

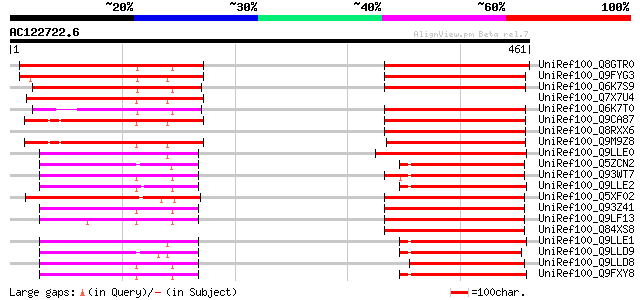
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.6 + phase: 0
(461 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GTR0 Sugar transporter [Citrus unshiu] 216 8e-55
UniRef100_Q9FYG3 F1N21.12 [Arabidopsis thaliana] 196 9e-49
UniRef100_Q6K7S9 Putative sugar transporter [Oryza sativa] 183 1e-44
UniRef100_Q7X7U4 Putative hexose transporter protein [Arabidopsi... 181 3e-44
UniRef100_Q6K7T0 Putative sugar transporter [Oryza sativa] 177 4e-43
UniRef100_Q9CA87 Putative sugar transporter; 77409-81599 [Arabid... 167 8e-40
UniRef100_Q8RXX6 Putative hexose transporter protein [Arabidopsi... 167 8e-40
UniRef100_Q9M9Z8 F20B17.24 [Arabidopsis thaliana] 166 2e-39
UniRef100_Q9LLE0 Hexose transporter [Solanum tuberosum] 125 3e-27
UniRef100_Q5ZCN2 Putative hexose transporter [Oryza sativa] 125 3e-27
UniRef100_Q93WT7 Hexose transporter pGlT [Olea europaea] 123 1e-26
UniRef100_Q9LLE2 Hexose transporter [Spinacia oleracea] 123 1e-26
UniRef100_Q5XF02 At1g05030 [Arabidopsis thaliana] 123 1e-26
UniRef100_Q93Z41 AT5g16150/T21H19_70 [Arabidopsis thaliana] 122 3e-26
UniRef100_Q9LF13 Sugar transporter-like protein [Arabidopsis tha... 122 3e-26
UniRef100_Q84XS8 Putative sugar transporter [Brassica rapa subsp... 121 4e-26
UniRef100_Q9LLE1 Hexose transporter [Nicotiana tabacum] 121 5e-26
UniRef100_Q9LLD9 Hexose transporter [Zea mays] 121 5e-26
UniRef100_Q9LLD8 Hexose transporter [Arabidopsis thaliana] 120 8e-26
UniRef100_Q9FXY8 Putative glucose translocator [Mesembryanthemum... 120 1e-25
>UniRef100_Q8GTR0 Sugar transporter [Citrus unshiu]
Length = 489
Score = 216 bits (551), Expect = 8e-55
Identities = 112/174 (64%), Positives = 129/174 (73%), Gaps = 10/174 (5%)
Query: 9 LLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEG 68
L+ N + E TNPSWKLS PHVLVAT++SFLFGYHLGVVNEPLESIS+DLGFNGNTLAEG
Sbjct: 32 LVQNGTEVENTNPSWKLSFPHVLVATLSSFLFGYHLGVVNEPLESISLDLGFNGNTLAEG 91
Query: 69 LVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS--NKQLVWHACRKIIC 126
LVVS+CLGGA G LSGWIAD VGRRRAFQLCALPMIIGA++S + L+ + +
Sbjct: 92 LVVSMCLGGAFIGSTLSGWIADGVGRRRAFQLCALPMIIGASISATTRNLIGMLLGRFVV 151
Query: 127 WNWLGLGPSRCCSLCDR--------AYGALIQIATCFGILGSLFIGIPVKEISG 172
+GLGP+ YGA IQIATC G++GSL IGIPVKEI+G
Sbjct: 152 GTGMGLGPTVAALYVTEVSPPFVRGTYGAFIQIATCLGLMGSLLIGIPVKEIAG 205
Score = 207 bits (527), Expect = 5e-52
Identities = 104/128 (81%), Positives = 114/128 (88%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+SM IQ +S+ +P + +LYLSVGGML+FV TFALGAGPVP LLL EIFPSRIRAKAM
Sbjct: 362 AVSMAIQVAASSSYIPGSASLYLSVGGMLMFVLTFALGAGPVPSLLLPEIFPSRIRAKAM 421
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A CMSVHWVINFFVGLLFLRLLE+LG QLLYS+F TFC+MAV FVKRNVVETKGKSLQEI
Sbjct: 422 AVCMSVHWVINFFVGLLFLRLLEQLGPQLLYSIFGTFCLMAVAFVKRNVVETKGKSLQEI 481
Query: 454 EIALLPQE 461
EIALLPQE
Sbjct: 482 EIALLPQE 489
>UniRef100_Q9FYG3 F1N21.12 [Arabidopsis thaliana]
Length = 493
Score = 196 bits (499), Expect = 9e-49
Identities = 105/176 (59%), Positives = 125/176 (70%), Gaps = 12/176 (6%)
Query: 9 LLDNFIDKE--TTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLA 66
LL+N +D E TTNPSWK SLPHVLVATI+SFLFGYHLGVVNEPLESIS DLGF+G+TLA
Sbjct: 32 LLENDVDNEMETTNPSWKCSLPHVLVATISSFLFGYHLGVVNEPLESISSDLGFSGDTLA 91
Query: 67 EGLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS--NKQLVWHACRKI 124
EGLVVS+CLGGA G L SG +AD GRRRAFQ+CALPMI+GA +S + L +
Sbjct: 92 EGLVVSVCLGGAFLGSLFSGGVADGFGRRRAFQICALPMILGAFVSGVSNSLAVMLLGRF 151
Query: 125 ICWNWLGLGPSRCC--------SLCDRAYGALIQIATCFGILGSLFIGIPVKEISG 172
+ +GLGP + YG+ IQIATC G++ +LFIGIPV I+G
Sbjct: 152 LVGTGMGLGPPVAALYVTEVSPAFVRGTYGSFIQIATCLGLMAALFIGIPVHNITG 207
Score = 196 bits (497), Expect = 2e-48
Identities = 99/125 (79%), Positives = 110/125 (87%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A +M +Q S+ LP AL LSVGG L+FV TFALGAGPVPGLLL EIFPSRIRAKAM
Sbjct: 366 AAAMALQVGATSSYLPHFSALCLSVGGTLVFVLTFALGAGPVPGLLLPEIFPSRIRAKAM 425
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
AFCMSVHWVINFFVGLLFLRLLEKLG +LLYSMF+TFC+MAV+FVKRNV+ETKGK+LQEI
Sbjct: 426 AFCMSVHWVINFFVGLLFLRLLEKLGPRLLYSMFSTFCLMAVMFVKRNVIETKGKTLQEI 485
Query: 454 EIALL 458
EI+LL
Sbjct: 486 EISLL 490
>UniRef100_Q6K7S9 Putative sugar transporter [Oryza sativa]
Length = 481
Score = 183 bits (464), Expect = 1e-44
Identities = 99/160 (61%), Positives = 111/160 (68%), Gaps = 10/160 (6%)
Query: 21 PSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALF 80
PSW++SLPHV VAT+TSFLFGYH GVVNEPLESIS DLGF GNTLAEGLVVSICLGGA
Sbjct: 36 PSWRMSLPHVCVATLTSFLFGYHSGVVNEPLESISTDLGFAGNTLAEGLVVSICLGGAFV 95
Query: 81 GCLLSGWIADAVGRRRAFQLCALPMIIGAAMS--NKQLVWHACRKIICWNWLGLGPSRCC 138
GCL SG IAD +GRRRAFQL ALPMIIGAA+S L + + +GLGP
Sbjct: 96 GCLFSGSIADGIGRRRAFQLSALPMIIGAAVSALTNSLEGMLLGRFLVGTGMGLGPPVAS 155
Query: 139 SLCDR--------AYGALIQIATCFGILGSLFIGIPVKEI 170
YG+ +QIATC GI+ SL IG PVK+I
Sbjct: 156 LYITEVSPPSVRGTYGSFVQIATCLGIVVSLLIGTPVKDI 195
Score = 177 bits (450), Expect = 4e-43
Identities = 87/125 (69%), Positives = 109/125 (86%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A +M +QA GA+ + ++YLSVGGMLLFV TF+LGAGPVPGLLL EIFP++IRAKAM
Sbjct: 354 AFAMGLQAVGANRHHLGSASVYLSVGGMLLFVLTFSLGAGPVPGLLLPEIFPNKIRAKAM 413
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A CMSVHWV+NFFV LLFLRLLE+LG Q+LY+MF++ C++A IFV+R+VVETKGK+LQEI
Sbjct: 414 ALCMSVHWVVNFFVSLLFLRLLEQLGPQVLYTMFSSACVVAAIFVRRHVVETKGKTLQEI 473
Query: 454 EIALL 458
E++LL
Sbjct: 474 EVSLL 478
>UniRef100_Q7X7U4 Putative hexose transporter protein [Arabidopsis thaliana]
Length = 338
Score = 181 bits (460), Expect = 3e-44
Identities = 97/167 (58%), Positives = 117/167 (69%), Gaps = 10/167 (5%)
Query: 16 KETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICL 75
K+ NPSWK SLPHVLVA++TS LFGYHLGVVNE LESIS+DLGF+GNT+AEGLVVS CL
Sbjct: 44 KDCGNPSWKRSLPHVLVASLTSLLFGYHLGVVNETLESISIDLGFSGNTIAEGLVVSTCL 103
Query: 76 GGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLGLG 133
GGA G L SG +AD VGRRRAFQL ALPMI+GA++ S + L+ + + +G+G
Sbjct: 104 GGAFIGSLFSGLVADGVGRRRAFQLSALPMIVGASVSASTESLMGMLLGRFLVGIGMGIG 163
Query: 134 PSRCC--------SLCDRAYGALIQIATCFGILGSLFIGIPVKEISG 172
PS + YG+ QIATC G+LGSLF GIP K+ G
Sbjct: 164 PSVTALYVTEVSPAYVRGTYGSSTQIATCIGLLGSLFAGIPAKDNLG 210
>UniRef100_Q6K7T0 Putative sugar transporter [Oryza sativa]
Length = 463
Score = 177 bits (450), Expect = 4e-43
Identities = 87/125 (69%), Positives = 109/125 (86%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A +M +QA GA+ + ++YLSVGGMLLFV TF+LGAGPVPGLLL EIFP++IRAKAM
Sbjct: 336 AFAMGLQAVGANRHHLGSASVYLSVGGMLLFVLTFSLGAGPVPGLLLPEIFPNKIRAKAM 395
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A CMSVHWV+NFFV LLFLRLLE+LG Q+LY+MF++ C++A IFV+R+VVETKGK+LQEI
Sbjct: 396 ALCMSVHWVVNFFVSLLFLRLLEQLGPQVLYTMFSSACVVAAIFVRRHVVETKGKTLQEI 455
Query: 454 EIALL 458
E++LL
Sbjct: 456 EVSLL 460
Score = 140 bits (353), Expect = 8e-32
Identities = 83/160 (51%), Positives = 95/160 (58%), Gaps = 28/160 (17%)
Query: 21 PSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALF 80
PSW++SLPHV VAT+TSFLFG F GNTLAEGLVVSICLGGA
Sbjct: 36 PSWRMSLPHVCVATLTSFLFG------------------FAGNTLAEGLVVSICLGGAFV 77
Query: 81 GCLLSGWIADAVGRRRAFQLCALPMIIGAAMS--NKQLVWHACRKIICWNWLGLGPSRCC 138
GCL SG IAD +GRRRAFQL ALPMIIGAA+S L + + +GLGP
Sbjct: 78 GCLFSGSIADGIGRRRAFQLSALPMIIGAAVSALTNSLEGMLLGRFLVGTGMGLGPPVAS 137
Query: 139 SLCDR--------AYGALIQIATCFGILGSLFIGIPVKEI 170
YG+ +QIATC GI+ SL IG PVK+I
Sbjct: 138 LYITEVSPPSVRGTYGSFVQIATCLGIVVSLLIGTPVKDI 177
>UniRef100_Q9CA87 Putative sugar transporter; 77409-81599 [Arabidopsis thaliana]
Length = 467
Score = 167 bits (422), Expect = 8e-40
Identities = 83/125 (66%), Positives = 105/125 (83%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+S+ +QA ++L G L+LSVGGMLLFV +FA GAGPVP LLL+EI P R+RA A+
Sbjct: 339 AVSLGLQAIAYTSLPSPFGTLFLSVGGMLLFVLSFATGAGPVPSLLLSEICPGRLRATAL 398
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A C++VHWVINFFVGLLFLR+LE+LG+ LL ++F FC++AVIFV++NVVETKGKSLQEI
Sbjct: 399 AVCLAVHWVINFFVGLLFLRMLEQLGSVLLNAIFGFFCVVAVIFVQKNVVETKGKSLQEI 458
Query: 454 EIALL 458
EI+LL
Sbjct: 459 EISLL 463
Score = 135 bits (339), Expect = 3e-30
Identities = 83/169 (49%), Positives = 103/169 (60%), Gaps = 12/169 (7%)
Query: 14 IDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSI 73
IDK + + +L A S LF H VVNE LESIS+DLGF+GNT+AEGLVVS
Sbjct: 16 IDKRVPSKEFLSALDKAETAA-NSCLFSGHR-VVNETLESISIDLGFSGNTIAEGLVVST 73
Query: 74 CLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLG 131
CLGGA G L SG +AD VGRRRAFQL ALPMI+GA++ S + L+ + + +G
Sbjct: 74 CLGGAFIGSLFSGLVADGVGRRRAFQLSALPMIVGASVSASTESLMGMLLGRFLVGIGMG 133
Query: 132 LGPSRCC--------SLCDRAYGALIQIATCFGILGSLFIGIPVKEISG 172
+GPS + YG+ QIATC G+LGSLF GIP K+ G
Sbjct: 134 IGPSVTALYVTEVSPAYVRGTYGSSTQIATCIGLLGSLFAGIPAKDNLG 182
>UniRef100_Q8RXX6 Putative hexose transporter protein [Arabidopsis thaliana]
Length = 363
Score = 167 bits (422), Expect = 8e-40
Identities = 83/125 (66%), Positives = 105/125 (83%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+S+ +QA ++L G L+LSVGGMLLFV +FA GAGPVP LLL+EI P R+RA A+
Sbjct: 235 AVSLGLQAIAYTSLPSPFGTLFLSVGGMLLFVLSFATGAGPVPSLLLSEICPGRLRATAL 294
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A C++VHWVINFFVGLLFLR+LE+LG+ LL ++F FC++AVIFV++NVVETKGKSLQEI
Sbjct: 295 AVCLAVHWVINFFVGLLFLRMLEQLGSVLLNAIFGFFCVVAVIFVQKNVVETKGKSLQEI 354
Query: 454 EIALL 458
EI+LL
Sbjct: 355 EISLL 359
Score = 43.9 bits (102), Expect = 0.010
Identities = 28/78 (35%), Positives = 40/78 (50%), Gaps = 10/78 (12%)
Query: 105 MIIGAAMS--NKQLVWHACRKIICWNWLGLGPSRCCSLCDRA--------YGALIQIATC 154
MI+GA++S + L+ + + +G+GPS YG+ QIATC
Sbjct: 1 MIVGASVSASTESLMGMLLGRFLVGIGMGIGPSVTALYVTEVSPAYVRGTYGSSTQIATC 60
Query: 155 FGILGSLFIGIPVKEISG 172
G+LGSLF GIP K+ G
Sbjct: 61 IGLLGSLFAGIPAKDNLG 78
>UniRef100_Q9M9Z8 F20B17.24 [Arabidopsis thaliana]
Length = 472
Score = 166 bits (419), Expect = 2e-39
Identities = 83/124 (66%), Positives = 104/124 (82%)
Query: 335 ISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMA 394
IS+ +QA ++L G L+LSVGGMLLFV +FA GAGPVP LLL+EI P R+RA A+A
Sbjct: 345 ISLGLQAIAYTSLPSPFGTLFLSVGGMLLFVLSFATGAGPVPSLLLSEICPGRLRATALA 404
Query: 395 FCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
C++VHWVINFFVGLLFLR+LE+LG+ LL ++F FC++AVIFV++NVVETKGKSLQEIE
Sbjct: 405 VCLAVHWVINFFVGLLFLRMLEQLGSVLLNAIFGFFCVVAVIFVQKNVVETKGKSLQEIE 464
Query: 455 IALL 458
I+LL
Sbjct: 465 ISLL 468
Score = 135 bits (339), Expect = 3e-30
Identities = 83/169 (49%), Positives = 103/169 (60%), Gaps = 12/169 (7%)
Query: 14 IDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSI 73
IDK + + +L A S LF H VVNE LESIS+DLGF+GNT+AEGLVVS
Sbjct: 16 IDKRVPSKEFLSALDKAETAA-NSCLFSGHR-VVNETLESISIDLGFSGNTIAEGLVVST 73
Query: 74 CLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLG 131
CLGGA G L SG +AD VGRRRAFQL ALPMI+GA++ S + L+ + + +G
Sbjct: 74 CLGGAFIGSLFSGLVADGVGRRRAFQLSALPMIVGASVSASTESLMGMLLGRFLVGIGMG 133
Query: 132 LGPSRCC--------SLCDRAYGALIQIATCFGILGSLFIGIPVKEISG 172
+GPS + YG+ QIATC G+LGSLF GIP K+ G
Sbjct: 134 IGPSVTALYVTEVSPAYVRGTYGSSTQIATCIGLLGSLFAGIPAKDNLG 182
>UniRef100_Q9LLE0 Hexose transporter [Solanum tuberosum]
Length = 470
Score = 125 bits (313), Expect = 3e-27
Identities = 66/134 (49%), Positives = 92/134 (68%)
Query: 326 LYIFCTV*AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFP 385
L I T A SM++ + + + T + L+V G +L+V +F+LGAGPVP LLL EIF
Sbjct: 335 LLISYTGMAASMMLLSLSFTWKVLTPYSGTLAVLGTVLYVLSFSLGAGPVPALLLPEIFA 394
Query: 386 SRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVET 445
SRIRAKA+A + VHW++NFF+GL FL ++ K G +Y FA C++AV+++ NVVET
Sbjct: 395 SRIRAKAVALSLGVHWIMNFFIGLYFLSIVTKFGISTVYMGFALSCLVAVVYITGNVVET 454
Query: 446 KGKSLQEIEIALLP 459
KG+SL+EIE L P
Sbjct: 455 KGRSLEEIERELSP 468
Score = 82.8 bits (203), Expect = 2e-14
Identities = 53/151 (35%), Positives = 76/151 (50%), Gaps = 10/151 (6%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN LE ++ DLG NT+ +G +VS L GA G G
Sbjct: 30 LPYVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENTVIQGWIVSTVLAGAFVGSFTGG 89
Query: 87 WIADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWLGLGPSRCC-------- 138
+AD GR + F L A+P+ +GA + A +G+G S
Sbjct: 90 VLADKFGRTKTFILDAIPLSVGAFLCTTAQSVQAMIIGRLLTGIGIGISSAIVPLYISEI 149
Query: 139 --SLCDRAYGALIQIATCFGILGSLFIGIPV 167
+ G + Q+ C GIL +L +G+P+
Sbjct: 150 SPTEIRGTLGTVNQLFICIGILVALVVGLPL 180
>UniRef100_Q5ZCN2 Putative hexose transporter [Oryza sativa]
Length = 513
Score = 125 bits (313), Expect = 3e-27
Identities = 63/111 (56%), Positives = 82/111 (73%), Gaps = 2/111 (1%)
Query: 347 LLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFF 406
L P +G L +V G +L+V +FALGAGPVP LLL EIF SRIRAKA+A + +HWV NFF
Sbjct: 401 LAPYSGPL--AVAGTVLYVLSFALGAGPVPALLLPEIFASRIRAKAVALSLGMHWVSNFF 458
Query: 407 VGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
+GL FL ++ K G +Y FA+ C +AV+++ NVVETKG+SL+EIE AL
Sbjct: 459 IGLYFLSVVNKFGISTVYLGFASVCALAVVYIAGNVVETKGRSLEEIERAL 509
Score = 88.2 bits (217), Expect = 4e-16
Identities = 59/154 (38%), Positives = 81/154 (52%), Gaps = 16/154 (10%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN LE ++ DLG + N + +G VVS L GA G G
Sbjct: 73 LPYVGVACLGAILFGYHLGVVNGALEYLAKDLGISENAVLQGWVVSTTLAGATAGSFTGG 132
Query: 87 WIADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWL-GLGPSRCCSLCD--- 142
+AD GR R F L A+P+ +GA +S H R +I L G+G +L
Sbjct: 133 ALADKFGRTRTFILDAIPLAVGAFLS---ATAHDVRTMIIGRLLAGIGIGISSALVPLYI 189
Query: 143 ---------RAYGALIQIATCFGILGSLFIGIPV 167
A G++ Q+ C GIL +L G+P+
Sbjct: 190 SEISPTEIRGALGSVNQLFICIGILAALVAGLPL 223
>UniRef100_Q93WT7 Hexose transporter pGlT [Olea europaea]
Length = 544
Score = 123 bits (309), Expect = 1e-26
Identities = 65/126 (51%), Positives = 91/126 (71%), Gaps = 4/126 (3%)
Query: 334 AISMIIQATGAS--TLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAK 391
A+SM++ + + TL P AG L +V G +L+V +F+LGAGPVP LLL EIF SRIRAK
Sbjct: 417 AVSMLLLSLTFTWKTLAPYAGTL--AVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAK 474
Query: 392 AMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQ 451
A+A + +HW+ NF +GL FL ++ K G +Y FA+ C++AV+++ NVVETKG+SL+
Sbjct: 475 AVALSLGMHWISNFVIGLYFLSVVTKFGISTVYLGFASVCLLAVMYIAGNVVETKGRSLE 534
Query: 452 EIEIAL 457
EIE AL
Sbjct: 535 EIERAL 540
Score = 80.1 bits (196), Expect = 1e-13
Identities = 50/151 (33%), Positives = 78/151 (51%), Gaps = 10/151 (6%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN LE ++ DLG N + +G VVS L GA G G
Sbjct: 104 LPYVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENAVLQGWVVSTLLAGATVGSFTGG 163
Query: 87 WIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLGLGPSRCCSLCDR- 143
+AD GR + F L A+P+ +GA + + + + +++ +G+ +
Sbjct: 164 SLADKFGRTKTFLLDAIPLAVGAFLCATAQNIETMIIGRLLAGIGIGISSAIVPLYISEI 223
Query: 144 -------AYGALIQIATCFGILGSLFIGIPV 167
G++ Q+ C GIL +L G+P+
Sbjct: 224 SPTEIRGTLGSVNQLFICIGILAALVAGLPL 254
>UniRef100_Q9LLE2 Hexose transporter [Spinacia oleracea]
Length = 551
Score = 123 bits (308), Expect = 1e-26
Identities = 61/113 (53%), Positives = 83/113 (72%), Gaps = 2/113 (1%)
Query: 347 LLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFF 406
L P +G L +V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+A + +HW NF
Sbjct: 439 LAPYSGTL--AVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVALSLGMHWASNFV 496
Query: 407 VGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLP 459
+GL FL ++ K G +Y FA+ C++AV+++ NVVETKG+SL+EIE+AL P
Sbjct: 497 IGLYFLSVVTKFGISKVYLGFASVCVLAVLYIAGNVVETKGRSLEEIELALSP 549
Score = 94.4 bits (233), Expect = 6e-18
Identities = 58/153 (37%), Positives = 84/153 (53%), Gaps = 14/153 (9%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN L+ +S DL GNT+ +G VVSI L GA G G
Sbjct: 111 LPYVGVACLGAILFGYHLGVVNGALDYLSADLAIAGNTVLQGWVVSILLAGATVGSFTGG 170
Query: 87 WIADAVGRRRAFQLCALPMIIGAAM----SNKQLVWHACRKIICWNWLGLGPSRCCSLCD 142
+AD GR + FQL A+P+ IGA + N Q++ +++C +G+ +
Sbjct: 171 SLADKFGRTKTFQLDAIPLAIGAYLCATAQNVQIM--MIGRLLCGIGIGISSALVPLYIS 228
Query: 143 R--------AYGALIQIATCFGILGSLFIGIPV 167
A G++ Q+ C GIL +L G+P+
Sbjct: 229 EISPTEIRGALGSVNQLFICIGILAALVAGLPL 261
>UniRef100_Q5XF02 At1g05030 [Arabidopsis thaliana]
Length = 524
Score = 123 bits (308), Expect = 1e-26
Identities = 68/168 (40%), Positives = 102/168 (60%), Gaps = 16/168 (9%)
Query: 15 DKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSIC 74
+K + + W + PHV VA++ +FLFGYH+GV+N P+ SI+ +LGF GN++ EGLVVSI
Sbjct: 68 EKFSADLGWLSAFPHVSVASMANFLFGYHIGVMNGPIVSIARELGFEGNSILEGLVVSIF 127
Query: 75 LGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWL-GLG 133
+ GA G +++G + D G RR FQ+ +P+I+GA +S + H+ +I+C +L GLG
Sbjct: 128 IAGAFIGSIVAGPLVDKFGYRRTFQIFTIPLILGALVSAQA---HSLDEILCGRFLVGLG 184
Query: 134 --------PSRCCSLCDRAY----GALIQIATCFGILGSLFIGIPVKE 169
P + Y G L QI TC GI+ SL +GIP ++
Sbjct: 185 IGVNTVLVPIYISEVAPTKYRGSLGTLCQIGTCLGIIFSLLLGIPAED 232
Score = 107 bits (267), Expect = 7e-22
Identities = 55/124 (44%), Positives = 78/124 (62%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+SM + L + LS+ G L+++F+FA+GAGPV GL++ E+ +R R K M
Sbjct: 394 AVSMFLIVYAVGFPLDEDLSQSLSILGTLMYIFSFAIGAGPVTGLIIPELSSNRTRGKIM 453
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
F SVHWV NF VGL FL L+EK G +Y+ F + ++A F VETKG+SL+EI
Sbjct: 454 GFSFSVHWVSNFLVGLFFLDLVEKYGVGTVYASFGSVSLLAAAFSHLFTVETKGRSLEEI 513
Query: 454 EIAL 457
E++L
Sbjct: 514 ELSL 517
>UniRef100_Q93Z41 AT5g16150/T21H19_70 [Arabidopsis thaliana]
Length = 546
Score = 122 bits (305), Expect = 3e-26
Identities = 61/124 (49%), Positives = 88/124 (70%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+SM++ + + A + L+V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+
Sbjct: 419 ALSMLLLSLSFTWKALAAYSGTLAVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAV 478
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A + +HW+ NF +GL FL ++ K G +Y FA C++AV+++ NVVETKG+SL+EI
Sbjct: 479 ALSLGMHWISNFVIGLYFLSVVTKFGISSVYLGFAGVCVLAVLYIAGNVVETKGRSLEEI 538
Query: 454 EIAL 457
E+AL
Sbjct: 539 ELAL 542
Score = 85.9 bits (211), Expect = 2e-15
Identities = 54/151 (35%), Positives = 80/151 (52%), Gaps = 10/151 (6%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP V VA + + LFGYHLGVVN LE ++ DLG NT+ +G +VS L GA G G
Sbjct: 106 LPFVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENTVLQGWIVSSLLAGATVGSFTGG 165
Query: 87 WIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLGLGPSRCCSLCDR- 143
+AD GR R FQL A+P+ IGA + + + + +++ +G+ +
Sbjct: 166 ALADKFGRTRTFQLDAIPLAIGAFLCATAQSVQTMIVGRLLAGIGIGISSAIVPLYISEI 225
Query: 144 -------AYGALIQIATCFGILGSLFIGIPV 167
A G++ Q+ C GIL +L G+P+
Sbjct: 226 SPTEIRGALGSVNQLFICIGILAALIAGLPL 256
>UniRef100_Q9LF13 Sugar transporter-like protein [Arabidopsis thaliana]
Length = 560
Score = 122 bits (305), Expect = 3e-26
Identities = 61/124 (49%), Positives = 88/124 (70%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+SM++ + + A + L+V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+
Sbjct: 433 ALSMLLLSLSFTWKALAAYSGTLAVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAV 492
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A + +HW+ NF +GL FL ++ K G +Y FA C++AV+++ NVVETKG+SL+EI
Sbjct: 493 ALSLGMHWISNFVIGLYFLSVVTKFGISSVYLGFAGVCVLAVLYIAGNVVETKGRSLEEI 552
Query: 454 EIAL 457
E+AL
Sbjct: 553 ELAL 556
Score = 76.3 bits (186), Expect = 2e-12
Identities = 54/165 (32%), Positives = 80/165 (47%), Gaps = 24/165 (14%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAE--------------GLVVS 72
LP V VA + + LFGYHLGVVN LE ++ DLG NT+ + G +VS
Sbjct: 106 LPFVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENTVLQGKYMMIHFFTPPVNGWIVS 165
Query: 73 ICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWL 130
L GA G G +AD GR R FQL A+P+ IGA + + + + +++ +
Sbjct: 166 SLLAGATVGSFTGGALADKFGRTRTFQLDAIPLAIGAFLCATAQSVQTMIVGRLLAGIGI 225
Query: 131 GLGPSRCCSLCDR--------AYGALIQIATCFGILGSLFIGIPV 167
G+ + A G++ Q+ C GIL +L G+P+
Sbjct: 226 GISSAIVPLYISEISPTEIRGALGSVNQLFICIGILAALIAGLPL 270
>UniRef100_Q84XS8 Putative sugar transporter [Brassica rapa subsp. pekinensis]
Length = 170
Score = 121 bits (304), Expect = 4e-26
Identities = 61/124 (49%), Positives = 88/124 (70%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+SM++ + + A + L+V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+
Sbjct: 44 ALSMLLLSLSFTWKALAAYSGTLAVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAV 103
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A + +HW+ NF +GL FL ++ K G +Y FA C++AV+++ NVVETKG+SL+EI
Sbjct: 104 ALSLGMHWISNFVIGLYFLSVVTKFGISSVYLGFAGVCVLAVMYIAGNVVETKGRSLEEI 163
Query: 454 EIAL 457
E+AL
Sbjct: 164 ELAL 167
>UniRef100_Q9LLE1 Hexose transporter [Nicotiana tabacum]
Length = 534
Score = 121 bits (303), Expect = 5e-26
Identities = 60/113 (53%), Positives = 83/113 (73%), Gaps = 2/113 (1%)
Query: 347 LLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFF 406
L P +G L +V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+A + +HW+ NFF
Sbjct: 422 LTPYSGTL--AVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVALSLGMHWISNFF 479
Query: 407 VGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLP 459
+GL FL ++ K G +Y FA+ C++AV+++ NVVETKG+SL++IE L P
Sbjct: 480 IGLYFLSIVTKFGISTVYLGFASVCLLAVMYIVGNVVETKGRSLEDIERELSP 532
Score = 80.9 bits (198), Expect = 7e-14
Identities = 53/151 (35%), Positives = 75/151 (49%), Gaps = 10/151 (6%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN LE ++ DLG NT+ +G +VS L GA G G
Sbjct: 94 LPYVGVACLGAILFGYHLGVVNGALEYLAKDLGIVENTVIQGWIVSSVLAGATVGSFTGG 153
Query: 87 WIADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWLGLGPSRCC-------- 138
+AD GR + F L A+P+ +GA + A +G+G S
Sbjct: 154 ALADKFGRTKTFVLDAIPLAVGAFLCTTAQSVQAMIIGRLLTGIGIGISSAIVPLYISEI 213
Query: 139 --SLCDRAYGALIQIATCFGILGSLFIGIPV 167
+ G + Q+ C GIL +L G+P+
Sbjct: 214 SPTEIRGTLGTVNQLFICIGILVALVAGLPL 244
>UniRef100_Q9LLD9 Hexose transporter [Zea mays]
Length = 542
Score = 121 bits (303), Expect = 5e-26
Identities = 61/108 (56%), Positives = 80/108 (73%), Gaps = 2/108 (1%)
Query: 347 LLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFF 406
L P +G L +V G +L+V +FALGAGPVP LLL EIF SRIRAKA+A + +HWV NFF
Sbjct: 430 LAPYSGTL--AVVGTVLYVLSFALGAGPVPALLLPEIFASRIRAKAVALSLGMHWVSNFF 487
Query: 407 VGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
+GL FL ++ K G +Y FA+ C +AV+++ NVVETKG+SL+EIE
Sbjct: 488 IGLYFLSVVSKFGISNVYLGFASVCALAVLYIAGNVVETKGRSLEEIE 535
Score = 82.4 bits (202), Expect = 2e-14
Identities = 57/154 (37%), Positives = 77/154 (49%), Gaps = 16/154 (10%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN LE ++ DLG N + +G VVS L GA G G
Sbjct: 102 LPYVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENAVLQGWVVSTSLAGATLGSFTGG 161
Query: 87 WIADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWL---GLGPSRCC----- 138
+AD GR R F L A+P+ +GA +S R +I L G+G S
Sbjct: 162 SLADKFGRTRTFILDAVPLALGAFLS---ATAQDIRTMIIGRLLAGIGIGVSSALVPLYI 218
Query: 139 -----SLCDRAYGALIQIATCFGILGSLFIGIPV 167
+ G + Q+ C GIL +L G+P+
Sbjct: 219 SEISPTEIRGTLGTVNQLFICIGILAALLAGLPL 252
>UniRef100_Q9LLD8 Hexose transporter [Arabidopsis thaliana]
Length = 515
Score = 120 bits (301), Expect = 8e-26
Identities = 57/102 (55%), Positives = 78/102 (75%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
L+V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+A + +HW+ NF +GL FL ++
Sbjct: 407 LAVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVALSLGMHWISNFVIGLYFLSVV 466
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
K G +Y FA C++AV+++ NVVETKG+SL+EIE+AL
Sbjct: 467 TKFGISSVYLGFAGVCVLAVLYIAGNVVETKGRSLEEIELAL 508
Score = 85.9 bits (211), Expect = 2e-15
Identities = 54/151 (35%), Positives = 80/151 (52%), Gaps = 10/151 (6%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP V VA + + LFGYHLGVVN LE ++ DLG NT+ +G +VS L GA G G
Sbjct: 72 LPFVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENTVLQGWIVSSLLAGATVGSFTGG 131
Query: 87 WIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLGLGPSRCCSLCDR- 143
+AD GR R FQL A+P+ IGA + + + + +++ +G+ +
Sbjct: 132 ALADKFGRTRTFQLDAIPLAIGAFLCATAQSVQTMIVGRLLAGIGIGISSAIVPLYISEI 191
Query: 144 -------AYGALIQIATCFGILGSLFIGIPV 167
A G++ Q+ C GIL +L G+P+
Sbjct: 192 SPTEIRGALGSVNQLFICIGILAALIAGLPL 222
>UniRef100_Q9FXY8 Putative glucose translocator [Mesembryanthemum crystallinum]
Length = 555
Score = 120 bits (300), Expect = 1e-25
Identities = 61/113 (53%), Positives = 82/113 (71%), Gaps = 2/113 (1%)
Query: 347 LLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFF 406
L P +G L +V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+A + +HW NF
Sbjct: 443 LAPYSGTL--AVLGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAVALSLGMHWASNFV 500
Query: 407 VGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALLP 459
+GL FL ++ K G +Y FA+ C++AV+++ NVVETKG+SL+EIE AL P
Sbjct: 501 IGLYFLSVVTKFGISRVYLGFASVCMLAVLYIAGNVVETKGRSLEEIERALSP 553
Score = 97.4 bits (241), Expect = 7e-19
Identities = 59/151 (39%), Positives = 84/151 (55%), Gaps = 10/151 (6%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP+V VA + + LFGYHLGVVN LE +S DLG GNT+ +G VVSI L GA G G
Sbjct: 115 LPYVGVACLGAILFGYHLGVVNGALEYLSPDLGIAGNTVLQGWVVSILLAGATVGSFTGG 174
Query: 87 WIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLGLGPSRCCSLCDR- 143
+AD GR R FQL A+P+ IGA + + + + +++C +G+ +
Sbjct: 175 SLADKFGRTRTFQLDAIPLAIGAYLCATAQSVQTMMIGRLLCGIGIGISSALVPLYISEI 234
Query: 144 -------AYGALIQIATCFGILGSLFIGIPV 167
A G++ Q+ C GIL +L G+P+
Sbjct: 235 SPTEIRGALGSVNQLFICIGILAALVAGLPL 265
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.341 0.151 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 703,221,237
Number of Sequences: 2790947
Number of extensions: 27551876
Number of successful extensions: 97681
Number of sequences better than 10.0: 1373
Number of HSP's better than 10.0 without gapping: 1003
Number of HSP's successfully gapped in prelim test: 370
Number of HSP's that attempted gapping in prelim test: 94476
Number of HSP's gapped (non-prelim): 2759
length of query: 461
length of database: 848,049,833
effective HSP length: 131
effective length of query: 330
effective length of database: 482,435,776
effective search space: 159203806080
effective search space used: 159203806080
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC122722.6


