

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.6 + phase: 0
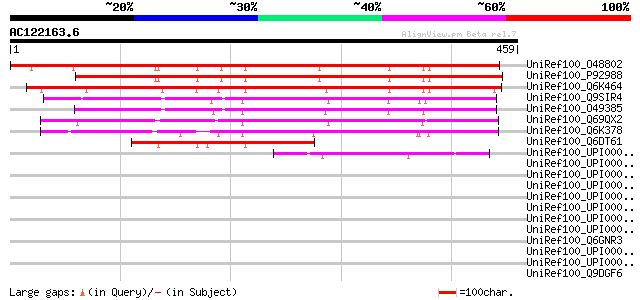
(459 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O48802 F2401.12 [Arabidopsis thaliana] 419 e-115
UniRef100_P92988 Putative cytoskeletal protein [Arabidopsis thal... 416 e-115
UniRef100_Q6K464 Putative octicosapeptide/Phox/Bem1p (PB1) domai... 402 e-110
UniRef100_Q9SIR4 Hypothetical protein At2g25290 [Arabidopsis tha... 302 1e-80
UniRef100_O49385 Hypothetical protein F10N7.120 [Arabidopsis tha... 298 2e-79
UniRef100_Q69QX2 Putative octicosapeptide/Phox/Bem1p (PB1) domai... 295 2e-78
UniRef100_Q6K378 Tetratricopeptide repeat protein-like [Oryza sa... 293 9e-78
UniRef100_Q6DT61 AT1G62390 [Arabidopsis lyrata] 176 1e-42
UniRef100_UPI00003069CB UPI00003069CB UniRef100 entry 118 4e-25
UniRef100_UPI0000366214 UPI0000366214 UniRef100 entry 41 0.082
UniRef100_UPI00002347AF UPI00002347AF UniRef100 entry 41 0.082
UniRef100_UPI00003A9BAA UPI00003A9BAA UniRef100 entry 40 0.11
UniRef100_UPI00000142CD UPI00000142CD UniRef100 entry 40 0.14
UniRef100_UPI00003AAE40 UPI00003AAE40 UniRef100 entry 40 0.14
UniRef100_UPI00003AAE3F UPI00003AAE3F UniRef100 entry 40 0.14
UniRef100_UPI000034DCC1 UPI000034DCC1 UniRef100 entry 40 0.14
UniRef100_Q6GNR3 MGC80940 protein [Xenopus laevis] 40 0.14
UniRef100_UPI000042DC3D UPI000042DC3D UniRef100 entry 40 0.18
UniRef100_UPI000031A5CC UPI000031A5CC UniRef100 entry 39 0.24
UniRef100_Q9DGF6 TRK-fused protein TFG [Xenopus laevis] 39 0.24
>UniRef100_O48802 F2401.12 [Arabidopsis thaliana]
Length = 751
Score = 419 bits (1076), Expect = e-115
Identities = 247/534 (46%), Positives = 323/534 (60%), Gaps = 91/534 (17%)
Query: 1 MRRSGSRRKKGNVSQPNS-------PSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEH 53
+ + G G+VS PN+ P N E S K L+ P
Sbjct: 209 VHKKGVTSPVGSVSLPNASNGKVERPQVVNPVTENGGSVSKGQASRVVLKPVSHSPKGSK 268
Query: 54 LKE---------NQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRS 104
++E + ++ +I WR LK VYD DIRL QMP+NC F+ LR+IV +FP S++
Sbjct: 269 VEELGSSSVAVVGKVQEKRIRWRPLKFVYDHDIRLGQMPVNCRFKELREIVSSRFPSSKA 328
Query: 105 VLIKYKDNDDDLVTITSTEELRFAES---CVY-------KTDSVEILKLYIVEVSPEHEP 154
VLIKYKDND DLVTITST EL+ AES C+ K+DSV +L+L++V+VSPE EP
Sbjct: 329 VLIKYKDNDGDLVTITSTAELKLAESAADCILTKEPDTDKSDSVGMLRLHVVDVSPEQEP 388
Query: 155 PLLKEEKEEENNEK------QKPLDCVLDEKMCTE-CNKVVEN-------------LEID 194
LL+EE+EE + P + + + ++ TE +K VE LE+D
Sbjct: 389 MLLEEEEEEVEEKPVIEEVISSPTESLSETEINTEKTDKEVEKEKASSSEDPETKELEMD 448
Query: 195 DWLYEFAQLFRSRVGTDK--YIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVA 252
DWL++FA LFR+ VG D +ID H+LG E CS+ALEETVTS++AQ L DKA KFQEVA
Sbjct: 449 DWLFDFAHLFRTHVGIDPDAHIDLHELGMELCSEALEETVTSEKAQPLFDKASAKFQEVA 508
Query: 253 ALAFFNWGNVHMCAARKFVRMDENENE--VLVMNESEFDFVQEKYYLAREKYEQAVVIKP 310
ALAFFNWGNVHMCAARK + +DE+ + V ++ +++V+E+Y LA+EKYEQA+ IKP
Sbjct: 509 ALAFFNWGNVHMCAARKRIPLDESAGKEVVAAQLQTAYEWVKERYTLAKEKYEQALSIKP 568
Query: 311 DFYEGLLAIGQQQFELAKLNWSFGIANKMDLG----KETLRLFDVAEEKMTAANDAWENL 366
DFYEGLLA+GQQQFE+AKL+WS+ +A K+D+ ETL LFD AE KM A + WE L
Sbjct: 569 DFYEGLLALGQQQFEMAKLHWSYLLAQKIDISGWDPSETLNLFDSAEAKMKDATEMWEKL 628
Query: 367 EKGKLGE------------------QGSVG-------------------MRSQIHLFWGN 389
E+ ++ + QG G MRSQIHLFWGN
Sbjct: 629 EEQRMDDLKNPNSNKKEEVSKRRKKQGGDGNEEVSETITAEEAAEQATAMRSQIHLFWGN 688
Query: 390 MLFERSQVEFKLGMSDWKKKLDASVERFKIAGASEADVSGILKKHCFNGNARDE 443
MLFERSQVE K+G W K LD++VERFK+AGASEAD++ ++K HC N A E
Sbjct: 689 MLFERSQVECKIGKDGWNKNLDSAVERFKLAGASEADIATVVKNHCSNEAAATE 742
>UniRef100_P92988 Putative cytoskeletal protein [Arabidopsis thaliana]
Length = 782
Score = 416 bits (1069), Expect = e-115
Identities = 231/463 (49%), Positives = 302/463 (64%), Gaps = 76/463 (16%)
Query: 60 EDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTI 119
++ +I WR LK VYD DIRL QMP+NC F+ LR+IV +FP S++VLIKYKDND DLVTI
Sbjct: 284 QEKRIRWRPLKFVYDHDIRLGQMPVNCRFKELREIVSSRFPSSKAVLIKYKDNDGDLVTI 343
Query: 120 TSTEELRFAES---CVY-------KTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEK- 168
TST EL+ AES C+ K+DSV +L+L++V+VSPE EP L+ +K + EK
Sbjct: 344 TSTAELKLAESAADCILTKEPDTDKSDSVGMLRLHVVDVSPEQEPMLVSRKKRRKMEEKP 403
Query: 169 ------QKPLDCVLDEKMCTE-CNKVVEN-------------LEIDDWLYEFAQLFRSRV 208
P + + + ++ TE +K VE LE+DDWL++FA LFR+ V
Sbjct: 404 VIEEVISSPTESLSETEINTEKTDKEVEKEKASSSEDPETKELEMDDWLFDFAHLFRTHV 463
Query: 209 GTDK--YIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNWGNVHMCA 266
G D +ID H+LG E CS+ALEETVTS++AQ L DKA KFQEVAALAFFNWGNVHMCA
Sbjct: 464 GIDPDAHIDLHELGMELCSEALEETVTSEKAQPLFDKASAKFQEVAALAFFNWGNVHMCA 523
Query: 267 ARKFVRMDENENE--VLVMNESEFDFVQEKYYLAREKYEQAVVIKPDFYEGLLAIGQQQF 324
ARK + +DE+ + V ++ +++V+E+Y LA+EKYEQA+ IKPDFYEGLLA+GQQQF
Sbjct: 524 ARKRIPLDESAGKEVVAAQLQTAYEWVKERYTLAKEKYEQALSIKPDFYEGLLALGQQQF 583
Query: 325 ELAKLNWSFGIANKMDLG----KETLRLFDVAEEKMTAANDAWENLEKGKLGE------- 373
E+AKL+WS+ +A K+D+ ETL LFD AE KM A + WE LE+ ++ +
Sbjct: 584 EMAKLHWSYLLAQKIDISGWDPSETLNLFDSAEAKMKDATEMWEKLEEQRMDDLKNPNSN 643
Query: 374 -----------QGSVG-------------------MRSQIHLFWGNMLFERSQVEFKLGM 403
QG G MRSQIHLFWGNMLFERSQVE K+G
Sbjct: 644 KKEEVSKRRKKQGGDGNEEVSETITAEEAAEQATAMRSQIHLFWGNMLFERSQVECKIGK 703
Query: 404 SDWKKKLDASVERFKIAGASEADVSGILKKHCFNGNARDEKIK 446
W K LD++VERFK+AGASEAD++ ++K HC N A E+++
Sbjct: 704 DGWNKNLDSAVERFKLAGASEADIATVVKNHCSNEAAATEEMR 746
>UniRef100_Q6K464 Putative octicosapeptide/Phox/Bem1p (PB1) domain-containing protein
[Oryza sativa]
Length = 544
Score = 402 bits (1032), Expect = e-110
Identities = 239/517 (46%), Positives = 315/517 (60%), Gaps = 87/517 (16%)
Query: 16 PNSPSSSNIAFE---PHPSPKKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQ--LK 70
P+ PS + F P + + + S+++ L P++ LK+ D K+ R LK
Sbjct: 6 PSPPSPKLVPFSNSPPSSANASAAESSQKVTPTPLVPSSLSLKDKALMDKKVVTRSRPLK 65
Query: 71 LVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAES 130
LVYD DIRLAQMP CSFR LR++V +FP S++VLIKYKD D DLVTIT + ELR AES
Sbjct: 66 LVYDHDIRLAQMPEKCSFRTLREVVASRFPSSKAVLIKYKDADGDLVTITCSAELRLAES 125
Query: 131 CVYKTDS------------VEILKLYIVEVSPEHEPPL-LKEEKEEENNE---------- 167
CV S + +L+L+IVEVSP+ EPP+ +EEK E++NE
Sbjct: 126 CVDIAGSEVIEDGARHGQKLPMLRLHIVEVSPDQEPPMPTEEEKLEQDNELLVKGEDNSP 185
Query: 168 -------------KQKPLDCVLDEKMCTECNKV------VENLEIDDWLYEFAQLFRSRV 208
KQ + V + + T K + EIDDWL +FA LFR++V
Sbjct: 186 HASAAVVTDAEVTKQDVENVVAEAEQNTLTGKKDCGHAECKEAEIDDWLLQFADLFRNQV 245
Query: 209 GTDK--YIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNWGNVHMCA 266
G D ++D H+LG E CS+ALEETVTS+EAQ L + A KFQEVAALA FNWGNVHMCA
Sbjct: 246 GVDADAHLDLHELGMELCSEALEETVTSEEAQALFEMAAAKFQEVAALALFNWGNVHMCA 305
Query: 267 ARKFVRMDENENEVLVMNE--SEFDFVQEKYYLAREKYEQAVVIKPDFYEGLLAIGQQQF 324
ARK + +DE+ + ++ + + +D+V+++Y LA KYE+A+ IKPDFYEGLLA+GQQ F
Sbjct: 306 ARKRIPLDESAPKKVMSAQLCTAYDWVRDRYALAGSKYEEALKIKPDFYEGLLALGQQHF 365
Query: 325 ELAKLNWSFGIANKMDLG----KETLRLFDVAEEKMTAANDAWENLEKGKLGE------- 373
E AKL+WSF +A+K+DL +T +LFD AE KM AA + WE +E+ ++ E
Sbjct: 366 ETAKLHWSFALADKVDLSAWDSSQTFKLFDSAEHKMRAATEMWEKVEEQRMAELKEPSSE 425
Query: 374 -------------QGSVG----------MRSQIHLFWGNMLFERSQVEFKLGMSDWKKKL 410
QG + MR QIHLFWGNMLFERSQVEFKL ++DWKK L
Sbjct: 426 ALKKRKKQHNADGQGELTPEEAAEQAAVMRQQIHLFWGNMLFERSQVEFKLDIADWKKNL 485
Query: 411 DASVERFKIAGASEADVSGILKKHCFN--GNARDEKI 445
DAS+ERFK+AGASE+D+S +LK H N D+KI
Sbjct: 486 DASIERFKLAGASESDISAVLKNHFSNTVSECEDKKI 522
>UniRef100_Q9SIR4 Hypothetical protein At2g25290 [Arabidopsis thaliana]
Length = 697
Score = 302 bits (774), Expect = 1e-80
Identities = 177/451 (39%), Positives = 268/451 (59%), Gaps = 47/451 (10%)
Query: 31 SPKKSDQISRRLREPKLGPTNEHLKENQ-KEDVKIAWRQLKLVYDDDIRLAQMPINCSFR 89
S +K I + E K+ ++ + ++ KED + R +KLV+ DDIR AQ+P++ S
Sbjct: 245 SGRKGKAIEEKKLEDKVAVMDKEVIASEIKEDATVT-RTVKLVHGDDIRWAQLPLDSSVV 303
Query: 90 LLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIVEVS 149
L+RD++K++FP + LIKY+D++ DLVTIT+T+ELR A S K S +LYI EVS
Sbjct: 304 LVRDVIKDRFPALKGFLIKYRDSEGDLVTITTTDELRLAASTREKLGS---FRLYIAEVS 360
Query: 150 PEHEPPLLKEEKEEENNEKQKPLDCVLDEKMC---TECNKVVENLEIDDWLYEFAQLFRS 206
P EP + +E ++ K V D E K +LE W+++FAQLF++
Sbjct: 361 PNQEPTYDVIDNDESTDKFAKGSSSVADNGSVGDFVESEKASTSLE--HWIFQFAQLFKN 418
Query: 207 RVG--TDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNWGNVHM 264
VG +D Y++ H+LG + ++A+E+ VT ++AQ+L D A KFQE+AALA FNWGNVHM
Sbjct: 419 HVGFDSDSYLELHNLGMKLYTEAMEDIVTGEDAQELFDIAADKFQEMAALAMFNWGNVHM 478
Query: 265 CAARKFVRMDENENEVLVMNESE--FDFVQEKYYLAREKYEQAVVIKPDFYEGLLAIGQQ 322
AR+ + E+ + ++ + E F++ + +Y A EKYE AV IK DFYE LLA+GQQ
Sbjct: 479 SKARRQIYFPEDGSRETILEKVEAGFEWAKNEYNKAAEKYEGAVKIKSDFYEALLALGQQ 538
Query: 323 QFELAKLNWSFGIANKMDL----GKETLRLFDVAEEKMTAANDAWENLEKG--------- 369
QFE AKL W ++ ++D+ ++ L+L++ AEE M WE +E+
Sbjct: 539 QFEQAKLCWYHALSGEVDIESDASQDVLKLYNKAEESMEKGMQIWEEMEERRLNGISNFD 598
Query: 370 -------KLGEQG-------------SVGMRSQIHLFWGNMLFERSQVEFKLGMSDWKKK 409
KLG G + M SQI+L WG++L+ERS VE+KLG+ W +
Sbjct: 599 KHKELLQKLGLDGIFSEASDEESAEQTANMSSQINLLWGSLLYERSIVEYKLGLPTWDEC 658
Query: 410 LDASVERFKIAGASEADVSGILKKHCFNGNA 440
L+ +VE+F++AGAS D++ ++K HC + NA
Sbjct: 659 LEVAVEKFELAGASATDIAVMVKNHCSSDNA 689
>UniRef100_O49385 Hypothetical protein F10N7.120 [Arabidopsis thaliana]
Length = 811
Score = 298 bits (763), Expect = 2e-79
Identities = 167/422 (39%), Positives = 253/422 (59%), Gaps = 45/422 (10%)
Query: 59 KEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVT 118
K++ R +KLV+ DDIR AQ+P++ + RL+RD+++++FP R LIKY+D + DLVT
Sbjct: 310 KKEGATVTRTIKLVHGDDIRWAQLPLDSTVRLVRDVIRDRFPALRGFLIKYRDTEGDLVT 369
Query: 119 ITSTEELRFAESCVYKTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEKQKPLDCVLDE 178
IT+T+ELR A S K S L+LYI EV+P+ EP E ++ K L + D
Sbjct: 370 ITTTDELRLAASTHDKLGS---LRLYIAEVNPDQEPTYDGMSNTESTDKVSKRLSSLADN 426
Query: 179 KMCTE---CNKVVENLEIDDWLYEFAQLFRSRVG--TDKYIDFHDLGTEFCSDALEETVT 233
E +K E +W+++FAQLF++ VG +D Y+D HDLG + ++A+E+ VT
Sbjct: 427 GSVGEYVGSDKASGCFE--NWIFQFAQLFKNHVGFDSDSYVDLHDLGMKLYTEAMEDAVT 484
Query: 234 SDEAQDLLDKAEFKFQEVAALAFFNWGNVHMCAARKFVRMDENENEVLVMN--ESEFDFV 291
++AQ+L A KFQE+ ALA NWGNVHM ARK V + E+ + ++ E+ F +
Sbjct: 485 GEDAQELFQIAADKFQEMGALALLNWGNVHMSKARKQVCIPEDASREAIIEAVEAAFVWT 544
Query: 292 QEKYYLAREKYEQAVVIKPDFYEGLLAIGQQQFELAKLNWSFGIANKMDL----GKETLR 347
Q +Y A EKYE+A+ +KPDFYE LLA+GQ+QFE AKL W + +K+DL +E L+
Sbjct: 545 QNEYNKAAEKYEEAIKVKPDFYEALLALGQEQFEHAKLCWYHALKSKVDLESEASQEVLK 604
Query: 348 LFDVAEEKMTAANDAWENLEKGKLGE-----------------------------QGSVG 378
L++ AE+ M WE +E+ +L + +
Sbjct: 605 LYNKAEDSMERGMQIWEEMEECRLNGISKLDKHKNMLRKLELDELFSEASEEETVEQTAN 664
Query: 379 MRSQIHLFWGNMLFERSQVEFKLGMSDWKKKLDASVERFKIAGASEADVSGILKKHCFNG 438
M SQI+L WG++L+ERS VE+KLG+ W + L+ +VE+F++AGAS D++ ++K HC +
Sbjct: 665 MSSQINLLWGSLLYERSIVEYKLGLPTWDECLEVAVEKFELAGASATDIAVMVKNHCSSE 724
Query: 439 NA 440
+A
Sbjct: 725 SA 726
>UniRef100_Q69QX2 Putative octicosapeptide/Phox/Bem1p (PB1) domain-/tetratricopeptide
repeat (TPR)-containing protein [Oryza sativa]
Length = 774
Score = 295 bits (754), Expect = 2e-78
Identities = 172/455 (37%), Positives = 257/455 (55%), Gaps = 47/455 (10%)
Query: 29 HPSPKKSDQISRRLREPKLGPTNEHLKENQKE-------DVKIAWRQLKLVYDDDIRLAQ 81
H + KK + + ++ H +EN+ E VK A + LKLV+ +DIR AQ
Sbjct: 274 HSAGKKIRRADAKKQKHSAMEPVHHAEENRHERYTETSVHVKEAMKDLKLVFGEDIRCAQ 333
Query: 82 MPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDSVEIL 141
MP NC+ LRDIV+ KFP +++LIKYKD + DLVTITS++ELR+A S D +
Sbjct: 334 MPANCNLSQLRDIVQNKFPSLKALLIKYKDKEGDLVTITSSDELRWAYSLA---DLEGPI 390
Query: 142 KLYIVEVSPEHEPPLLKEEKEEENNEKQKPLDCVLDEKMCTECNKVVENLEIDDWLYEFA 201
+LYIV V P E + + +K + + + N IDDW+ +FA
Sbjct: 391 RLYIVAVDPAQELGVDVVRRRSSFASLEKAYYSMSENGSSRHDDD--HNCSIDDWMIQFA 448
Query: 202 QLFRSRVG--TDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNW 259
+LF++ +G +D Y+D HDLG +A+E+TV S+EAQ++ AE KFQE+AALA FNW
Sbjct: 449 RLFKNHLGFDSDSYLDLHDLGMRLYYEAMEDTVASEEAQEIFQVAELKFQEMAALALFNW 508
Query: 260 GNVHMCAARKFVRMDENENEVLVMNESE--FDFVQEKYYLAREKYEQAVVIKPDFYEGLL 317
GNVHM +ARK + ++ + ++ + + +++ +Y A KY +AV KPDF+EGL+
Sbjct: 509 GNVHMASARKRPPLSDDASMECILEQVKVAYEWACAEYAKAGAKYGEAVKTKPDFFEGLI 568
Query: 318 AIGQQQFELAKLNWSFGIANKMDLGKETLRLFDVAEEKMTAANDAWENLEKGKLG----- 372
A+GQQQFE AKL W + +A K+D+G E L LF+ AE+ M WE +E +L
Sbjct: 569 ALGQQQFEQAKLCWYYALACKIDMGTEVLGLFNHAEDNMEKGMGMWEGMENTRLRGLSKP 628
Query: 373 -------------------------EQGSVGMRSQIHLFWGNMLFERSQVEFKLGMSDWK 407
EQ S +RS +++ WG +L+ERS VEF LG+ W+
Sbjct: 629 SKEKIIFEKMGIDGYMKDMSSDEAFEQAS-SIRSHVNILWGTILYERSVVEFILGLPSWE 687
Query: 408 KKLDASVERFKIAGASEADVSGILKKHCFNGNARD 442
+ L ++E+FK GAS AD++ ++K H N ++
Sbjct: 688 ESLTVAIEKFKTGGASPADINVMVKNHSANETTQE 722
>UniRef100_Q6K378 Tetratricopeptide repeat protein-like [Oryza sativa]
Length = 872
Score = 293 bits (749), Expect = 9e-78
Identities = 175/458 (38%), Positives = 262/458 (56%), Gaps = 62/458 (13%)
Query: 29 HPSPKKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQLKLVYDDDIRLAQMPINCSF 88
H + Q R +P + ++H +N K+ K A + +K V DDIR+ +P + +
Sbjct: 381 HIGRGEDKQEKRSTLKPTIHGRDKH--KNHKDVNKRAMKSVKFVCGDDIRIVVIPEHITL 438
Query: 89 RLLRDIVKEKF-PISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIVE 147
L DI + K+ P +S+L+K+ D + DLVTITSTEELR+ E + D ++ ++LYI E
Sbjct: 439 MQLMDIARYKYTPHLKSILLKFMDKEGDLVTITSTEELRWVE----ELDPLKPVRLYIKE 494
Query: 148 VSPEHE------PPLLKEEKEEENNEKQKPLDCVLDEKMCTECNKV----VENLEIDDWL 197
VSP+ E P K E N+ +EC +N DDW+
Sbjct: 495 VSPDREITRDLVMPTTSYSKLERNHNSM------------SECGSSRHGGEKNSYTDDWM 542
Query: 198 YEFAQLFRSRVG--TDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALA 255
+FA+LF+ VG +D Y+D DLG +A+EET+TS+EAQ++ AE KFQE+AALA
Sbjct: 543 VQFARLFKYHVGFDSDAYVDLRDLGMRLYYEAMEETITSEEAQEIFQSAEAKFQEMAALA 602
Query: 256 FFNWGNVHMCAARKFVRM--DENENEVLVMNESEFDFVQEKYYLAREKYEQAVVIKPDFY 313
FNWGNVHM A+K + + D ++ +L+ ++ +++ +Y A +K+E+AV +KPDFY
Sbjct: 603 LFNWGNVHMSRAKKRLLLSDDASQESILLQVKNAYEWACAEYVKAGKKFEEAVDVKPDFY 662
Query: 314 EGLLAIGQQQFELAKLNWSFGIANKMDLGKETLRLFDVAEEKMTAANDAWENLE----KG 369
EGL+A+GQQQFE AKL+W + A K+ +G E L LF+ AE+ M + WE +E KG
Sbjct: 663 EGLIALGQQQFEQAKLSWRYADACKIGMGTEVLELFNHAEDNMEKGIEMWEGIEYLRVKG 722
Query: 370 ------------KLGEQGSV-------------GMRSQIHLFWGNMLFERSQVEFKLGMS 404
KLG G + MRSQ+++ WG +L+ERS VEFKLG+S
Sbjct: 723 LSKSKKEKVLLDKLGLNGHLKEFSADEAFEQASNMRSQLNISWGTILYERSVVEFKLGLS 782
Query: 405 DWKKKLDASVERFKIAGASEADVSGILKKHCFNGNARD 442
W++ L ++E+FK GAS D+S ++K HC N ++
Sbjct: 783 SWEESLTEAIEKFKTGGASLPDISVMIKNHCANEKTQE 820
>UniRef100_Q6DT61 AT1G62390 [Arabidopsis lyrata]
Length = 199
Score = 176 bits (446), Expect = 1e-42
Identities = 100/198 (50%), Positives = 127/198 (63%), Gaps = 32/198 (16%)
Query: 111 DNDDDLVTITSTEELRFAESCVY----------KTDSVEILKLYIVEVSPEHEPPLLKEE 160
DND DLVTITST EL+ AES K+DSV +L+L++V+VSPE EP LL+EE
Sbjct: 1 DNDGDLVTITSTAELKLAESAADSLLTKEPDTDKSDSVGMLRLHVVDVSPEQEPMLLEEE 60
Query: 161 KEEENNEK------QKPLDCVLD--------------EKMCTECNKVVENLEIDDWLYEF 200
+EE + P + V + EK + + + LE+DDWL++F
Sbjct: 61 EEEVEEKPVVEEIISSPTESVSETEINNEKGDKEVEKEKASSSEDPETKELEMDDWLFDF 120
Query: 201 AQLFRSRVGTDK--YIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFN 258
A LFR+ VG D +ID H+LG E CS+ALEETVTS++AQ L DKA KFQEVAALAFFN
Sbjct: 121 AHLFRTHVGIDPDAHIDLHELGMELCSEALEETVTSEKAQPLFDKASAKFQEVAALAFFN 180
Query: 259 WGNVHMCAARKFVRMDEN 276
WGNVHMCAARK + +DE+
Sbjct: 181 WGNVHMCAARKRIPLDES 198
>UniRef100_UPI00003069CB UPI00003069CB UniRef100 entry
Length = 237
Score = 118 bits (295), Expect = 4e-25
Identities = 78/229 (34%), Positives = 119/229 (51%), Gaps = 37/229 (16%)
Query: 240 LLDKAEFKFQEVAALAFFNWGNVHMCAARKFVRMDENENEVL---------VMNESEFDF 290
+LD A KFQE AAL+ FNWGNVHMCAARK MD + L V D
Sbjct: 3 VLDDAAKKFQEAAALSLFNWGNVHMCAARK--AMDGGRDPPLEEGGPPGAAVATADNRDE 60
Query: 291 VQEKYYLAREKYEQAVVIKPDFYEGLLAIGQQQFELAKLNW-SFGIANKMDLGKETLRLF 349
V ++ A+ +YE A+V+KPDF++ +A+ Q+++E A+L + G++ + +
Sbjct: 61 VVDRLDKAKGRYEAALVVKPDFHDATIALAQRRYECARLLCAAAGLSGAGESPAKGNGDS 120
Query: 350 DVAEEKMTAA------------------------NDAWENLEKGKLGEQGSVGMRSQIHL 385
D AE + + A + + ++G+ E MR+Q+ +
Sbjct: 121 DEAEAEFSGACSDFDAVLAILPEDPPKPPEPVEKKEPEQEKKEGEEEEAEQPSMRAQVQV 180
Query: 386 FWGNMLFERSQVEFKLGMSDWKKKLDASVERFKIAGASEADVSGILKKH 434
WGN LFE+SQ+ +LG +W+ LD +VE+FK AG +EAD+ LK H
Sbjct: 181 MWGNTLFEQSQMRARLG-KEWRPLLDVAVEKFKGAGCAEADIEQALKVH 228
>UniRef100_UPI0000366214 UPI0000366214 UniRef100 entry
Length = 355
Score = 40.8 bits (94), Expect = 0.082
Identities = 24/76 (31%), Positives = 41/76 (53%), Gaps = 11/76 (14%)
Query: 72 VYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESC 131
++++DI ++ + +++ + + K + V IKYKD DDDL+TI + +L FA C
Sbjct: 26 IHNEDITYDELVL-----MMQRVFRGKLQSNDEVTIKYKDEDDDLITIFDSSDLSFAIQC 80
Query: 132 VYKTDSVEILKLYIVE 147
ILKL + E
Sbjct: 81 ------SRILKLTLFE 90
>UniRef100_UPI00002347AF UPI00002347AF UniRef100 entry
Length = 913
Score = 40.8 bits (94), Expect = 0.082
Identities = 36/129 (27%), Positives = 58/129 (44%), Gaps = 14/129 (10%)
Query: 12 NVSQPNSPSSSNIAFEPHPS-----PKKSDQISRRLREPKLGPTNEHLKENQKEDVKIAW 66
N SQ NSP++ ++ + S P+ SD R PT+ E ++E
Sbjct: 758 NRSQNNSPTNQSLPIRTNTSHAFHEPQYSD--GRAAAPLSDQPTSPLSHEPEEEPFMPTQ 815
Query: 67 RQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPI-------SRSVLIKYKDNDDDLVTI 119
+ K+ +D++ + N FR L D V K S++V ++Y+D D D VTI
Sbjct: 816 LKAKVNFDENYVTLVISSNIGFRTLTDRVDAKLARFTNRSIGSKTVRLRYQDEDGDFVTI 875
Query: 120 TSTEELRFA 128
S E ++ A
Sbjct: 876 DSDEAVQLA 884
>UniRef100_UPI00003A9BAA UPI00003A9BAA UniRef100 entry
Length = 395
Score = 40.4 bits (93), Expect = 0.11
Identities = 31/109 (28%), Positives = 54/109 (49%), Gaps = 16/109 (14%)
Query: 72 VYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESC 131
++++DI ++ + +++ + + K + V IKYKD D DL+TI + +L FA C
Sbjct: 26 IHNEDITYDELVL-----MMQRVFRGKLLSNDEVTIKYKDEDGDLITIFDSSDLSFAIQC 80
Query: 132 VYKTDSVEILKLYI-VEVSP----EHEPPLLKEEKEEENNEKQKPLDCV 175
ILKL + V P ++ L+ E E N+ + LDC+
Sbjct: 81 ------SRILKLTLFVNGQPRPLESNQVKYLRRELIELRNKVNRLLDCL 123
>UniRef100_UPI00000142CD UPI00000142CD UniRef100 entry
Length = 404
Score = 40.0 bits (92), Expect = 0.14
Identities = 19/60 (31%), Positives = 35/60 (57%), Gaps = 5/60 (8%)
Query: 72 VYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESC 131
++++DI ++ + +++ + + K + V IKYKD DDDL+TI + +L FA C
Sbjct: 26 IHNEDITYDELVL-----MMQRVFRGKLQSNDEVTIKYKDEDDDLITIFDSSDLSFAIQC 80
>UniRef100_UPI00003AAE40 UPI00003AAE40 UniRef100 entry
Length = 1006
Score = 40.0 bits (92), Expect = 0.14
Identities = 39/164 (23%), Positives = 70/164 (41%), Gaps = 9/164 (5%)
Query: 12 NVSQPNSPSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQLKL 71
N P + S P P +S + +++ P + P NE EN E VK R+L L
Sbjct: 529 NTKSPVELNKSYDVESPSPLLMQSKSMHQQMDTPSVPPANEQFLENSFEKVK---RRLDL 585
Query: 72 VYDD---DIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLV--TITSTEELR 126
DD D + + + + + ++K+P++ + K +D + I T+ L
Sbjct: 586 DTDDCQKDNNPCVVTVGMEEQERQWLQEQKYPVTSVYITKNAASDSSMAKEEILKTKMLA 645
Query: 127 FAESCVYKTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEKQK 170
F E + + +L I+ E E L++E EE+ + +K
Sbjct: 646 F-EEMRKRLEEQHAQQLSILIAEQEREQEKLQKELEEQERKGKK 688
>UniRef100_UPI00003AAE3F UPI00003AAE3F UniRef100 entry
Length = 986
Score = 40.0 bits (92), Expect = 0.14
Identities = 39/164 (23%), Positives = 70/164 (41%), Gaps = 9/164 (5%)
Query: 12 NVSQPNSPSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQLKL 71
N P + S P P +S + +++ P + P NE EN E VK R+L L
Sbjct: 529 NTKSPVELNKSYDVESPSPLLMQSKSMHQQMDTPSVPPANEQFLENSFEKVK---RRLDL 585
Query: 72 VYDD---DIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLV--TITSTEELR 126
DD D + + + + + ++K+P++ + K +D + I T+ L
Sbjct: 586 DTDDCQKDNNPCVVTVGMEEQERQWLQEQKYPVTSVYITKNAASDSSMAKEEILKTKMLA 645
Query: 127 FAESCVYKTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEKQK 170
F E + + +L I+ E E L++E EE+ + +K
Sbjct: 646 F-EEMRKRLEEQHAQQLSILIAEQEREQEKLQKELEEQERKGKK 688
>UniRef100_UPI000034DCC1 UPI000034DCC1 UniRef100 entry
Length = 461
Score = 40.0 bits (92), Expect = 0.14
Identities = 19/60 (31%), Positives = 35/60 (57%), Gaps = 5/60 (8%)
Query: 72 VYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESC 131
++++DI ++ + +++ + + K + V IKYKD DDDL+TI + +L FA C
Sbjct: 26 IHNEDITYDELVL-----MMQRVFRGKLQSNDEVTIKYKDEDDDLITIFDSSDLSFAIQC 80
>UniRef100_Q6GNR3 MGC80940 protein [Xenopus laevis]
Length = 398
Score = 40.0 bits (92), Expect = 0.14
Identities = 31/109 (28%), Positives = 53/109 (48%), Gaps = 16/109 (14%)
Query: 72 VYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESC 131
++++DI ++ + +++ + + K + V IKYKD D DL+TI + +L FA C
Sbjct: 26 IHNEDITYDELVL-----MMQRVFRGKLLTNDEVTIKYKDEDGDLITIFDSSDLSFAIQC 80
Query: 132 VYKTDSVEILKLYI-VEVSP----EHEPPLLKEEKEEENNEKQKPLDCV 175
ILKL + V P + L+ E E N+ + LDC+
Sbjct: 81 ------SRILKLTLFVNGQPRPLESSQVKYLRRELIELRNKVNRLLDCL 123
>UniRef100_UPI000042DC3D UPI000042DC3D UniRef100 entry
Length = 805
Score = 39.7 bits (91), Expect = 0.18
Identities = 52/234 (22%), Positives = 94/234 (39%), Gaps = 14/234 (5%)
Query: 16 PNSPSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQLKLVYDD 75
P + S ++ P KKS + ++++ KL ++H K +D K+ Q + + D
Sbjct: 318 PETKSDKDLKDLPSKEKKKSFEKPPKVKKEKLPKQSKHSKAEIPDDQKLTAFQERYLEDA 377
Query: 76 DI--RLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVY 133
+ + P N F L +D + E P S + + +N D+ + S + +
Sbjct: 378 TLLFEFVENP-NVRFLLPKDAICEVLPASTMINAENGNNSDNRDILMSFIWIHNQKEVDE 436
Query: 134 KTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEKQKPLDCVLDEKMCTECNKVVENLEI 193
+E K + E E ++EKEEE E++K + L+++ E NK + +
Sbjct: 437 YEKKLEEYK----KKKAEEEEKKRQKEKEEEEEEERKNKE-QLEKEKSNEKNKEGDESKS 491
Query: 194 DDWLYEFAQLFRSRVGTDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFK 247
L E + G K ++ D T E SD+ Q + E K
Sbjct: 492 VQSLDEVQE------GQSKDVEMGDNTTNTVKSENEVIEISDDVQATEEPEEIK 539
>UniRef100_UPI000031A5CC UPI000031A5CC UniRef100 entry
Length = 259
Score = 39.3 bits (90), Expect = 0.24
Identities = 50/220 (22%), Positives = 98/220 (43%), Gaps = 36/220 (16%)
Query: 1 MRRSGSRRKKGNVSQPNSPSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEHLKENQKE 60
M++ + KK ++ NI S KK+ +SR ++ K+ T + +N+K
Sbjct: 1 MKKKKKKTKKNKLNFKKKKFKLNIKSNYKKSKKKN--LSRNFKKRKIKNTPKVKIKNKKN 58
Query: 61 DVKIAWRQLKLVYDDDIRLAQMPINCSFRL-----LRDIVKEKFPISRSVLIKYKDNDDD 115
VKI + L+ + +AQ + SF++ L + +++ F + + +YKD
Sbjct: 59 KVKIISKNKSLI--SKLAIAQENFSLSFKININFSLENFIQKFFDSISNKIQEYKD---- 112
Query: 116 LVTITSTEELR--FAESCVYKTDSVEILKLYIVEVSPEHEPPLL----KEEKEEENNEKQ 169
T EE R E + K +S++I + E + + LL +E KEEE E++
Sbjct: 113 ----TRLEEKRRIKIEKAILKQESIKIER----EKKQKEQLILLEAKKRELKEEERLERE 164
Query: 170 KPLD---------CVLDEKMCTECNKVVENLEIDDWLYEF 200
+ D ++ ++ + K +E L+I+ + +F
Sbjct: 165 RTKDIKIFLKKEQALIRKEQAEKQKKFLEELKIEKQIEKF 204
>UniRef100_Q9DGF6 TRK-fused protein TFG [Xenopus laevis]
Length = 409
Score = 39.3 bits (90), Expect = 0.24
Identities = 31/109 (28%), Positives = 52/109 (47%), Gaps = 16/109 (14%)
Query: 72 VYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESC 131
++++DI ++ + +++ + + K + V IKYKD D DL+TI + +L FA C
Sbjct: 26 IHNEDITYDELVL-----MMQRVFRGKLLTNDEVTIKYKDEDGDLITIFDSSDLSFAIQC 80
Query: 132 VYKTDSVEILKLYI-VEVSP----EHEPPLLKEEKEEENNEKQKPLDCV 175
ILKL + V P + L+ E E N+ LDC+
Sbjct: 81 ------SRILKLTLFVNGQPRPLESSQVKYLRRELIELRNKVNSLLDCL 123
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 767,337,081
Number of Sequences: 2790947
Number of extensions: 33003953
Number of successful extensions: 112277
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 95
Number of HSP's that attempted gapping in prelim test: 112063
Number of HSP's gapped (non-prelim): 267
length of query: 459
length of database: 848,049,833
effective HSP length: 131
effective length of query: 328
effective length of database: 482,435,776
effective search space: 158238934528
effective search space used: 158238934528
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC122163.6


