

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122724.2 + phase: 0
(255 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
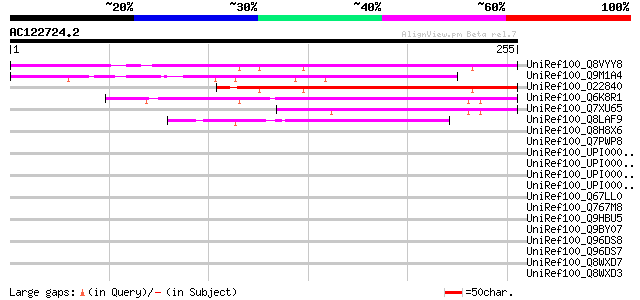
UniRef100_Q8VYY8 Hypothetical protein At2g43630 [Arabidopsis tha... 137 2e-31
UniRef100_Q9M1A4 Hypothetical protein T16L24.190 [Arabidopsis th... 119 1e-25
UniRef100_O22840 Hypothetical protein At2g43630 [Arabidopsis tha... 119 1e-25
UniRef100_Q6K8R1 Putative glycine-rich protein [Oryza sativa] 89 8e-17
UniRef100_Q7XU65 OSJNBb0091E11.5 protein [Oryza sativa] 86 7e-16
UniRef100_Q8LAF9 Hypothetical protein At5g20130 [Arabidopsis tha... 53 9e-06
UniRef100_Q8H8X6 Hypothetical protein OSJNBa0062C05.24 [Oryza sa... 43 0.009
UniRef100_Q7PWP8 ENSANGP00000013932 [Anopheles gambiae str. PEST] 43 0.009
UniRef100_UPI00003693C6 UPI00003693C6 UniRef100 entry 42 0.012
UniRef100_UPI00003693C5 UPI00003693C5 UniRef100 entry 42 0.012
UniRef100_UPI00003693C4 UPI00003693C4 UniRef100 entry 42 0.012
UniRef100_UPI00003693C3 UPI00003693C3 UniRef100 entry 42 0.012
UniRef100_Q67LL0 Hypothetical protein [Symbiobacterium thermophi... 42 0.012
UniRef100_Q767M8 Corneodesmosin [Sus scrofa] 42 0.012
UniRef100_Q9HBU5 Sodium bicarbonate cotransporter-like protein [... 42 0.012
UniRef100_Q9BY07 Sodium bicarbonate cotransporter NBC4a [Homo sa... 42 0.012
UniRef100_Q96DS8 Sodium bicarbonate cotransporter NBC4c [Homo sa... 42 0.012
UniRef100_Q96DS7 Sodium bicarbonate cotransporter NBC4d [Homo sa... 42 0.012
UniRef100_Q8WXD7 Sodium bicarbonate cotransporter NBC4e [Homo sa... 42 0.012
UniRef100_Q8WXD3 Sodium bicarbonate cotransporter 4f [Homo sapiens] 42 0.012
>UniRef100_Q8VYY8 Hypothetical protein At2g43630 [Arabidopsis thaliana]
Length = 274
Score = 137 bits (346), Expect = 2e-31
Identities = 95/274 (34%), Positives = 147/274 (52%), Gaps = 31/274 (11%)
Query: 1 MNAIQVAASNPAIYVRKNYQPHHFLSTPKGLSMNANPLSRTFLIPRAKLSSGLYCVRQSA 60
M+ Q P+ Y + +P+ S P S+ + RT RA + S
Sbjct: 1 MSFTQANCFRPSYYPARITRPNCISSVPIRSSVRFDHFPRTSFTLRATAAV-------ST 53
Query: 61 TTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSED-SQRKSLEEAMKS----- 114
+P+L + + +P KS++ VCL GGK D S++ S K++E+AM
Sbjct: 54 QFSPLL-----DHRRRLPTGKSKQSSAVCLFGGKDKPDGSDEISPWKAIEKAMGKKSVED 108
Query: 115 -LQEKIQKGEY--SGGSGSKPPGGRGRGGGGGNSD------GSEEGSTGGMSDETAQIVF 165
L+E+IQK ++ + G+ PP G G GGGGGN + G E+G G++DET Q+V
Sbjct: 109 MLREQIQKKDFYDTDSGGNMPPRGGGSGGGGGNGEERPEGSGGEDGGLAGIADETLQVVL 168
Query: 166 ATIGFMFVYIYVINGVELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDE 225
AT+GF+F+Y Y+I G EL KLARD+I++L+G ++VRL RA + F +K++ Q DE
Sbjct: 169 ATLGFIFLYTYIITGEELVKLARDYIRFLMGRPKTVRLTRAMDSWNGFLEKMSRQRVYDE 228
Query: 226 DGLETG----PLRWNIADFYRDVLRNYMKPNSNE 255
LE P ++ + YR V++ Y+ NS+E
Sbjct: 229 YWLEKAIINTPTWYDSPEKYRRVIKAYVDSNSDE 262
>UniRef100_Q9M1A4 Hypothetical protein T16L24.190 [Arabidopsis thaliana]
Length = 246
Score = 119 bits (297), Expect = 1e-25
Identities = 84/238 (35%), Positives = 127/238 (53%), Gaps = 29/238 (12%)
Query: 1 MNAIQVAASNPAIYVRKNYQPHHFLSTP--KGLSMNANPLSRTFLIPRAKLSSGLYCVRQ 58
M++ Q P+++ + Q H S P L + PL + L RA SS +
Sbjct: 1 MSSTQANLCRPSLFCARTTQTRHVSSAPFMSSLRFDYRPLPK--LAIRASASSSM----- 53
Query: 59 SATTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSED--SQRKSLEEAM--KS 114
S+ +P+ N Q PV VCL GGK + S + S +++E+AM KS
Sbjct: 54 SSQFSPLQNHRCRNQRQG-PV--------VCLLGGKDKSNGSNELSSTWEAIEKAMGKKS 104
Query: 115 LQEKIQKGEYSGGSGSKPPGGRGRGGGG----GNSDGSEEGSTGGMS---DETAQIVFAT 167
+++ +++ +G PP GRG GGGG N G G GG++ DET Q+V AT
Sbjct: 105 VEDMLREQIQKKDTGGIPPRGRGGGGGGRNGGNNGSGGSSGEDGGLASFGDETLQVVLAT 164
Query: 168 IGFMFVYIYVINGVELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDE 225
+GF+F+Y Y+ING EL +LARD+I+YL+G +SVRL R SRF++K++ + +E
Sbjct: 165 LGFIFLYFYIINGEELFRLARDYIRYLIGRPKSVRLTRVMEGWSRFFEKMSRKKVYNE 222
>UniRef100_O22840 Hypothetical protein At2g43630 [Arabidopsis thaliana]
Length = 174
Score = 119 bits (297), Expect = 1e-25
Identities = 67/163 (41%), Positives = 102/163 (62%), Gaps = 15/163 (9%)
Query: 105 RKSLEEAMKSLQEKIQKGEY--SGGSGSKPPGGRGRGGGGGNSD------GSEEGSTGGM 156
+KS+E+ L+E+IQK ++ + G+ PP G G GGGGGN + G E+G G+
Sbjct: 3 KKSVEDM---LREQIQKKDFYDTDSGGNMPPRGGGSGGGGGNGEERPEGSGGEDGGLAGI 59
Query: 157 SDETAQIVFATIGFMFVYIYVINGVELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKK 216
+DET Q+V AT+GF+F+Y Y+I G EL KLARD+I++L+G ++VRL RA + F +K
Sbjct: 60 ADETLQVVLATLGFIFLYTYIITGEELVKLARDYIRFLMGRPKTVRLTRAMDSWNGFLEK 119
Query: 217 ITGQNEVDEDGLETG----PLRWNIADFYRDVLRNYMKPNSNE 255
++ Q DE LE P ++ + YR V++ Y+ NS+E
Sbjct: 120 MSRQRVYDEYWLEKAIINTPTWYDSPEKYRRVIKAYVDSNSDE 162
>UniRef100_Q6K8R1 Putative glycine-rich protein [Oryza sativa]
Length = 250
Score = 89.4 bits (220), Expect = 8e-17
Identities = 72/228 (31%), Positives = 105/228 (45%), Gaps = 27/228 (11%)
Query: 49 LSSGLYCVRQSATTAPVLK--VYPSNLTQSVPVLKSQKPRH-VCLAGGKGMMDNSEDSQR 105
LS Y + + P L+ VYP + V V SQK +C + GK + D
Sbjct: 28 LSHPQYSLTTHSVRFPKLQKQVYP----RLVLVAASQKKLPPLCASSGKVNPEAENDPFM 83
Query: 106 KSLEEAMKS----------LQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+SL++AM L+E+I K G G G R R GG G+S G E+ S
Sbjct: 84 ESLKKAMDDAKKPRPIQDLLKEQIAKLREQGSGGGG--GNRNRRGGSGDSGGPEDESFKE 141
Query: 156 MSDETAQIVFATIGFMFVYIYVINGVELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYK 215
DE Q++ AT+ F+ VYI++I G EL +LARD+ +YL+ G ++ RLKRA K F +
Sbjct: 142 SLDELVQVILATVAFILVYIHIIRGEELYRLARDYTRYLVTGKRTARLKRAMQKWRNFSE 201
Query: 216 KITGQNEVDEDGLE----TGPLRW----NIADFYRDVLRNYMKPNSNE 255
ED E + P W ++ R +P++ E
Sbjct: 202 SFMQSEGSQEDQYERAATSKPTWWQQPQKFVHLMEELCRGNWRPHAQE 249
>UniRef100_Q7XU65 OSJNBb0091E11.5 protein [Oryza sativa]
Length = 337
Score = 86.3 bits (212), Expect = 7e-16
Identities = 49/136 (36%), Positives = 73/136 (53%), Gaps = 15/136 (11%)
Query: 135 GRGRGGGGGNSDGSEEGSTGGMSDET--------AQIVFATIGFMFVYIYVINGVELTKL 186
G G GG GG + S EGS GG DE+ Q++ ATI F+ +YI++I G EL +L
Sbjct: 201 GSGSGGNGGGNKNSHEGSGGGSEDESLTESLYEMVQVLLATIAFILMYIHIIRGEELYRL 260
Query: 187 ARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDEDGLE---TGPLRW----NIAD 239
ARD+ +YL+ G ++ RLKRA F + IT ++ V E E + P+ W
Sbjct: 261 ARDYTRYLVTGKRTSRLKRAMLNWHNFCEGITNKDSVQESTFERSTSEPMWWQQPLKFVH 320
Query: 240 FYRDVLRNYMKPNSNE 255
++ R Y +P++ E
Sbjct: 321 RIEELYRGYFRPHAQE 336
>UniRef100_Q8LAF9 Hypothetical protein At5g20130 [Arabidopsis thaliana]
Length = 202
Score = 52.8 bits (125), Expect = 9e-06
Identities = 45/149 (30%), Positives = 64/149 (42%), Gaps = 15/149 (10%)
Query: 80 LKSQKPRHVCLAGGKGMMDNSEDSQRKSLEEAM-------KSLQEKIQKGEYSGGSGSKP 132
+ K R CL G + E+ RKSLE A+ + ++I+K E SGG
Sbjct: 39 ITKSKGRFSCLFSGG---NQREEQARKSLESALGGKKNEFEKWDKEIKKREESGGGN--- 92
Query: 133 PGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVINGVELTKLARDFIK 192
GG+G GGGGG G S E QI F + + VY+ V G + + +
Sbjct: 93 -GGKG-GGGGGWFGGGGWFSGDHFWKEAQQIAFTLLAILAVYMVVAKGEVMAAFVLNPLL 150
Query: 193 YLLGGTQSVRLKRASYKCSRFYKKITGQN 221
Y L GT+ +S R K++G N
Sbjct: 151 YALRGTREGLSSLSSKLMGREASKVSGDN 179
>UniRef100_Q8H8X6 Hypothetical protein OSJNBa0062C05.24 [Oryza sativa]
Length = 467
Score = 42.7 bits (99), Expect = 0.009
Identities = 26/71 (36%), Positives = 34/71 (47%), Gaps = 5/71 (7%)
Query: 92 GGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEG 151
GG+G M E+ Q K + + +K +KI K +GG G GG G GG + GS
Sbjct: 28 GGQGQMHRGEEKQHKPVLKKVKEKVKKI-KNTIAGGGG----GGHGGNNGGERASGSSSS 82
Query: 152 STGGMSDETAQ 162
S G D AQ
Sbjct: 83 SEEGEDDVAAQ 93
>UniRef100_Q7PWP8 ENSANGP00000013932 [Anopheles gambiae str. PEST]
Length = 405
Score = 42.7 bits (99), Expect = 0.009
Identities = 34/104 (32%), Positives = 42/104 (39%), Gaps = 5/104 (4%)
Query: 56 VRQSATTAPVLKVYPSNLTQSVPV-LKSQKPRHVCLAGGKGMMDNSEDSQRKSLEEAMKS 114
V + PV + YP + + VPV + + P H +GG G S A S
Sbjct: 272 VIEKPVPVPVDRPYPVYIEKEVPVTVVKEVPVH---SGGGGGGGGYGGGAGHSSASASAS 328
Query: 115 LQEKIQKGEYSGGSGSKPPGGRGRGGGG-GNSDGSEEGSTGGMS 157
G + GG G GG G GGGG GN G G GG S
Sbjct: 329 AHASASAGSFGGGYGGSNGGGYGGGGGGSGNVGGGYGGGHGGGS 372
>UniRef100_UPI00003693C6 UPI00003693C6 UniRef100 entry
Length = 1120
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 427 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 470
>UniRef100_UPI00003693C5 UPI00003693C5 UniRef100 entry
Length = 1073
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 427 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 470
>UniRef100_UPI00003693C4 UPI00003693C4 UniRef100 entry
Length = 1136
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 427 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 470
>UniRef100_UPI00003693C3 UPI00003693C3 UniRef100 entry
Length = 1039
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 427 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 470
>UniRef100_Q67LL0 Hypothetical protein [Symbiobacterium thermophilum]
Length = 134
Score = 42.4 bits (98), Expect = 0.012
Identities = 37/112 (33%), Positives = 47/112 (41%), Gaps = 17/112 (15%)
Query: 52 GLYCVRQ---SATTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSEDSQRKSL 108
G VRQ S A VL V ++ +P + + + GG ED +++
Sbjct: 33 GTVRVRQRLFSLMLALVLAVGAVTVSGCMPQQQPESQQQHSQGGG-------EDREKQER 85
Query: 109 EEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDET 160
EE K +EK E GG G K G G G GGG G GS GG D T
Sbjct: 86 EEGEKDQEEK----ETGGGKGGKSGGSGGSGQGGGQQQG---GSGGGSQDST 130
>UniRef100_Q767M8 Corneodesmosin [Sus scrofa]
Length = 262
Score = 42.4 bits (98), Expect = 0.012
Identities = 27/77 (35%), Positives = 30/77 (38%), Gaps = 9/77 (11%)
Query: 85 PRHVCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGN 144
P CL G G S S S ++ S SG SG G G GGG G
Sbjct: 51 PNDPCLLGKGGSSSFSSHSSSSSSSSSISSP---------SGSSGGSHGGSSGSGGGSGG 101
Query: 145 SDGSEEGSTGGMSDETA 161
S GS GS+GG A
Sbjct: 102 SSGSSSGSSGGSGGSIA 118
>UniRef100_Q9HBU5 Sodium bicarbonate cotransporter-like protein [Homo sapiens]
Length = 1074
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 428 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 471
>UniRef100_Q9BY07 Sodium bicarbonate cotransporter NBC4a [Homo sapiens]
Length = 1137
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 428 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 471
>UniRef100_Q96DS8 Sodium bicarbonate cotransporter NBC4c [Homo sapiens]
Length = 1121
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 428 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 471
>UniRef100_Q96DS7 Sodium bicarbonate cotransporter NBC4d [Homo sapiens]
Length = 1040
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 428 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 471
>UniRef100_Q8WXD7 Sodium bicarbonate cotransporter NBC4e [Homo sapiens]
Length = 1051
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 428 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 471
>UniRef100_Q8WXD3 Sodium bicarbonate cotransporter 4f [Homo sapiens]
Length = 760
Score = 42.4 bits (98), Expect = 0.012
Identities = 21/44 (47%), Positives = 23/44 (51%)
Query: 114 SLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SL E Q GG G P GG G GGGGG+ G+ G GG S
Sbjct: 428 SLAELGQMNGSVGGGGGAPGGGNGGGGGGGSGGGAGSGGAGGTS 471
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 454,947,166
Number of Sequences: 2790947
Number of extensions: 20339378
Number of successful extensions: 208676
Number of sequences better than 10.0: 1645
Number of HSP's better than 10.0 without gapping: 912
Number of HSP's successfully gapped in prelim test: 819
Number of HSP's that attempted gapping in prelim test: 166631
Number of HSP's gapped (non-prelim): 21712
length of query: 255
length of database: 848,049,833
effective HSP length: 125
effective length of query: 130
effective length of database: 499,181,458
effective search space: 64893589540
effective search space used: 64893589540
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC122724.2


