ I KWKEEGH++ RKY+LNR++RELRK KRYKHALE+CE
Sbjct: 59 GRDTLGGRLLSLVYTKRSAVVTIRKWKEEGHSV-RKYELNRIVRELRKIKRYKHALEICE 117
Query: 94 WMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNL 153
WM +Q DIKL GDYAV LDLI+K+RGLNSAEKFFED+PD+MRG CT+LLH+YVQN L
Sbjct: 118 WMVVQEDIKLQAGDYAVHLDLISKIRGLNSAEKFFEDMPDQMRGHAACTSLLHSYVQNKL 177
Query: 154 TNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNTSPDVVTFNLL 213
++KAEAL KM ECGFL+S +PYN M+S+YIS G+ EKVP L +ELK+ TSPD+VT+NL
Sbjct: 178 SDKAEALFEKMGECGFLKSCLPYNHMLSMYISRGQFEKVPVLIKELKIRTSPDIVTYNLW 237
Query: 214 LTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEME 273
LTA AS NDVE AE+V L+ K+ K++PDWVTYS LTNLY + D +EKA L+EME
Sbjct: 238 LTAFASGNDVEGAEKVYLKAKEEKLNPDWVTYSVLTNLYAKT----DNVEKARLALREME 293
Query: 274 KRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFA 333
K S++ RVAY+SL+SLHAN+G+ D VN W K+K+ F KM+D EY+ MIS++VKLG+F
Sbjct: 294 KLVSKKNRVAYASLISLHANLGDKDGVNLTWKKVKSSFKKMNDAEYLSMISAVVKLGEFE 353
Query: 334 GVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLT 393
+ LY EWESVSGT D R+ NL+L Y+++ ++ + E F ++VEKG+ SYS+WE+LT
Sbjct: 354 QAKGLYDEWESVSGTGDARIPNLILAEYMNRDEVLLGEKFYERIVEKGINPSYSTWEILT 413
Query: 394 RGYLKKKDVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAG 453
YLK+KD++K L FGKAI SVK+W + RLV+ A ++EQ +++GAE+L+ +L+ AG
Sbjct: 414 WAYLKRKDMEKVLDCFGKAIDSVKKWTVNVRLVKGACKELEEQGNVKGAEKLMTLLQKAG 473
Query: 454 HVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLLDLTSKMCVSDVSGILS 512
+VNT +YN L+TYA AG+M L+V ERM KDNV+LD+ET L+ LTS+M V+++S +S
Sbjct: 474 YVNTQLYNSLLRTYAKAGEMALIVEERMAKDNVELDEETKELIRLTSQMRVTEISSTIS 532
>UniRef100_O22714 F8A5.28 protein [Arabidopsis thaliana]
Length = 491
Score = 273 bits (698), Expect = 9e-72
Identities = 152/440 (34%), Positives = 253/440 (56%), Gaps = 12/440 (2%)
Query: 69 KYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFF 128
K+++ I++LR Y AL++ E M+ + K V D A+ LDL+ K R + + E +F
Sbjct: 55 KWEVGDTIKKLRNRGLYYPALKLSEVMEERGMNKTVS-DQAIHLDLVAKAREITAGENYF 113
Query: 129 EDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGK 188
DLP+ + + T +LL+ Y + LT KAE L++KM E S + YN +M+LY G+
Sbjct: 114 VDLPETSKTELTYGSLLNCYCKELLTEKAEGLLNKMKELNITPSSMSYNSLMTLYTKTGE 173
Query: 189 LEKVPKLFEELKV-NTSPDVVTFNLLLTACASENDVETAERVLLQLKK-AKVDPDWVTYS 246
EKVP + +ELK N PD T+N+ + A A+ ND+ ERV+ ++ + +V PDW TYS
Sbjct: 174 TEKVPAMIQELKAENVMPDSYTYNVWMRALAATNDISGVERVIEEMNRDGRVAPDWTTYS 233
Query: 247 TLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGK 306
+ ++Y+ +A + EKA L+E+E + ++ AY L++L+ +G + EV RIW
Sbjct: 234 NMASIYV-DAGLSQKAEKA---LQELEMKNTQRDFTAYQFLITLYGRLGKLTEVYRIWRS 289
Query: 307 MKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQ 366
++ K S+ Y+ MI LVKL D G E L+KEW++ T D+R+ N+L+ +Y +G
Sbjct: 290 LRLAIPKTSNVAYLNMIQVLVKLNDLPGAETLFKEWQANCSTYDIRIVNVLIGAYAQEGL 349
Query: 367 MEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVK----QWIPD 422
++ A + +G L+ +WE+ Y+K D+ + L KA+S K +W+P
Sbjct: 350 IQKANELKEKAPRRGGKLNAKTWEIFMDYYVKSGDMARALECMSKAVSIGKGDGGKWLPS 409
Query: 423 PRLVQEAFTVIQEQAHIEGAEQLLVILRN-AGHVNTNIYNLFLKTYAAAGKMPLVVAERM 481
P V+ + +++ + GAE LL IL+N ++ I+ ++TYAAAGK + R+
Sbjct: 410 PETVRALMSYFEQKKDVNGAENLLEILKNGTDNIGAEIFEPLIRTYAAAGKSHPAMRRRL 469
Query: 482 KKDNVQLDKETHRLLDLTSK 501
K +NV++++ T +LLD S+
Sbjct: 470 KMENVEVNEATKKLLDEVSQ 489
>UniRef100_O23677 T7I23.8 protein [Arabidopsis thaliana]
Length = 524
Score = 256 bits (653), Expect = 1e-66
Identities = 145/418 (34%), Positives = 240/418 (56%), Gaps = 10/418 (2%)
Query: 48 PKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQND-IKLVEG 106
P+ A +N+W++ G L K++L R+++ELRK KR ALEV +WM + + +L
Sbjct: 79 PELGAASVLNQWEKAGRKLT-KWELCRVVKELRKYKRANQALEVYDWMNNRGERFRLSAS 137
Query: 107 DYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSE 166
D A+QLDLI KVRG+ AE+FF LP+ + + +LL+AYV+ KAEAL++ M +
Sbjct: 138 DAAIQLDLIGKVRGIPDAEEFFLQLPENFKDRRVYGSLLNAYVRAKSREKAEALLNTMRD 197
Query: 167 CGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV-NTSPDVVTFNLLLTACASENDVET 225
G+ P+P+N MM+LY++ + +KV + E+K + D+ ++N+ L++C S VE
Sbjct: 198 KGYALHPLPFNVMMTLYMNLREYDKVDAMVFEMKQKDIRLDIYSYNIWLSSCGSLGSVEK 257
Query: 226 AERVLLQLKK-AKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAY 284
E V Q+K + P+W T+ST+ +YI+ EKA L+++E R + R+ Y
Sbjct: 258 MELVYQQMKSDVSIYPNWTTFSTMATMYIKMGET----EKAEDALRKVEARITGRNRIPY 313
Query: 285 SSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWES 344
LLSL+ ++GN E+ R+W K+ + + Y ++SSLV++GD G E +Y+EW
Sbjct: 314 HYLLSLYGSLGNKKELYRVWHVYKSVVPSIPNLGYHALVSSLVRMGDIEGAEKVYEEWLP 373
Query: 345 VSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKK 404
V + D R+ NLL+ +YV Q+E AE + +VE G S S+WE+L G+ +K+ + +
Sbjct: 374 VKSSYDPRIPNLLMNAYVKNDQLETAEGLFDHMVEMGGKPSSSTWEILAVGHTRKRCISE 433
Query: 405 FLHYFGKAISS--VKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIY 460
L A S+ W P ++ F + +E++ + E +L +LR +G + Y
Sbjct: 434 ALTCLRNAFSAEGSSNWRPKVLMLSGFFKLCEEESDVTSKEAVLELLRQSGDLEDKSY 491
>UniRef100_Q8LPS6 At1g02150/T7I23.8 [Arabidopsis thaliana]
Length = 524
Score = 255 bits (651), Expect = 2e-66
Identities = 144/418 (34%), Positives = 240/418 (56%), Gaps = 10/418 (2%)
Query: 48 PKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQND-IKLVEG 106
P+ A +N+W++ G L K++L R+++ELRK KR A+EV +WM + + +L
Sbjct: 79 PELGAASVLNQWEKAGRKLT-KWELCRVVKELRKYKRANQAIEVYDWMNNRGERFRLSAS 137
Query: 107 DYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSE 166
D A+QLDLI KVRG+ AE+FF LP+ + + +LL+AYV+ KAEAL++ M +
Sbjct: 138 DAAIQLDLIGKVRGIPDAEEFFLQLPENFKDRRVYGSLLNAYVRAKSREKAEALLNTMRD 197
Query: 167 CGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV-NTSPDVVTFNLLLTACASENDVET 225
G+ P+P+N MM+LY++ + +KV + E+K + D+ ++N+ L++C S VE
Sbjct: 198 KGYALHPLPFNVMMTLYMNLREYDKVDAMVFEMKQKDIRLDIYSYNIWLSSCGSLGSVEK 257
Query: 226 AERVLLQLKK-AKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAY 284
E V Q+K + P+W T+ST+ +YI+ EKA L+++E R + R+ Y
Sbjct: 258 MELVYQQMKSDVSIYPNWTTFSTMATMYIKMGET----EKAEDALRKVEARITGRNRIPY 313
Query: 285 SSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWES 344
LLSL+ ++GN E+ R+W K+ + + Y ++SSLV++GD G E +Y+EW
Sbjct: 314 HYLLSLYGSLGNKKELYRVWHVYKSVVPSIPNLGYHALVSSLVRMGDIEGAEKVYEEWLP 373
Query: 345 VSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKK 404
V + D R+ NLL+ +YV Q+E AE + +VE G S S+WE+L G+ +K+ + +
Sbjct: 374 VKSSYDPRIPNLLMNAYVKNDQLETAEGLFDHMVEMGGKPSSSTWEILAVGHTRKRCISE 433
Query: 405 FLHYFGKAISS--VKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIY 460
L A S+ W P ++ F + +E++ + E +L +LR +G + Y
Sbjct: 434 ALTCLRNAFSAEGSSNWRPKVLMLSGFFKLCEEESDVTSKEAVLELLRQSGDLEDKSY 491
>UniRef100_Q7XN02 OSJNBb0038F03.9 protein [Oryza sativa]
Length = 511
Score = 247 bits (631), Expect = 5e-64
Identities = 145/413 (35%), Positives = 236/413 (57%), Gaps = 9/413 (2%)
Query: 61 EEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQND-IKLVEGDYAVQLDLITKVR 119
+EG K++L R+ RELRK +R+ AL+V +WM + D L D A+QLDLI KVR
Sbjct: 99 DEGERRLDKWELCRIARELRKFRRFNLALQVYDWMTERRDRFSLSSSDMAIQLDLIAKVR 158
Query: 120 GLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRM 179
G++ AE++FE+LPD ++ + T +LL+ Y Q + K E+ +M + GF +P+N +
Sbjct: 159 GVSHAEEYFEELPDPLKDKRTYGSLLNVYAQAMMKEKTESTFEQMRKKGFATDTLPFNVL 218
Query: 180 MSLYISNGKLEKVPKLFEEL-KVNTSPDVVTFNLLLTACASENDVETAERVLLQL-KKAK 237
M+ Y+ + EKV L +E+ + N + DV T+N+ + +CA+ D + E+V Q+ +
Sbjct: 219 MNFYVDAEEAEKVSILIDEMMERNVAFDVCTYNIWIKSCAAMQDADAMEQVFNQMIRDET 278
Query: 238 VDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNV 297
V +W TY+TL +++I+ + EKA +LKE EKRT+ + + L++L++++G
Sbjct: 279 VVANWTTYTTLASMHIKLGNS----EKAEESLKEAEKRTTGREKKCFHYLMTLYSHLGKK 334
Query: 298 DEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLL 357
+EV R+W KA F + + Y ++S+LV+LGD G E LY+EW S S + D + N+L
Sbjct: 335 EEVYRVWNWYKATFPTIHNLGYQEVLSALVRLGDIEGAELLYEEWASKSSSFDPKTMNIL 394
Query: 358 LTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGK--AISS 415
L Y +G + AE N+ VEKG ++WE+L YLK + L K A++S
Sbjct: 395 LAWYAREGFVTKAEQTLNRFVEKGGNPKPNTWEILGTAYLKDGQSSEALSCLEKATAVAS 454
Query: 416 VKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYA 468
+W P P V+ +E+ E A++L+ +LR+ Y + TYA
Sbjct: 455 PSKWRPRPTNVESLLANFKEKNDAESADRLMNVLRSRRCEENEEYKSLINTYA 507
>UniRef100_Q8LNF8 Putative leaf protein [Oryza sativa]
Length = 545
Score = 241 bits (615), Expect = 4e-62
Identities = 153/468 (32%), Positives = 250/468 (52%), Gaps = 15/468 (3%)
Query: 36 DTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWM 95
D+L RR+ + P+ + +W K ++ +I+ L + +R+ AL++ WM
Sbjct: 81 DSLFRRVAAAADPRLPLSPVLEQWCLAEERPIAKPEIQSIIKYLCRRRRFSQALQLSMWM 140
Query: 96 KLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTN 155
+ + L GD A +L+LITKV GL+ A ++F+ +PD+++ Q +LL Y +
Sbjct: 141 TERLHLHLSPGDVAYRLELITKVHGLDRAVEYFDSMPDQLKQQQCYGSLLKCYAEAKCVE 200
Query: 156 KAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVN-TSPDVVTFNLLL 214
KAE L KM G + S YN MM LY+ +G++E+V + ++ + DV T + L+
Sbjct: 201 KAEELFEKMRGMG-MASSYAYNVMMRLYLQDGQVERVHSMHRTMEESGIVADVFTTDTLV 259
Query: 215 TACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEK 274
A D+E E+VL + W +Y+T+ + +++ E+A +E EK
Sbjct: 260 AAYVVAEDIEAIEKVLEKADTCNDLMTWHSYATIGKVLMQSGME----ERALQAFQESEK 315
Query: 275 RTSRET-RVAYSSLLSLHANMGNVDEVNRIW----GKMKACFCKMSDDEYVCMISSLVKL 329
+ ++++ RVAY LL+++A++G EV+RIW K+ A C + Y+C IS L+K+
Sbjct: 316 KIAKKSNRVAYGFLLTMYADLGMNSEVDRIWDVYKSKVPASAC---NSMYMCRISVLLKM 372
Query: 330 GDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSW 389
D G E Y+EWES +D R+ NLLLT+Y +G ME AE +Q V+KG ++W
Sbjct: 373 NDIVGAEKAYEEWESKHVYHDSRLINLLLTAYCKEGLMEKAEALVDQFVKKGRTPFGNTW 432
Query: 390 ELLTRGYLKKKDVKKFLHYFGKAISS-VKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVI 448
L GY K K KA++S +W PD V + EQ ++E AE++ +
Sbjct: 433 YKLAGGYFKVGQASKAADLTKKALASGSNEWTPDLTNVLMSLNYFAEQKNVEAAEEMASL 492
Query: 449 LRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLL 496
L+ +IY+ LKTY AGK + +RMKKD ++ D+ET ++L
Sbjct: 493 LQRLITPTRDIYHGLLKTYVNAGKPVSDLLDRMKKDGMEADEETEKIL 540
>UniRef100_Q9SKU6 Hypothetical protein At2g20710 [Arabidopsis thaliana]
Length = 490
Score = 237 bits (604), Expect = 7e-61
Identities = 148/436 (33%), Positives = 233/436 (52%), Gaps = 10/436 (2%)
Query: 25 GVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKR 84
G T S DTL RR+ P S + ++ W ++G+ L + +L+ +I+ LRK R
Sbjct: 27 GKTTPSPLDPYDTLQRRVARSGDPSASIIKVLDGWLDQGN-LVKTSELHSIIKMLRKFSR 85
Query: 85 YKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTAL 144
+ HAL++ +WM ++ EGD A++LDLI KV GL AEKFFE +P + R AL
Sbjct: 86 FSHALQISDWMSEHRVHEISEGDVAIRLDLIAKVGGLGEAEKFFETIPMERRNYHLYGAL 145
Query: 145 LHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNT- 203
L+ Y + +KAE + +M E GFL+ +PYN M++LY+ GK V KL E++ T
Sbjct: 146 LNCYASKKVLHKAEQVFQEMKELGFLKGCLPYNVMLNLYVRTGKYTMVEKLLREMEDETV 205
Query: 204 SPDVVTFNLLLTACASENDVETAERVLLQLKKAK-VDPDWVTYSTLTNLYIRNASVDDCL 262
PD+ T N L A + +DVE E+ L++ + + + DW TY+ N YI+
Sbjct: 206 KPDIFTVNTRLHAYSVVSDVEGMEKFLMRCEADQGLHLDWRTYADTANGYIKAG----LT 261
Query: 263 EKAASTLKEMEKRTSRETRV-AYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVC 321
EKA L++ E+ + + R AY L+S + G +EV R+W K + Y+
Sbjct: 262 EKALEMLRKSEQMVNAQKRKHAYEVLMSFYGAAGKKEEVYRLWSLYKE-LDGFYNTGYIS 320
Query: 322 MISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKG 381
+IS+L+K+ D VE + +EWE+ D+R+ +LL+T Y +G ME AE N LV+K
Sbjct: 321 VISALLKMDDIEEVEKIMEEWEAGHSLFDIRIPHLLITGYCKKGMMEKAEEVVNILVQKW 380
Query: 382 VCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVKQ-WIPDPRLVQEAFTVIQEQAHIE 440
S+WE L GY ++K + + +AI K W P ++ ++ Q +E
Sbjct: 381 RVEDTSTWERLALGYKMAGKMEKAVEKWKRAIEVSKPGWRPHQVVLMSCVDYLEGQRDME 440
Query: 441 GAEQLLVILRNAGHVN 456
G ++L +L GH++
Sbjct: 441 GLRKILRLLSERGHIS 456
>UniRef100_Q6K8X7 Pentatricopeptide (PPR) repeat-containing protein-like [Oryza
sativa]
Length = 517
Score = 236 bits (603), Expect = 9e-61
Identities = 146/488 (29%), Positives = 250/488 (50%), Gaps = 17/488 (3%)
Query: 27 TAASSSSGGDTLGRRLLSLVYPKRS----AVIAINKW-KEEGHTLPRKYQLNRMIRELRK 81
TA GGD L R LL + P++S A A+ +W +E G P +L R + LR+
Sbjct: 38 TAREDGDGGDLLSRCLLRI--PRKSGRAAAAAAVERWARERGRVSPP--ELRRDVVRLRR 93
Query: 82 NKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTC 141
+RY+ ALE+ WM ND +L D+ V+LDLI KV G + AE+++ L +
Sbjct: 94 ARRYEQALEILSWMDSHNDFRLSPSDHMVRLDLIAKVHGTSQAEEYYRKLSTAASKKAAS 153
Query: 142 TALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV 201
LLH YV KAE M+++ G P +N +M LY++ + EKV + +K
Sbjct: 154 FPLLHCYVTERNVQKAETFMAELQRYGLPVDPHSFNEIMKLYVATCQYEKVLSVIYLMKR 213
Query: 202 NTSP-DVVTFNLLLTACASENDVETAERVLLQ-LKKAKVDPDWVTYSTLTNLYIRNASVD 259
N P +V+++N+ + ACA + + + + + L V+ W TY TL N++ +
Sbjct: 214 NNIPRNVLSYNIWMNACAEVSGLASVQSAFKEMLNDDMVEVGWSTYCTLANIFKKYGQSS 273
Query: 260 DCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEY 319
KA + L+ E + S R+ YS +++ +A + + D V R+W K ++ Y
Sbjct: 274 ----KALACLRTAETKLSSTGRLGYSFIMTCYAALNDRDGVIRLWEASKIVPGRIPAANY 329
Query: 320 VCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVE 379
+ + L+K+GD E + WE+ S +DVRVSN+LL +YV G +E AE ++E
Sbjct: 330 MSAMVCLIKVGDIGRAEWTFGSWEAESKKHDVRVSNVLLGAYVRNGWIEKAERLHLHMLE 389
Query: 380 KGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVK--QWIPDPRLVQEAFTVIQEQA 437
KG +Y +WE+L G+++ K + K ++ K +S +K W P L++ +EQ
Sbjct: 390 KGAHPNYKTWEILMEGFVQSKQMDKAVNAMKKGLSLLKTCHWRPPLELLEAIAKYFEEQG 449
Query: 438 HIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLLD 497
+E A++ + +L+ + +Y L Y A +P + + + D + +D+E +L+
Sbjct: 450 SVEDADRFIKVLQKFNLTSLPLYKSLLGAYINADIVPQNIPQMIAGDQIDMDEEMDQLII 509
Query: 498 LTSKMCVS 505
SK+ ++
Sbjct: 510 RASKIDIT 517
>UniRef100_Q8LNF7 Putative leaf protein [Oryza sativa]
Length = 513
Score = 236 bits (602), Expect = 1e-60
Identities = 153/473 (32%), Positives = 252/473 (52%), Gaps = 22/473 (4%)
Query: 34 GGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCE 93
G D+L RR+ P+ + +W K +L +++ LR+ R+ ALE+
Sbjct: 48 GEDSLFRRVAG-ADPRIPLAPVLEQWWLAEERPVSKPELQSLVKYLRRRCRFSQALELSM 106
Query: 94 WMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNL 153
WM + + L GD A +L+LI+KV GL+ A ++F+ +P+++R +LL Y +
Sbjct: 107 WMTERRHLHLSPGDVAYRLELISKVHGLDKAVEYFDAVPNQLRELQCYGSLLRCYAEAER 166
Query: 154 TNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVN-TSPDVVTFNL 212
KAE L M G S YN MM+LY G++E+V +++ ++ PD+ T +
Sbjct: 167 VEKAEELFENMRGMGMANS-YAYNAMMNLYSQIGQVERVDSMYKAMEEGGIVPDIFTIDN 225
Query: 213 LLTACASENDVETAERVLLQLKKAKVDP--DWVTYSTLTNLYIRNASVDDCLEKAASTLK 270
L++A A DVE E+VL +KA + W +++ + ++++ E+A +
Sbjct: 226 LVSAYADVEDVEAIEKVL---EKASCNNLMSWHSFAIVGKVFMKAGMQ----ERALQAFQ 278
Query: 271 EMEKRTS--RETRVAYSSLLSLHANMGNVDEVNRIW----GKMKACFCKMSDDEYVCMIS 324
E EKR + ++ RVAY LL+++A++ EV+RIW K+ A C Y+C IS
Sbjct: 279 ESEKRITARKDGRVAYGFLLTMYADLQMDSEVDRIWDVYRSKVPASACNTM---YMCRIS 335
Query: 325 SLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCL 384
L+K+ D G E Y+EWES +D R+ N+LLT+Y +G ME AE +Q ++KG L
Sbjct: 336 VLLKMNDIVGAEKAYEEWESKHVYHDSRLINILLTAYCKEGLMEKAEALVDQFIKKGRTL 395
Query: 385 SYSSWELLTRGYLKKKDVKKFLHYFGKAISSV-KQWIPDPRLVQEAFTVIQEQAHIEGAE 443
++W L GY K V K KA++S +W PD V + EQ ++E AE
Sbjct: 396 FSNTWYKLAGGYFKVGQVSKAADLTKKALASASNEWKPDLANVLMSINYFAEQKNVEAAE 455
Query: 444 QLLVILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLL 496
++ +L+ + ++Y+ LKTY AG+ + +RMKKD ++ D+ET ++L
Sbjct: 456 EMASLLQRLVPLTRDVYHGLLKTYVNAGEPASDLLDRMKKDGIEADEETDKIL 508
>UniRef100_Q6ZJE1 Putative DNA-binding protein [Oryza sativa]
Length = 506
Score = 233 bits (594), Expect = 1e-59
Identities = 146/461 (31%), Positives = 243/461 (52%), Gaps = 21/461 (4%)
Query: 16 HLSTEPARRGVTAASS--SSGGDTLGRR----------LLSLVYPKRSAVIAINKWKEEG 63
H ST R AASS S G + GR+ L L +P V I +W +
Sbjct: 31 HGSTPALIRAAAAASSPASPRGHSGGRKPARPPSLQSTLWPLGHPGTLLVPEIERWAAKP 90
Query: 64 HTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNS 123
R +L R+++ELRK +R++ ALEV EWM + +K + D+AV LDLI ++ G ++
Sbjct: 91 GNRLRHVELERIVKELRKRRRHRQALEVSEWMNAKGHVKFLPKDHAVHLDLIGEIHGSSA 150
Query: 124 AEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLY 183
AE +F +LPDK + + ALL+ Y + L K+ A KM E GF+ S +PYN +M LY
Sbjct: 151 AETYFNNLPDKDKTEKPYGALLNCYTRELLVEKSLAHFQKMKELGFVFSTLPYNNIMGLY 210
Query: 184 ISNGKLEKVPKLFEELKVN-TSPDVVTFNLLLTACASENDVETAERVLLQLK-KAKVDPD 241
+ G+ EKVP + E+K N PD ++ + + + + D E L +++ + K+ D
Sbjct: 211 TNLGQHEKVPSVIAEMKSNGIVPDNFSYRICINSYGTRADFFGMENTLEEMECEPKIVVD 270
Query: 242 WVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVN 301
W TY+ + + YI+ + EKA S LK+ E + + + +Y+ L+SL+ ++G+ EVN
Sbjct: 271 WNTYAVVASNYIKG----NIREKAFSALKKAEAKINIKDSDSYNHLISLYGHLGDKSEVN 326
Query: 302 RIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSY 361
R+W + + + +Y M++ LVKL + E L KEWES D +V N+LLT Y
Sbjct: 327 RLWALQMSNCNRHINKDYTTMLAVLVKLNEIEEAEVLLKEWESSGNAFDFQVPNVLLTGY 386
Query: 362 VDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAI---SSVKQ 418
+ ++ AE + ++KG +SW ++ GY +K D K A+ +
Sbjct: 387 RQKDLLDKAEALLDDFLKKGKMPPSTSWAIVAAGYAEKGDAAKAYELTKNALCVYAPNTG 446
Query: 419 WIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNI 459
WIP P +++ + ++ +E E + +L+ A +N+++
Sbjct: 447 WIPRPGMIEMILKYLGDEGDVEEVEIFVDLLKVAVPLNSDM 487
>UniRef100_Q84JR3 Hypothetical protein At4g21705 [Arabidopsis thaliana]
Length = 492
Score = 221 bits (563), Expect = 4e-56
Identities = 139/470 (29%), Positives = 234/470 (49%), Gaps = 11/470 (2%)
Query: 37 TLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMK 96
TL ++ L PK S + W + G + +L R++ +LR+ KR+ HALEV +WM
Sbjct: 26 TLYSKISPLGDPKSSVYPELQNWVQCGKKVSVA-ELIRIVHDLRRRKRFLHALEVSKWMN 84
Query: 97 LQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNK 156
++AV LDLI +V G +AE++FE+L ++ + T ALL+ YV+ K
Sbjct: 85 ETGVCVFSPTEHAVHLDLIGRVYGFVTAEEYFENLKEQYKNDKTYGALLNCYVRQQNVEK 144
Query: 157 AEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELK-VNTSPDVVTFNLLLT 215
+ KM E GF+ S + YN +M LY + G+ EKVPK+ EE+K N +PD ++ + +
Sbjct: 145 SLLHFEKMKEMGFVTSSLTYNNIMCLYTNIGQHEKVPKVLEEMKEENVAPDNYSYRICIN 204
Query: 216 ACASENDVETAERVLLQLKKAK-VDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEK 274
A + D+E L +++ + + DW TY+ YI DC ++A LK E
Sbjct: 205 AFGAMYDLERIGGTLRDMERRQDITMDWNTYAVAAKFYIDGG---DC-DRAVELLKMSEN 260
Query: 275 RTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAG 334
R ++ Y+ L++L+A +G EV R+W K + + +Y+ ++ SLVK+
Sbjct: 261 RLEKKDGEGYNHLITLYARLGKKIEVLRLWDLEKDVCKRRINQDYLTVLQSLVKIDALVE 320
Query: 335 VENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTR 394
E + EW+S D RV N ++ Y+ + E AE L +G + SWEL+
Sbjct: 321 AEEVLTEWKSSGNCYDFRVPNTVIRGYIGKSMEEKAEAMLEDLARRGKATTPESWELVAT 380
Query: 395 GYLKKKDVKKFLHYFGKAIS---SVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRN 451
Y +K ++ A+ ++W P LV + + ++ ++ E + LRN
Sbjct: 381 AYAEKGTLENAFKCMKTALGVEVGSRKWRPGLTLVTSVLSWVGDEGSLKEVESFVASLRN 440
Query: 452 AGHVNTNIYNLFLKTYAAAGKMPL-VVAERMKKDNVQLDKETHRLLDLTS 500
VN +Y+ +K G + + +RMK D +++D+ET +L S
Sbjct: 441 CIGVNKQMYHALVKADIREGGRNIDTLLQRMKDDKIEIDEETTVILSTRS 490
>UniRef100_Q93WC5 AT4g01990/T7B11_26 [Arabidopsis thaliana]
Length = 502
Score = 219 bits (558), Expect = 1e-55
Identities = 135/451 (29%), Positives = 242/451 (52%), Gaps = 12/451 (2%)
Query: 56 INKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLI 115
+N++ EG + +K+ L R ++LRK ++ + ALE+ EWM+ + +I D+A++L+LI
Sbjct: 60 LNQFVMEGVPV-KKHDLIRYAKDLRKFRQPQRALEIFEWME-RKEIAFTGSDHAIRLNLI 117
Query: 116 TKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVP 175
K +GL +AE +F L D ++ Q T +LL+ Y KA+A M + + + +P
Sbjct: 118 AKSKGLEAAETYFNSLDDSIKNQSTYGSLLNCYCVEKEEVKAKAHFENMVDLNHVSNSLP 177
Query: 176 YNRMMSLYISNGKLEKVPKLFEELKVNT-SPDVVTFNLLLTACASENDVETAERVLLQLK 234
+N +M++Y+ G+ EKVP L +K + +P +T+++ + +C S D++ E+VL ++K
Sbjct: 178 FNNLMAMYMGLGQPEKVPALVVAMKEKSITPCDITYSMWIQSCGSLKDLDGVEKVLDEMK 237
Query: 235 -KAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHAN 293
+ + W T++ L +YI+ KA LK +E + + R Y L++L+
Sbjct: 238 AEGEGIFSWNTFANLAAIYIKVGLYG----KAEEALKSLENNMNPDVRDCYHFLINLYTG 293
Query: 294 MGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRV 353
+ N EV R+W +K + +++ Y+ M+ +L KL D GV+ ++ EWES T D+R+
Sbjct: 294 IANASEVYRVWDLLKKRYPNVNNSSYLTMLRALSKLDDIDGVKKVFAEWESTCWTYDMRM 353
Query: 354 SNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAI 413
+N+ ++SY+ Q E AE N ++K + +LL LK L +F A+
Sbjct: 354 ANVAISSYLKQNMYEEAEAVFNGAMKKCKGQFSKARQLLMMHLLKNDQADLALKHFEAAV 413
Query: 414 -SSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAGK 472
K W L+ F +E ++GAE+ L +++ Y L +KTY AAGK
Sbjct: 414 LDQDKNWTWSSELISSFFLHFEEAKDVDGAEEFCKTLTKWSPLSSETYTLLMKTYLAAGK 473
Query: 473 MPLVVAERMKKDNVQLDKETHRLLDLTSKMC 503
+ +R+++ + +D+E LL SK+C
Sbjct: 474 ACPDMKKRLEEQGILVDEEQECLL---SKIC 501
>UniRef100_Q94CZ7 P0682B08.20 protein [Oryza sativa]
Length = 504
Score = 219 bits (558), Expect = 1e-55
Identities = 133/461 (28%), Positives = 241/461 (51%), Gaps = 12/461 (2%)
Query: 43 LSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIK 102
LS P V + +W E L + Q+ +R RK K+ KHAL++ +WM+ + +
Sbjct: 47 LSAHLPSGGMVEELGRWLRERRPLSEE-QVLFCVRRFRKFKQNKHALQLMDWMEARG-VN 104
Query: 103 LVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMS 162
L +A++LDL++K+ G+++AE++F LPD R + T + LL+ Y ++ + K L
Sbjct: 105 LELKHHALRLDLVSKLNGIHAAEEYFGSLPDIFRSKQTYSTLLNCYAEHRMAEKGLELYE 164
Query: 163 KMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVN-TSPDVVTFNLLLTACASEN 221
M + + YN +M LY+ + EK+P +++ + P+ ++ +L + N
Sbjct: 165 NMKAMNIVSDILVYNNLMCLYLKTDQPEKIPTTVVKMQESGIQPNKFSYFVLTESYIMMN 224
Query: 222 DVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETR 281
D+E+AE+VL +L++ P W Y+TL N Y + D KA TLK+ E+ +
Sbjct: 225 DIESAEKVLKELQEVNSVP-WSLYATLANGYNKLQQFD----KAEFTLKKAEEVLDKHDV 279
Query: 282 VAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKE 341
++ LLS +AN GN+ EV RIW +K+ F K ++ Y+ M+ +L KL DF ++ +++E
Sbjct: 280 FSWHCLLSHYANSGNLSEVKRIWESLKSAFKKCTNRSYLVMLKALKKLDDFDTLQQIFQE 339
Query: 342 WESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKD 401
WES D+++ N+++ +Y+D+G ++ AE Q +Y ++ + YL+K
Sbjct: 340 WESSHEHYDMKIPNIIIQAYLDKGMVDKAEAM-RQTTMAQDHSNYRTFCIFAEFYLEKSK 398
Query: 402 VKKFLHYFGKAISSVK--QWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNI 459
+ + L + A VK W+P+ +LV ++ ++G E L+N G ++
Sbjct: 399 MNEALQVWKDAKKMVKGQDWVPE-KLVNRYLKHFEDSKDVDGMETFCECLKNLGRLDAEA 457
Query: 460 YNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLLDLTS 500
Y ++TY + G+ + +RM+ D V + E L S
Sbjct: 458 YEALIRTYISVGRTNPSIPQRMEVDRVDIRPEMFESLKAIS 498
>UniRef100_Q7XV52 OSJNBa0086B14.3 protein [Oryza sativa]
Length = 430
Score = 211 bits (537), Expect = 4e-53
Identities = 127/370 (34%), Positives = 197/370 (52%), Gaps = 14/370 (3%)
Query: 22 ARRGVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRK 81
A T A D L R+ L KRSA A+ +W EG +L + R+L +
Sbjct: 62 AAAAATPADVGGDEDDLRSRVFRLRLAKRSATAALERWAGEGRAASAA-ELRGIARDLSR 120
Query: 82 NKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTC 141
RYKHALEV EWMK ++ L E DY +++DLIT+V G N+AE FFE LP ++
Sbjct: 121 AGRYKHALEVAEWMKTHHESDLSENDYGMRIDLITRVFGANAAEDFFEKLPACVQSLEAY 180
Query: 142 TALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELK- 200
TALLH+Y ++ +T+KAE L +M + + + YN MM+LYIS G+L+KVP + EELK
Sbjct: 181 TALLHSYARSKMTDKAERLFKRMKDANLSMNILVYNEMMTLYISVGELDKVPVIAEELKR 240
Query: 201 VNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPD----WVTYSTLTNLYIRNA 256
SPD+ T+NL ++A A+ D+E + +L ++ K DP+ W Y L +Y+
Sbjct: 241 QKFSPDLFTYNLRISASAASMDLEGFKGILDEMSK---DPNSNEGWKLYQNLAVIYVDAG 297
Query: 257 SVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSD 316
L + ++L E E + SR + Y L+ LH +GN D + IW M +M+
Sbjct: 298 Q----LVGSGNSLVEAEAKISRREWITYDFLVILHTGLGNRDRIKDIWKSMLMTSQRMTS 353
Query: 317 DEYVCMISSLVKLGDFAGVENLYKEWE-SVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCN 375
Y+C++SS + G + +W+ S + D+ N L ++++ G + A F
Sbjct: 354 RNYICVLSSYLMCGQLKDAGEVIDQWQRSKAPEFDISACNRLFDAFLNAGFTDTANSFRE 413
Query: 376 QLVEKGVCLS 385
+++K L+
Sbjct: 414 LMLQKSCILT 423
>UniRef100_Q94B59 Hypothetical protein T5E8_250 [Arabidopsis thaliana]
Length = 409
Score = 209 bits (533), Expect = 1e-52
Identities = 126/354 (35%), Positives = 200/354 (55%), Gaps = 10/354 (2%)
Query: 25 GVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKR 84
G + S D L R+ L PKRSA + KW EG+ + +L + +ELR+ +R
Sbjct: 44 GNSLVEESEEKDDLKSRIFRLRLPKRSATTVLEKWIGEGNQMTIN-ELREISKELRRTRR 102
Query: 85 YKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTAL 144
YKHALEV EWM + K+ + DYA ++DLI+KV G+++AE++FE L + T T+L
Sbjct: 103 YKHALEVTEWMVQHEESKISDADYASRIDLISKVFGIDAAERYFEGLDIDSKTAETYTSL 162
Query: 145 LHAYVQNNLTNKAEALMSKMSECGFLR-SPVPYNRMMSLYISNGKLEKVPKLFEELK-VN 202
LHAY + T +AEAL ++ E L + YN MM+LY+S G++EKVP++ E LK
Sbjct: 163 LHAYAASKQTERAEALFKRIIESDSLTFGAITYNEMMTLYMSVGQVEKVPEVIEVLKQKK 222
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLK-KAKVDPDWVTYSTLTNLYIRNASVDDC 261
SPD+ T+NL L++CA+ +++ ++L +++ A + WV Y LT++YI ++ V +
Sbjct: 223 VSPDIFTYNLWLSSCAATFNIDELRKILEEMRHDASSNEGWVRYIDLTSIYINSSRVTN- 281
Query: 262 LEKAASTLK-EMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYV 320
A STL E EK S+ + Y L+ LH +GN +++IW ++ +S Y+
Sbjct: 282 ---AESTLPVEAEKSISQREWITYDFLMILHTGLGNKVMIDQIWKSLRNTNQILSSRSYI 338
Query: 321 CMISSLVKLGDFAGVENLYKEW-ESVSGTNDVRVSNLLLTSYVDQGQMEMAEIF 373
C++SS + LG E + +W ES + D +L ++ D G +A F
Sbjct: 339 CVLSSYLMLGHLREAEEIIHQWKESKTTEFDASACLRILNAFRDVGLEGIASGF 392
>UniRef100_Q8LCU1 Hypothetical protein [Arabidopsis thaliana]
Length = 409
Score = 209 bits (532), Expect = 2e-52
Identities = 126/354 (35%), Positives = 200/354 (55%), Gaps = 10/354 (2%)
Query: 25 GVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKR 84
G + S D L R+ L PKRSA + KW EG+ + +L + +ELR+ +R
Sbjct: 44 GNSLVEESEEKDDLKSRIFRLRLPKRSATTVLEKWIGEGNQMTIN-ELREISKELRRTRR 102
Query: 85 YKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTAL 144
YKHALEV EWM + K+ + DYA ++DLI+KV G+++AE++FE L + T T+L
Sbjct: 103 YKHALEVTEWMVQHEESKISDADYASRIDLISKVFGIDAAERYFEGLHIDSKTAETYTSL 162
Query: 145 LHAYVQNNLTNKAEALMSKMSECGFLR-SPVPYNRMMSLYISNGKLEKVPKLFEELK-VN 202
LHAY + T +AEAL ++ E L + YN MM+LY+S G++EKVP++ E LK
Sbjct: 163 LHAYAASKQTERAEALFKRIIESDSLTFGAITYNEMMTLYMSVGQVEKVPEVIEVLKQKK 222
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLK-KAKVDPDWVTYSTLTNLYIRNASVDDC 261
SPD+ T+NL L++CA+ +++ ++L +++ A + WV Y LT++YI ++ V +
Sbjct: 223 VSPDIFTYNLWLSSCAATFNIDELRKILEEMRHDASSNEGWVRYIDLTSIYINSSRVTN- 281
Query: 262 LEKAASTLK-EMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYV 320
A STL E EK S+ + Y L+ LH +GN +++IW ++ +S Y+
Sbjct: 282 ---AESTLPVEAEKSISQREWITYDFLMILHTGLGNKVMIDQIWKSLRNTNQILSSRSYI 338
Query: 321 CMISSLVKLGDFAGVENLYKEW-ESVSGTNDVRVSNLLLTSYVDQGQMEMAEIF 373
C++SS + LG E + +W ES + D +L ++ D G +A F
Sbjct: 339 CVLSSYLMLGHLREAEEIIHQWKESKTTEFDASACLRILNAFRDVGLEGIASGF 392
>UniRef100_Q9FZ24 T6A9.6 protein [Arabidopsis thaliana]
Length = 537
Score = 206 bits (525), Expect = 1e-51
Identities = 140/481 (29%), Positives = 247/481 (51%), Gaps = 13/481 (2%)
Query: 23 RRGVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKN 82
R V A + +S L ++L L + +N++ EG T+ RK L R + LRK
Sbjct: 58 RTSVAAPTVASRQRELYKKLSMLSVTGGTVAETLNQFIMEGITV-RKDDLFRCAKTLRKF 116
Query: 83 KRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDL-PDKMRGQPTC 141
+R +HA E+ +WM+ + + D+A+ LDLI K +GL +AE +F +L P Q T
Sbjct: 117 RRPQHAFEIFDWME-KRKMTFSVSDHAICLDLIGKTKGLEAAENYFNNLDPSAKNHQSTY 175
Query: 142 TALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV 201
AL++ Y KA+A M E F+ + +P+N MMS+Y+ + EKVP L + +K
Sbjct: 176 GALMNCYCVELEEEKAKAHFEIMDELNFVNNSLPFNNMMSMYMRLSQPEKVPVLVDAMKQ 235
Query: 202 N-TSPDVVTFNLLLTACASENDVETAERVLLQL-KKAKVDPDWVTYSTLTNLYIRNASVD 259
SP VT+++ + +C S ND++ E+++ ++ K ++ W T+S L +Y +
Sbjct: 236 RGISPCGVTYSIWMQSCGSLNDLDGLEKIIDEMGKDSEAKTTWNTFSNLAAIYTKAG--- 292
Query: 260 DCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEY 319
EKA S LK ME++ + R ++ L+SL+A + EV R+W +K ++++ Y
Sbjct: 293 -LYEKADSALKSMEEKMNPNNRDSHHFLMSLYAGISKGPEVYRVWESLKKARPEVNNLSY 351
Query: 320 VCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVE 379
+ M+ ++ KLGD G++ ++ EWES D+R++N+ + +Y+ E AE + ++
Sbjct: 352 LVMLQAMSKLGDLDGIKKIFTEWESKCWAYDMRLANIAINTYLKGNMYEEAEKILDGAMK 411
Query: 380 KGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAIS----SVKQWIPDPRLVQEAFTVIQE 435
K + +LL L+ + + A+S + +W LV F ++
Sbjct: 412 KSKGPFSKARQLLMIHLLENDKADLAMKHLEAAVSDSAENKDEWGWSSELVSLFFLHFEK 471
Query: 436 QAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRL 495
++GAE IL N +++ +KTYAAA K + ER+ + +++ +E L
Sbjct: 472 AKDVDGAEDFCKILSNWKPLDSETMTFLIKTYAAAEKTSPDMRERLSQQQIEVSEEIQDL 531
Query: 496 L 496
L
Sbjct: 532 L 532
>UniRef100_Q8VYY9 Hypothetical protein At1g02370 [Arabidopsis thaliana]
Length = 537
Score = 206 bits (525), Expect = 1e-51
Identities = 140/481 (29%), Positives = 247/481 (51%), Gaps = 13/481 (2%)
Query: 23 RRGVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKN 82
R V A + +S L ++L L + +N++ EG T+ RK L R + LRK
Sbjct: 58 RTSVAAPTVASRQRELYKKLSMLSVTGGTVAETLNQFIMEGITV-RKDDLFRCAKTLRKF 116
Query: 83 KRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDL-PDKMRGQPTC 141
+R +HA E+ +WM+ + + D+A+ LDLI K +GL +AE +F +L P Q T
Sbjct: 117 RRPQHAFEIFDWME-KRKMTFSVSDHAICLDLIGKTKGLEAAENYFNNLDPSAKNHQSTY 175
Query: 142 TALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV 201
AL++ Y KA+A M E F+ + +P+N MMS+Y+ + EKVP L + +K
Sbjct: 176 GALMNCYCVELEEEKAKAHFEIMDELNFVNNSLPFNNMMSMYMRLSQPEKVPVLVDAIKQ 235
Query: 202 N-TSPDVVTFNLLLTACASENDVETAERVLLQL-KKAKVDPDWVTYSTLTNLYIRNASVD 259
SP VT+++ + +C S ND++ E+++ ++ K ++ W T+S L +Y +
Sbjct: 236 RGISPCGVTYSIWMQSCGSLNDLDGLEKIIDEMGKDSEAKTTWNTFSNLAAIYTKAG--- 292
Query: 260 DCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEY 319
EKA S LK ME++ + R ++ L+SL+A + EV R+W +K ++++ Y
Sbjct: 293 -LYEKADSALKSMEEKMNPNNRDSHHFLMSLYAGISKGPEVYRVWESLKKARPEVNNLSY 351
Query: 320 VCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVE 379
+ M+ ++ KLGD G++ ++ EWES D+R++N+ + +Y+ E AE + ++
Sbjct: 352 LVMLQAMSKLGDLDGIKKIFTEWESKCWAYDMRLANIAINTYLKGNMYEEAEKILDGAMK 411
Query: 380 KGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAIS----SVKQWIPDPRLVQEAFTVIQE 435
K + +LL L+ + + A+S + +W LV F ++
Sbjct: 412 KSKGPFSKARQLLMIHLLENDKADLAMKHLEAAVSDSAENKDEWGWSSELVSLFFLHFEK 471
Query: 436 QAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRL 495
++GAE IL N +++ +KTYAAA K + ER+ + +++ +E L
Sbjct: 472 AKDVDGAEDFCKILSNWKPLDSETMTFLIKTYAAAEKTSPDMRERLSQQQIEVSEEIQDL 531
Query: 496 L 496
L
Sbjct: 532 L 532
>UniRef100_Q9FY70 Hypothetical protein T5E8_250 [Arabidopsis thaliana]
Length = 402
Score = 195 bits (495), Expect = 3e-48
Identities = 122/354 (34%), Positives = 196/354 (54%), Gaps = 17/354 (4%)
Query: 25 GVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKR 84
G + S D L R+ L PKRSA + KW EG+ + +L + +ELR+ +R
Sbjct: 44 GNSLVEESEEKDDLKSRIFRLRLPKRSATTVLEKWIGEGNQMTIN-ELREISKELRRTRR 102
Query: 85 YKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTAL 144
YKHALE + K+ + DYA ++DLI+KV G+++AE++FE L + T T+L
Sbjct: 103 YKHALE-------HEESKISDADYASRIDLISKVFGIDAAERYFEGLDIDSKTAETYTSL 155
Query: 145 LHAYVQNNLTNKAEALMSKMSECGFLR-SPVPYNRMMSLYISNGKLEKVPKLFEELK-VN 202
LHAY + T +AEAL ++ E L + YN MM+LY+S G++EKVP++ E LK
Sbjct: 156 LHAYAASKQTERAEALFKRIIESDSLTFGAITYNEMMTLYMSVGQVEKVPEVIEVLKQKK 215
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLK-KAKVDPDWVTYSTLTNLYIRNASVDDC 261
SPD+ T+NL L++CA+ +++ ++L +++ A + WV Y LT++YI ++ V +
Sbjct: 216 VSPDIFTYNLWLSSCAATFNIDELRKILEEMRHDASSNEGWVRYIDLTSIYINSSRVTN- 274
Query: 262 LEKAASTLK-EMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYV 320
A STL E EK S+ + Y L+ LH +GN +++IW ++ +S Y+
Sbjct: 275 ---AESTLPVEAEKSISQREWITYDFLMILHTGLGNKVMIDQIWKSLRNTNQILSSRSYI 331
Query: 321 CMISSLVKLGDFAGVENLYKEW-ESVSGTNDVRVSNLLLTSYVDQGQMEMAEIF 373
C++SS + LG E + +W ES + D +L ++ D G +A F
Sbjct: 332 CVLSSYLMLGHLREAEEIIHQWKESKTTEFDASACLRILNAFRDVGLEGIASGF 385
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 806,630,233
Number of Sequences: 2790947
Number of extensions: 32140738
Number of successful extensions: 105671
Number of sequences better than 10.0: 997
Number of HSP's better than 10.0 without gapping: 442
Number of HSP's successfully gapped in prelim test: 570
Number of HSP's that attempted gapping in prelim test: 97797
Number of HSP's gapped (non-prelim): 3784
length of query: 512
length of database: 848,049,833
effective HSP length: 132
effective length of query: 380
effective length of database: 479,644,829
effective search space: 182265035020
effective search space used: 182265035020
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC122725.14


