

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.4 + phase: 0
(307 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
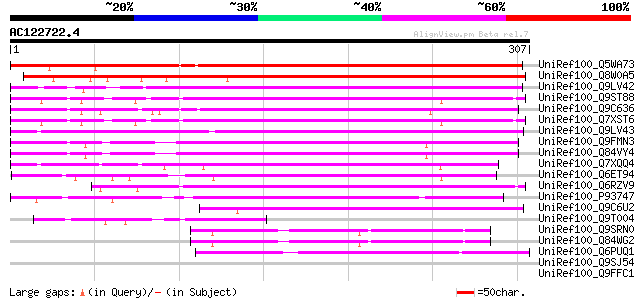
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5WA73 Hypothetical protein P0681F10.43 [Oryza sativa] 285 1e-75
UniRef100_Q8W0A5 P0452F10.18 protein [Oryza sativa] 252 1e-65
UniRef100_Q9LV42 Emb|CAB53482.1 [Arabidopsis thaliana] 224 2e-57
UniRef100_Q9ST88 CAA30379.1 protein [Oryza sativa] 214 3e-54
UniRef100_Q9C636 Hypothetical protein F2G19.25 [Arabidopsis thal... 214 3e-54
UniRef100_Q7XST6 OSJNBa0039K24.20 protein [Oryza sativa] 213 4e-54
UniRef100_Q9LV43 Arabidopsis thaliana genomic DNA, chromosome 3,... 208 2e-52
UniRef100_Q9FMN3 Emb|CAB53482.1 [Arabidopsis thaliana] 196 5e-49
UniRef100_Q84VY4 At5g42860 [Arabidopsis thaliana] 195 1e-48
UniRef100_Q7XQQ4 OSJNBa0084A10.1 protein [Oryza sativa] 184 3e-45
UniRef100_Q6ET94 Hypothetical protein OJ1725_H08.8-1 [Oryza sativa] 184 3e-45
UniRef100_Q6RZV9 Hypothetical protein G9-6 [Musa acuminata] 183 6e-45
UniRef100_P93747 Hypothetical protein At2g41990 [Arabidopsis tha... 162 1e-38
UniRef100_Q9C6U2 Hypothetical protein T8G24.9 [Arabidopsis thali... 73 1e-11
UniRef100_Q9T004 Hypothetical protein T12J5.40 [Arabidopsis thal... 50 6e-05
UniRef100_Q9SRN0 T19F11.6 protein [Arabidopsis thaliana] 49 1e-04
UniRef100_Q84WG2 Putative VAMP protein SEC22 [Arabidopsis thaliana] 49 2e-04
UniRef100_Q6PUQ1 Hypothetical protein [Petunia integrifolia subs... 49 2e-04
UniRef100_Q9SJ54 Putative harpin-induced protein [Arabidopsis th... 45 0.002
UniRef100_Q9FFC1 Gb|AAF18257.1 [Arabidopsis thaliana] 45 0.003
>UniRef100_Q5WA73 Hypothetical protein P0681F10.43 [Oryza sativa]
Length = 322
Score = 285 bits (728), Expect = 1e-75
Identities = 155/318 (48%), Positives = 208/318 (64%), Gaps = 17/318 (5%)
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQ--------TPYYVQSPSRESSHDGDKSSTMQPSPM-- 49
MH K++SDVTS PSSP RS K+ YYVQSPSRES G KSS+MQ +P+
Sbjct: 2 MHAKSESDVTSLAPSSPPRSPKRGGGVGVGGANYYVQSPSRESHDGGYKSSSMQATPVYN 61
Query: 50 ---DSPSHQSYGHHSRASSSSRISGGYNSSFLGRKVNRK-VNDKGWIGSKVIEEENGGYG 105
+SPSH SYG HSR+SS SR SG RK +NDKGW VIEEE G Y
Sbjct: 62 SPNESPSHPSYGRHSRSSSVSRFSGTLRDGSRKAGGERKALNDKGWPECNVIEEE-GPYE 120
Query: 106 DFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGS 165
D GD G+SRR Q + + FVL+F++FC IIW A+ Y+P + VKSLT+ +FY GEG+
Sbjct: 121 DLAGD-TGLSRRCQIVLGFLCFVLLFTVFCLIIWGAARPYEPDVVVKSLTMDDFYAGEGT 179
Query: 166 DVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSR 225
D +GVPTK++T+NCS+ + V+NPA+ FGI+V++ + L+YS++++ G++++Y+Q RKS
Sbjct: 180 DHSGVPTKLVTLNCSLHIAVYNPASMFGIHVTTGPIRLLYSEISIGVGQVRRYYQPRKSH 239
Query: 226 RTVSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCS 285
R V+ + G+KVPLYGAG + S G + LTL F++ SRG V+GKLVR H V+C
Sbjct: 240 RLVTAVVHGNKVPLYGAGGGLMLSSSGGAVPLTLDFDLTSRGYVIGKLVRVTHKVHVTCP 299
Query: 286 IAVDPHNNKYIKLKENEC 303
I VD K IK + C
Sbjct: 300 IVVDAKKTKPIKFSKKAC 317
>UniRef100_Q8W0A5 P0452F10.18 protein [Oryza sativa]
Length = 321
Score = 252 bits (643), Expect = 1e-65
Identities = 145/320 (45%), Positives = 196/320 (60%), Gaps = 23/320 (7%)
Query: 9 VTSFDPSSP-RSQKQTP--YYVQ-SPSRES-SHDGDKSSTMQP------SPMDSPSHQS- 56
+TS SSP RS K YYVQ SPSR+S DGDKS + Q SP+DSPSH S
Sbjct: 1 MTSLATSSPSRSPKAAAAAYYVQISPSRDSHDDDGDKSPSTQATPVYNNSPLDSPSHHSS 60
Query: 57 -YGHHSRASSSSRISGGYNSS-----FLGRKVNRKVNDKGW--IGSKVIEEENGGYGDFD 108
+G HSR SS+SR SG S+ G R++ KGW + + + EEE G Y +FD
Sbjct: 61 SFGRHSRVSSASRFSGNLRSASGRSRLGGGGGRRRLGAKGWRDVDAIIDEEEEGAYDEFD 120
Query: 109 GDGNGVSRRVQFFVAVVGF---VLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGS 165
D +G + V F L F+L C I+W + HYKP + VKSLTV NFY GEG
Sbjct: 121 DDDDGGYEPSRCCVLAFRFSLLALAFTLVCLIVWGIARHYKPGVLVKSLTVGNFYAGEGI 180
Query: 166 DVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSR 225
D TGVPTK++T+NCS+++ VHNP+T FGI+VSS S+ +++S + +A G+L+K++Q R S
Sbjct: 181 DRTGVPTKLVTMNCSLQINVHNPSTMFGIHVSSTSIQILFSQIAIANGQLEKFYQPRSSH 240
Query: 226 RTVSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCS 285
S + G K+PLYGAG A S G + LTL V++RG V+GKLVR HT+RV C
Sbjct: 241 HVASAIVHGEKIPLYGAGETFALSNAGGAVPLTLDLVVRARGYVIGKLVRVTHTKRVKCP 300
Query: 286 IAVDPHNNKYIKLKENECRY 305
+ +D ++K I+ ++ C Y
Sbjct: 301 VVIDSGSSKPIRFIQSACSY 320
>UniRef100_Q9LV42 Emb|CAB53482.1 [Arabidopsis thaliana]
Length = 298
Score = 224 bits (572), Expect = 2e-57
Identities = 130/310 (41%), Positives = 186/310 (59%), Gaps = 23/310 (7%)
Query: 1 MHIKTDSDVTSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSS-----TMQPSPMDSPSHQ 55
M+ K+DSDVTS D SSP K+ YYVQSPSR+S DKSS T Q +P +SPSH
Sbjct: 3 MYPKSDSDVTSLDLSSP---KRPTYYVQSPSRDS----DKSSSVALTTHQTTPTESPSHP 55
Query: 56 SYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVS 115
S +SR+S G F K RK + W + E +G Y D D GVS
Sbjct: 56 SI--------ASRVSNGGGGGFRW-KGRRKYHGGIWWPADKEEGGDGRYEDLYEDNRGVS 106
Query: 116 R-RVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKM 174
+ + VV + +F L C +++ AS P + +K + V +FY+GEGSD TGVPTK+
Sbjct: 107 IVTCRLILGVVATLSIFFLLCSVLFGASQSSPPIVYIKGVNVRSFYYGEGSDNTGVPTKI 166
Query: 175 LTVNCSMRMLVHNPATFFGIYVSSKSVNLMYS-DLTVATGELKKYHQQRKSRRTVSVNIQ 233
+ V CS+ + HNP+T FGI+VSS +V+L+YS T+A LK YHQ ++S T +N+
Sbjct: 167 MNVKCSVVITTHNPSTLFGIHVSSTAVSLIYSRQFTLANARLKSYHQPKQSNHTSRINLI 226
Query: 234 GSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDPHNN 293
GSKVPLYGAGA++ +S ++G + + L FE++SR N+VG LV+++H + +SC + +N
Sbjct: 227 GSKVPLYGAGAELVASDNSGGVPVKLEFEIRSRANIVGNLVKSRHRKHLSCFFFISSYNT 286
Query: 294 KYIKLKENEC 303
++K C
Sbjct: 287 TFVKFTHKTC 296
>UniRef100_Q9ST88 CAA30379.1 protein [Oryza sativa]
Length = 835
Score = 214 bits (544), Expect = 3e-54
Identities = 139/324 (42%), Positives = 189/324 (57%), Gaps = 29/324 (8%)
Query: 1 MHIKTDSDVTSFDPSSP----RSQKQTP-YYVQSPSRESSHDGDKS-----STMQPSPMD 50
MH KTDS+VTS PSSP S+ P YYVQSPSR+S HDG+K+ ST SPM
Sbjct: 521 MHAKTDSEVTSLAPSSPPRSPTSRGGRPVYYVQSPSRDS-HDGEKTATSVHSTPALSPMG 579
Query: 51 SPSHQSYGHHSRASSSSRISGGY----NSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGD 106
SP H R SSSSR SG + S GRK KGW VIEEE G D
Sbjct: 580 SPRHSV----GRDSSSSRFSGHPKRKGDKSSSGRK--GAPAGKGWQEIGVIEEE--GLLD 631
Query: 107 FDGDGNGVSRRVQFF-VAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGS 165
+ + G+ +R ++F + V+GFV++FS F ++W AS KPQ+ +KS+T NF G+
Sbjct: 632 DEDERRGIPKRCKYFLIFVLGFVVLFSFFALVLWGASRSQKPQIVIKSITFENFIIQAGT 691
Query: 166 DVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSR 225
D + VPT M T N ++++ N TFFGI+V++ L YS LT+A+G+L K++Q R SR
Sbjct: 692 DASLVPTDMATTNSTVKLTYRNTGTFFGIHVTADPFTLSYSQLTLASGDLNKFYQARSSR 751
Query: 226 RTVSVNIQGSKVPLYGAGADVASSVDNGK---ISLTLVFEVKSRGNVVGKLVRTKHTQRV 282
RTVSV + G+KVPLYG G + + +G + + L V SR V+G LV+ K T+ +
Sbjct: 752 RTVSVGVMGNKVPLYGGGPTLTAGKGSGSMAPVPMILRTTVHSRAYVLGALVKPKFTRAI 811
Query: 283 SCSIAVDPHN-NKYIKLKENECRY 305
C + ++P NK I L + C Y
Sbjct: 812 ECKVLMNPAKLNKPISL-DKSCIY 834
>UniRef100_Q9C636 Hypothetical protein F2G19.25 [Arabidopsis thaliana]
Length = 342
Score = 214 bits (544), Expect = 3e-54
Identities = 140/341 (41%), Positives = 192/341 (56%), Gaps = 45/341 (13%)
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKS-----STMQPSPMDSP-- 52
MH KTDS+VTS SSP RS ++ YYVQSPSR+S HDG+K+ ST SPM SP
Sbjct: 1 MHAKTDSEVTSLAASSPARSPRRPVYYVQSPSRDS-HDGEKTATSFHSTPVLSPMGSPPH 59
Query: 53 SHQSYGHHSRASSSSRISGGYNSSFLGRKVN------RKVN--DKGWIGSKVIEEENGGY 104
SH S G HSR SSSSR SG RKVN RK + +K W VIEEE G
Sbjct: 60 SHSSMGRHSRESSSSRFSGSLKPG--SRKVNPNDGSKRKGHGGEKQWKECAVIEEE-GLL 116
Query: 105 GDFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
D D DG GV RR +VGF ++F F I++ A+ KP+++VKS+T G
Sbjct: 117 DDGDRDG-GVPRRCYVLAFIVGFFILFGFFSLILYGAAKPMKPKITVKSITFETLKIQAG 175
Query: 165 SDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKS 224
D GV T M+T+N ++RML N TFFG++V+S ++L +S + + +G +KK++Q RKS
Sbjct: 176 QDAGGVGTDMITMNATLRMLYRNTGTFFGVHVTSTPIDLSFSQIKIGSGSVKKFYQGRKS 235
Query: 225 RRTVSVNIQGSKVPLYGAGADVA-----------------------SSVDNGKISLTLVF 261
RTV V++ G K+PLYG+G+ + + +TL F
Sbjct: 236 ERTVLVHVIGEKIPLYGSGSTLLPPAPPAPLPKPKKKKGAPVPIPDPPAPPAPVPMTLSF 295
Query: 262 EVKSRGNVVGKLVRTKHTQRVSCSIAVDPHN-NKYIKLKEN 301
V+SR V+GKLV+ K +++ C I + N NK+I + +N
Sbjct: 296 VVRSRAYVLGKLVQPKFYKKIECDINFEHKNLNKHIVITKN 336
>UniRef100_Q7XST6 OSJNBa0039K24.20 protein [Oryza sativa]
Length = 315
Score = 213 bits (543), Expect = 4e-54
Identities = 139/324 (42%), Positives = 189/324 (57%), Gaps = 29/324 (8%)
Query: 1 MHIKTDSDVTSFDPSSP----RSQKQTP-YYVQSPSRESSHDGDKS-----STMQPSPMD 50
MH KTDS+VTS PSSP S+ P YYVQSPSR+S HDG+K+ ST SPM
Sbjct: 1 MHAKTDSEVTSLAPSSPPRSPTSRGGRPVYYVQSPSRDS-HDGEKTATSVHSTPALSPMG 59
Query: 51 SPSHQSYGHHSRASSSSRISGGY----NSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGD 106
SP H R SSSSR SG + S GRK KGW VIEEE G D
Sbjct: 60 SPRHSV----GRDSSSSRFSGHPKRKGDKSSSGRK--GAPAGKGWQEIGVIEEE--GLLD 111
Query: 107 FDGDGNGVSRRVQFF-VAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGS 165
+ + G+ +R ++F + V+GFV++FS F ++W AS KPQ+ +KS+T NF G+
Sbjct: 112 DEDERRGIPKRCKYFLIFVLGFVVLFSFFALVLWGASRSQKPQIVIKSITFDNFIIQAGT 171
Query: 166 DVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSR 225
D + VPT M T N ++++ N TFFGI+V++ L YS LT+A+G+L K++Q R SR
Sbjct: 172 DASLVPTDMATTNSTVKLTYRNTGTFFGIHVTADPFTLSYSQLTLASGDLNKFYQARSSR 231
Query: 226 RTVSVNIQGSKVPLYGAGADVASSVDNGK---ISLTLVFEVKSRGNVVGKLVRTKHTQRV 282
RTVSV + G+KVPLYG G + + +G + + L V SR V+G LV+ K T+ +
Sbjct: 232 RTVSVGVMGNKVPLYGGGPTLTAGKGSGSMAPVPMILRTTVHSRAYVLGALVKPKFTRAI 291
Query: 283 SCSIAVDPHN-NKYIKLKENECRY 305
C + ++P NK I L + C Y
Sbjct: 292 ECKVLMNPAKLNKPISL-DKSCIY 314
>UniRef100_Q9LV43 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MOB24
[Arabidopsis thaliana]
Length = 306
Score = 208 bits (529), Expect = 2e-52
Identities = 122/306 (39%), Positives = 182/306 (58%), Gaps = 7/306 (2%)
Query: 1 MHIKTDSDVTSFDPSSPRSQKQTPYYVQSPSRESS-HDGDKSSTMQPSPMDSPSHQSYGH 59
M ++S+ TS D S S + YYV+SPS S DG+KSS+ + +
Sbjct: 2 MDATSESERTSLDLSI--SSPKQAYYVESPSSVSQLDDGEKSSSSASLIQVHTPNYTILS 59
Query: 60 HSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVSRRVQ 119
SR SSSSR S G + K + + ++ W E+ Y D G G R V
Sbjct: 60 ESRLSSSSRTSNGTSGMGFRWKGSSRRSNMYWPEKPYTINEDEVYDDNRGLSVGQCRAV- 118
Query: 120 FFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNC 179
+ ++G V+VFS+FC ++W AS + P +SVKS+ +H+FY+GEG D TGV TK+L+ N
Sbjct: 119 --LVILGTVVVFSVFCSVLWGASHPFSPIVSVKSVDIHSFYYGEGIDRTGVATKILSFNS 176
Query: 180 SMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPL 239
S+++ + +PA +FGI+VSS + L +S LT+ATG+LK Y+Q RKS+ V + G++VPL
Sbjct: 177 SVKVTIDSPAPYFGIHVSSSTFKLTFSALTLATGQLKSYYQPRKSKHISIVKLTGAEVPL 236
Query: 240 YGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAV-DPHNNKYIKL 298
YGAG +A+S GK+ + L FE++SRGN++GKLV++KH VSCS + +K I+
Sbjct: 237 YGAGPHLAASDKKGKVPVKLEFEIRSRGNLLGKLVKSKHENHVSCSFFISSSKTSKPIEF 296
Query: 299 KENECR 304
C+
Sbjct: 297 THKTCK 302
>UniRef100_Q9FMN3 Emb|CAB53482.1 [Arabidopsis thaliana]
Length = 320
Score = 196 bits (499), Expect = 5e-49
Identities = 121/331 (36%), Positives = 178/331 (53%), Gaps = 47/331 (14%)
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKSST------MQPSPMDSPS 53
MH KTDS+VTS SSP RS ++ Y+VQSPSR+S HDG+K++T + SPM SP
Sbjct: 1 MHAKTDSEVTSLSASSPTRSPRRPAYFVQSPSRDS-HDGEKTATSFHSTPVLTSPMGSPP 59
Query: 54 HQSYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNG 113
H HS +S S+I+G G K + EE G D D +
Sbjct: 60 HS----HSSSSRFSKINGSKRKGHAGEKQFAMI------------EEEGLLDDGDREQEA 103
Query: 114 VSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTK 173
+ RR +VGF L+F+ F I++AA+ KP++SVKS+T G D G+ T
Sbjct: 104 LPRRCYVLAFIVGFSLLFAFFSLILYAAAKPQKPKISVKSITFEQLKVQAGQDAGGIGTD 163
Query: 174 MLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQ 233
M+T+N ++RML N TFFG++V+S ++L +S +T+ +G +KK++Q RKS+RTV VN+
Sbjct: 164 MITMNATLRMLYRNTGTFFGVHVTSSPIDLSFSQITIGSGSIKKFYQSRKSQRTVVVNVL 223
Query: 234 GSKVPLYGAGAD----------------------VASSVDNGKISLTLVFEVKSRGNVVG 271
G K+PLYG+G+ V + + L F V+SR V+G
Sbjct: 224 GDKIPLYGSGSTLVPPPPPAPIPKPKKKKGPIVIVEPPAPPAPVPMRLNFTVRSRAYVLG 283
Query: 272 KLVRTKHTQRVSCSIAVDPHN-NKYIKLKEN 301
KLV+ K +R+ C I + +K+I + N
Sbjct: 284 KLVQPKFYKRIVCLINFEHKKLSKHIPITNN 314
>UniRef100_Q84VY4 At5g42860 [Arabidopsis thaliana]
Length = 320
Score = 195 bits (495), Expect = 1e-48
Identities = 121/331 (36%), Positives = 177/331 (52%), Gaps = 47/331 (14%)
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKSST------MQPSPMDSPS 53
MH KTDS+VTS SSP RS ++ Y+VQSPSR+S HDG+K++T + SPM SP
Sbjct: 1 MHAKTDSEVTSLSASSPTRSPRRPAYFVQSPSRDS-HDGEKTATSFHSTPVLTSPMGSPP 59
Query: 54 HQSYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNG 113
H HS +S S+I+G G K + EE G D D +
Sbjct: 60 HS----HSSSSRFSKINGSKRKGHAGEKQFAMI------------EEEGLLDDGDREQEA 103
Query: 114 VSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTK 173
+ RR +VGF L+F+ F I++AA+ KP++SVKS+T G D G+ T
Sbjct: 104 LPRRCYVLAFIVGFSLLFAFFSLILYAAAKPQKPKISVKSITFEQLKVQAGQDAGGIGTD 163
Query: 174 MLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQ 233
M+T+N ++RML N TFFG +V+S ++L +S +T+ +G +KK++Q RKS+RTV VN+
Sbjct: 164 MITMNATLRMLYRNTGTFFGXHVTSSPIDLSFSQITIGSGSIKKFYQSRKSQRTVVVNVL 223
Query: 234 GSKVPLYGAGAD----------------------VASSVDNGKISLTLVFEVKSRGNVVG 271
G K+PLYG+G+ V + + L F V+SR V+G
Sbjct: 224 GDKIPLYGSGSTLVPPPPPAPIPKPKKKKGPIVIVEPPAPPAPVPMRLNFTVRSRAYVLG 283
Query: 272 KLVRTKHTQRVSCSIAVDPHN-NKYIKLKEN 301
KLV+ K +R+ C I + +K+I + N
Sbjct: 284 KLVQPKFYKRIVCLINFEHKKLSKHIPITNN 314
>UniRef100_Q7XQQ4 OSJNBa0084A10.1 protein [Oryza sativa]
Length = 490
Score = 184 bits (467), Expect = 3e-45
Identities = 118/323 (36%), Positives = 171/323 (52%), Gaps = 39/323 (12%)
Query: 1 MHIKTDSDVTSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSSTMQPSPMDSPSHQSYGHH 60
+H KTDS+VTS +S S + YYVQSPS + + +S+ SP SP +S G+H
Sbjct: 156 LHAKTDSEVTSSMAAS--SPPRAAYYVQSPSHDDGENKTAASSFHSSPAASPP-RSLGNH 212
Query: 61 SRASSSSRISGGYNSSFLGRKVNRKVNDKG---------------------WIGSKVIEE 99
SR SSSSR S + S R+ D G W+ IEE
Sbjct: 213 SRESSSSRFSAAKSGS--SRRTAAAGGDGGKGGVAAGRGGGGGGGGGRRSPWMKEAAIEE 270
Query: 100 ENGGYGDFDGDGNG--------VSRRVQFFVAVVG-FVLVFSLFCFIIWAASLHYKPQLS 150
E D D DG G + RR ++ + VG F +F F I+W AS + KP +S
Sbjct: 271 EGLLMEDDDADGGGGGGGGFSSLPRRWRYALGFVGAFFALFFFFALILWGASHNQKPVVS 330
Query: 151 VKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTV 210
+ S+T HNF G+D + VPT++ TVN ++RM N +FFG++V+++ + L Y L +
Sbjct: 331 INSITFHNFVIQAGTDASLVPTELSTVNATVRMTFRNTGSFFGVHVTAEPLTLYYYQLLM 390
Query: 211 ATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAGADVASSVDNG----KISLTLVFEVKSR 266
A+G +K ++Q RKS R V+V + G KVPLYG G+ ++S+ G + L L +SR
Sbjct: 391 ASGNVKYFYQSRKSSRHVAVAVVGDKVPLYGGGSGLSSTPVKGAPPAPVPLQLAVRFRSR 450
Query: 267 GNVVGKLVRTKHTQRVSCSIAVD 289
V+GKLV+ K V CS+ +D
Sbjct: 451 AFVLGKLVKPKFLTNVQCSVRLD 473
>UniRef100_Q6ET94 Hypothetical protein OJ1725_H08.8-1 [Oryza sativa]
Length = 337
Score = 184 bits (466), Expect = 3e-45
Identities = 116/323 (35%), Positives = 174/323 (52%), Gaps = 47/323 (14%)
Query: 2 HIKTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHD------------------GDKSS 42
H KTDSDVTS PSSP RS +++ YYV SP+ +SH +K S
Sbjct: 4 HAKTDSDVTSLAPSSPPRSPRRSAYYVLSPA--ASHPDVVVASGGAGGGGGGVAAAEKMS 61
Query: 43 TMQPSPMDSPSHQSYGH------HSRASSSSRI--SGGYNSSFLGRKVNRKVNDKGWIGS 94
+P +SP H Y H HSR SS+ R+ S S R++ GS
Sbjct: 62 FAGSTPAESPLHYHYHHSGAAVHHSRESSTGRLLFSDQLRSGAAAGVPWRRLAQGSGAGS 121
Query: 95 KVIEEENGGYGDFDGDGNGVSRRVQ----FFVAVVGFVLVFSLFCFIIWAASLHYKPQLS 150
GD D D G++ + + FV VF+ F ++W AS YKP +
Sbjct: 122 ---------VGDDDDDEGGLAGAASQWRCYALGAFAFVAVFAFFLLVLWGASKSYKPHVV 172
Query: 151 VKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTV 210
VKS+ ++ G+D TGVPTKM++VN ++R+ N TFF ++V+S +L Y DLTV
Sbjct: 173 VKSVVFETYHIQGGTDRTGVPTKMMSVNATVRLRFRNRGTFFSLHVTSTPFHLFYDDLTV 232
Query: 211 ATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAGADVASSVDNGK-----ISLTLVFEVKS 265
ATG + +++Q R+S R V+V++ G +VPLYGAGA++ S +NG+ + + + F +++
Sbjct: 233 ATGHMAEFYQPRRSGRVVTVSVVGKQVPLYGAGAELHSKPNNGRLGPAVVPVRMAFVLRA 292
Query: 266 RGNVVGKLVRTKHTQRVSCSIAV 288
R +++G LVR+K +RV C + V
Sbjct: 293 RAHILGLLVRSKFYRRVLCRLDV 315
>UniRef100_Q6RZV9 Hypothetical protein G9-6 [Musa acuminata]
Length = 267
Score = 183 bits (464), Expect = 6e-45
Identities = 113/269 (42%), Positives = 152/269 (56%), Gaps = 15/269 (5%)
Query: 49 MDSP--SHQSYGHHSRASSSSRISGGYN--------SSFLGRKVNRKVNDKGWIGSKVIE 98
M SP SH S G SR SSSSR SGG SS RK R +DK W IE
Sbjct: 1 MASPRHSHSSVGPQSRDSSSSRFSGGIKPGGGPRKMSSHDDRKGGRSKSDKAWKECATIE 60
Query: 99 EENGGYGDFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHN 158
EE G + + + G+ RR V ++GFV++FS F I+W AS KP++++KS+ N
Sbjct: 61 EE--GLLEDEREEKGLPRRCYSLVFLLGFVILFSFFALILWGASRSQKPEITMKSVRFEN 118
Query: 159 FYFGEGSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKY 218
F G+D + VPT M T+N ++R N TFFG++VS+ L Y LT+A GE+ +
Sbjct: 119 FIIQAGTDASLVPTDMATLNSTVRFSFRNTGTFFGVHVSATPFVLSYDQLTLAGGEMNNF 178
Query: 219 HQQRKSRRTVSVNIQGSKVPLYGAGADVASSVDN-GKISLTLVFEVKSRGNVVGKLVRTK 277
+Q RKS+R V V + G KVPLYG G +ASS + + L+L F VKSR V+GKLV+ K
Sbjct: 179 YQSRKSQRNVEVVVLGKKVPLYGGGPSLASSPGSVATVPLSLSFMVKSRAYVLGKLVKPK 238
Query: 278 HTQRVSCSIAVDPHN-NKYIKLKENECRY 305
V C + + P N + LK N C+Y
Sbjct: 239 FEIGVQCKVVMKPAQLNTRVSLK-NSCQY 266
>UniRef100_P93747 Hypothetical protein At2g41990 [Arabidopsis thaliana]
Length = 297
Score = 162 bits (410), Expect = 1e-38
Identities = 103/302 (34%), Positives = 167/302 (55%), Gaps = 28/302 (9%)
Query: 1 MHIKTDSDVTSFDP---SSPRSQKQTPYYVQSPSRESSHDGDKSSTMQP-SPMDSPSHQS 56
MH KTDS+ TS D S PRS + YYVQSPS +HD +K S S M SP+H
Sbjct: 1 MHAKTDSEATSIDAAALSPPRSAIRPLYYVQSPS---NHDVEKMSFGSGCSLMGSPTHPH 57
Query: 57 YGH-----HSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDG 111
Y H HSR SS+SR S S+ + R+ + G +++ G GD
Sbjct: 58 YYHCSPIHHSRESSTSRFSDRALLSYKSIRERRRYINDG-------DDKTDG-----GDD 105
Query: 112 NGVSRRVQFFV-AVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
+ R V+ +V ++ + +F++F I+W AS Y P+++VK + V + G+D++GV
Sbjct: 106 DDPFRNVRLYVWLLLSVIFLFTVFSLILWGASKSYPPKVTVKGMLVRDLNLQAGNDLSGV 165
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSV 230
PT ML++N ++R+ NP+TFF ++V++ + L YS+L +++GE+ K+ R V
Sbjct: 166 PTDMLSLNSTVRIYYRNPSTFFAVHVTASPLLLHYSNLLLSSGEMNKFTVGRNGETNVVT 225
Query: 231 NIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDP 290
+QG ++PLYG V+ +D + L L + S+ ++G+LV +K R+ CS +D
Sbjct: 226 VVQGHQIPLYGG---VSFHLDTLSLPLNLTIVLHSKAYILGRLVTSKFYTRIICSFTLDA 282
Query: 291 HN 292
++
Sbjct: 283 NH 284
>UniRef100_Q9C6U2 Hypothetical protein T8G24.9 [Arabidopsis thaliana]
Length = 271
Score = 72.8 bits (177), Expect = 1e-11
Identities = 48/196 (24%), Positives = 88/196 (44%), Gaps = 4/196 (2%)
Query: 113 GVSRRVQFFVAVVGFVLVFSL--FCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
G S + V VG+ +FSL + + A+ P +S + + F EG D GV
Sbjct: 73 GTSNSSWWIVLQVGWRFLFSLGVALLVFYIATQPPHPNISFRIGRFNQFMLEEGVDSHGV 132
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSV 230
TK LT NCS ++++ N + FG+++ S+ + L A + K + T +
Sbjct: 133 STKFLTFNCSTKLIIDNKSNVFGLHIHPPSIKFFFGPLNFAKAQGPKLYGLSHESTTFQL 192
Query: 231 NIQGSKVPLYGAGADVASS-VDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAV- 288
I + +YGAG ++ + + L L + S VV ++ K+ +V C + +
Sbjct: 193 YIATTNRAMYGAGTEMNDMLLSRAGLPLILRTSIISDYRVVWNIINPKYHHKVECLLLLA 252
Query: 289 DPHNNKYIKLKENECR 304
D + ++ + +CR
Sbjct: 253 DKERHSHVTMIREKCR 268
>UniRef100_Q9T004 Hypothetical protein T12J5.40 [Arabidopsis thaliana]
Length = 163
Score = 50.4 bits (119), Expect = 6e-05
Identities = 45/147 (30%), Positives = 70/147 (47%), Gaps = 21/147 (14%)
Query: 15 SSPRSQKQTPYYVQSPSRESSHDGDKSSTMQP-SPMDSPSHQ-----SYGHHSRASSSS- 67
SSP++ ++ Y V SP D DK ST SP SP + ++ HHS A SSS
Sbjct: 8 SSPQNTRKPVYVVHSPPNT---DVDKISTGSGFSPFGSPLNDQGQVSNFQHHSVAESSSY 64
Query: 68 -RISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVSRRVQFFVAVV- 125
R SG + + +V+ + + E+E+ Y + DG R +F+ ++
Sbjct: 65 PRSSGPLRNEYSSVQVHD-------LDRRTHEDED--YDEMDGPDEKRRRITRFYSCLLF 115
Query: 126 GFVLVFSLFCFIIWAASLHYKPQLSVK 152
VL F+LFC I+W S + P ++K
Sbjct: 116 TLVLAFTLFCLILWGVSKSFAPIATLK 142
>UniRef100_Q9SRN0 T19F11.6 protein [Arabidopsis thaliana]
Length = 209
Score = 49.3 bits (116), Expect = 1e-04
Identities = 47/183 (25%), Positives = 79/183 (42%), Gaps = 14/183 (7%)
Query: 108 DGDGNGVSRRV---QFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
D + +G SRR + F +++ + + L +IWA KP+ ++ TV+ F
Sbjct: 3 DCENHGHSRRKLIRRIFWSIIFVLFIIFLTILLIWAILQPSKPRFILQDATVYAF----- 57
Query: 165 SDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQR 222
+V+G P +LT N + + NP GIY V Y +T T + +Q
Sbjct: 58 -NVSGNPPNLLTSNFQITLSSRNPNNKIGIYYDRLDVYATYRSQQITFPT-SIPPTYQGH 115
Query: 223 KSRRTVSVNIQGSKVPLYG-AGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQR 281
K S + G+ VP+ G + + DNG + L + + + R VG + K+
Sbjct: 116 KDVDIWSPFVYGTSVPIAPFNGVSLDTDKDNGVVLLIIRADGRVRWK-VGTFITGKYHLH 174
Query: 282 VSC 284
V C
Sbjct: 175 VKC 177
>UniRef100_Q84WG2 Putative VAMP protein SEC22 [Arabidopsis thaliana]
Length = 209
Score = 48.9 bits (115), Expect = 2e-04
Identities = 47/183 (25%), Positives = 79/183 (42%), Gaps = 14/183 (7%)
Query: 108 DGDGNGVSRRV---QFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
D + +G SRR + F +++ + + L +IWA KP+ ++ TV+ F
Sbjct: 3 DCENHGHSRRKLIRRIFWSIIFVLFIIFLTILLIWAILQPSKPRFILQDATVYAF----- 57
Query: 165 SDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQR 222
+V+G P +LT N + + NP GIY V Y +T T + +Q
Sbjct: 58 -NVSGNPPNILTSNFQITLSSRNPNNKIGIYYDRLDVYATYRSQQITFPT-SIPPTYQGH 115
Query: 223 KSRRTVSVNIQGSKVPLYG-AGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQR 281
K S + G+ VP+ G + + DNG + L + + + R VG + K+
Sbjct: 116 KDVDIWSPFVYGTSVPIAPFNGVSLDTDKDNGVVLLIIRADGRVRWK-VGTFITGKYHLH 174
Query: 282 VSC 284
V C
Sbjct: 175 VKC 177
>UniRef100_Q6PUQ1 Hypothetical protein [Petunia integrifolia subsp. inflata]
Length = 234
Score = 48.5 bits (114), Expect = 2e-04
Identities = 47/200 (23%), Positives = 83/200 (41%), Gaps = 12/200 (6%)
Query: 111 GNGVSRRVQFFVAVVGFVLVF-SLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTG 169
GN R + V+ F++V F F + P +SL V F +
Sbjct: 42 GNCCIRCICCCYCVLFFIIVLLGAFAFYFYTMYQPKLPTYKFQSLEVKEFGYQ------- 94
Query: 170 VPTKMLTVNCSMRMLVHNPATFFG-IYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTV 228
P LT + + + NP IY SVN+ Y+D T+ +G L +HQ K+ +
Sbjct: 95 -PDFSLTTDIIVTIRAENPNKAISLIYGKESSVNVTYTDSTICSGTLPSFHQGHKNVTMM 153
Query: 229 SVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAV 288
+ ++G G + + NGKI L + +V + VG+ + +CS+ V
Sbjct: 154 QIELKGKSQFGSGLYEAMQDNEKNGKIPLLVKLKVPVQ-VYVGEYPLRQFNVLANCSLVV 212
Query: 289 DP-HNNKYIKLKENECRYNI 307
D K +++ +N+ + +
Sbjct: 213 DNLKPGKKVEIIKNDVTFKL 232
>UniRef100_Q9SJ54 Putative harpin-induced protein [Arabidopsis thaliana]
Length = 210
Score = 45.1 bits (105), Expect = 0.002
Identities = 42/184 (22%), Positives = 76/184 (40%), Gaps = 15/184 (8%)
Query: 109 GDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVT 168
G G G + R+ ++GF+++ + F++W KP+ ++ TV+ F +
Sbjct: 12 GGGGGTASRI--CGVIIGFIIIVLITIFLVWIILQPTKPRFILQDATVYAFNLSQ----- 64
Query: 169 GVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQRKSRR 226
+LT N + + N + GIY V Y +T+ T + +Q K
Sbjct: 65 ---PNLLTSNFQITIASRNRNSRIGIYYDRLHVYATYRNQQITLRTA-IPPTYQGHKEDN 120
Query: 227 TVSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGN-VVGKLVRTKHTQRVSCS 285
S + G+ VP+ A VA + + +TL+ R VG L+ K+ V C
Sbjct: 121 VWSPFVYGNSVPIAPFNA-VALGDEQNRGFVTLIIRADGRVRWKVGTLITGKYHLHVRCQ 179
Query: 286 IAVD 289
++
Sbjct: 180 AFIN 183
>UniRef100_Q9FFC1 Gb|AAF18257.1 [Arabidopsis thaliana]
Length = 207
Score = 44.7 bits (104), Expect = 0.003
Identities = 39/166 (23%), Positives = 72/166 (42%), Gaps = 18/166 (10%)
Query: 121 FVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCS 180
F+A VGF+ I W + K + +V++ +V NF + ++ T T+
Sbjct: 38 FMAAVGFL--------ITWLETKPKKLRYTVENASVQNFNLTNDNHMSA--TFQFTIQS- 86
Query: 181 MRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLY 240
HNP +Y SS + + + D T+A ++ +HQ R + + + + V +
Sbjct: 87 -----HNPNHRISVYYSSVEIFVKFKDQTLAFDTVEPFHQPRMNVKQIDETLIAENVAVS 141
Query: 241 GA-GADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCS 285
+ G D+ S GKI + + + R VG + T ++ CS
Sbjct: 142 KSNGKDLRSQNSLGKIGFEVFVKARVRFK-VGIWKSSHRTAKIKCS 186
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 527,089,280
Number of Sequences: 2790947
Number of extensions: 22383531
Number of successful extensions: 77368
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 97
Number of HSP's that attempted gapping in prelim test: 76468
Number of HSP's gapped (non-prelim): 659
length of query: 307
length of database: 848,049,833
effective HSP length: 127
effective length of query: 180
effective length of database: 493,599,564
effective search space: 88847921520
effective search space used: 88847921520
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC122722.4


