

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.9 - phase: 0
(380 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
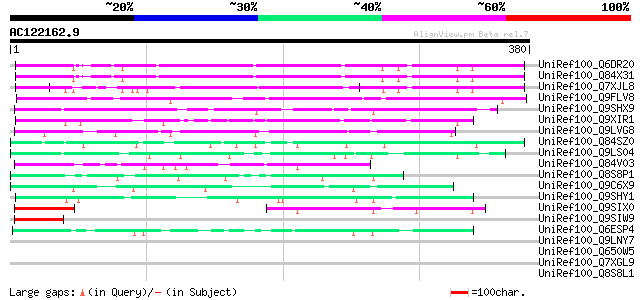
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6DR20 Hypothetical protein [Arabidopsis thaliana] 120 5e-26
UniRef100_Q84X31 Hypothetical protein At2g17036/F6P23.4 [Arabido... 119 2e-25
UniRef100_Q7XJL8 At2g17030 protein [Arabidopsis thaliana] 103 1e-20
UniRef100_Q9FLV8 Gb|AAD22308.1 [Arabidopsis thaliana] 93 1e-17
UniRef100_Q9SHX9 F1E22.13 [Arabidopsis thaliana] 80 9e-14
UniRef100_Q9XIR1 F13O11.14 protein [Arabidopsis thaliana] 70 7e-11
UniRef100_Q9LVG8 Gb|AAF18598.1 [Arabidopsis thaliana] 69 3e-10
UniRef100_Q84SZ0 Hypothetical protein OSJNBa0087C10.17 [Oryza sa... 59 2e-07
UniRef100_Q9LS04 Gb|AAC14516.2 [Arabidopsis thaliana] 59 3e-07
UniRef100_Q84V03 Hypothetical protein At2g16365/F16F14.28 [Arabi... 56 1e-06
UniRef100_Q8S8P1 Hypothetical protein At2g26160 [Arabidopsis tha... 56 2e-06
UniRef100_Q9C6X9 Hypothetical protein T7O23.22 [Arabidopsis thal... 52 2e-05
UniRef100_Q9SHY1 F1E22.11 [Arabidopsis thaliana] 52 4e-05
UniRef100_Q9SIX0 Hypothetical protein At2g16290 [Arabidopsis tha... 51 6e-05
UniRef100_Q9SIW9 Hypothetical protein At2g16300 [Arabidopsis tha... 50 1e-04
UniRef100_Q6ESP4 Hypothetical protein P0684A08.38 [Oryza sativa] 48 5e-04
UniRef100_Q9LNY7 F9C16.31 [Arabidopsis thaliana] 46 0.002
UniRef100_Q650W5 Hypothetical protein OSJNBa0070E11.6 [Oryza sat... 46 0.002
UniRef100_Q7XGL9 Hypothetical protein [Oryza sativa] 45 0.003
UniRef100_Q8S8L1 Hypothetical protein At2g24255 [Arabidopsis tha... 44 0.007
>UniRef100_Q6DR20 Hypothetical protein [Arabidopsis thaliana]
Length = 404
Score = 120 bits (302), Expect = 5e-26
Identities = 123/400 (30%), Positives = 191/400 (47%), Gaps = 45/400 (11%)
Query: 5 DWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNH----HNILPFKFPLLKCED 60
DW+ LPKDLL LIS+ +++ DLI+FRS+CS+WRS++ P HN LPF FP
Sbjct: 3 DWATLPKDLLDLISKCLESSFDLIQFRSVCSSWRSAAGPKRLLWAHN-LPF-FP-----S 55
Query: 61 NSIKYNDNEISSSSNHHPSQL----EEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPR 116
+ + N I + H S L E Q W+++ N + K+ L+K
Sbjct: 56 DDKPFLSNVILRVA--HQSILLIKPNEPQCEADLFGWIVKVWDNIYVSRKM--TLLKPLS 111
Query: 117 PQQEETLAH-PRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTCHGKDP 175
+ H PR+FD +K +V L + L ++ H L V D
Sbjct: 112 SSRNYFPQHLPRIFDMSKFTVRELCREVKLYHPDYYCVPGHTALELELGKTVVKYLNDDK 171
Query: 176 LILGSLGYCSYRPL-LFRDRDEHWKPI-PKMSSLYGDTCVFKGQLYAVHQSGQTVTIGPD 233
+L L Y L +FR D W I + S D +F G+ +A+ +G+TV +
Sbjct: 172 FVL--LTILEYGKLAVFRSWDREWTVINDYIPSRCQDLIMFDGRFFAIDYNGRTVVVDYS 229
Query: 234 S-SVELAAQPLGHDSPGGNKMLVESEGRLLLLGIEESSN---YFSIDFFELDEK-----K 284
S + LAA PL G K L+ES G + L+ IE N F+ F+ + K
Sbjct: 230 SFKLTLAANPL--IGGGDKKFLIESCGEMFLVDIEFCLNEKPEFTGGFYSYFNETTVSYK 287
Query: 285 KKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSASASDL---CIQKGNCAIFI--DE 339
K+ +L+ E+EK+WV++ + GD++FF+G +FSAS +D+ C+ G+ F +E
Sbjct: 288 FKFFKLV---EREKRWVEVEDLGDKMFFLGDDSTFSASTADIIPRCVGTGSFVFFYTHEE 344
Query: 340 SVLHNNNMVRGKRVFHLDQDRL-SRGSKYLNLFL-PPEWI 377
S++ ++ G F + L ++ +Y LF PP WI
Sbjct: 345 SLVVMDDRNLGVFDFRSGKTELVNKLPEYAKLFWPPPPWI 384
>UniRef100_Q84X31 Hypothetical protein At2g17036/F6P23.4 [Arabidopsis thaliana]
Length = 404
Score = 119 bits (297), Expect = 2e-25
Identities = 122/400 (30%), Positives = 190/400 (47%), Gaps = 45/400 (11%)
Query: 5 DWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNH----HNILPFKFPLLKCED 60
DW+ LPKDLL IS+ +++ DLI+FRS+CS+WRS++ P HN LPF FP
Sbjct: 3 DWATLPKDLLDXISKCLESSFDLIQFRSVCSSWRSAAGPKRLLWAHN-LPF-FP-----S 55
Query: 61 NSIKYNDNEISSSSNHHPSQL----EEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPR 116
+ + N I + H S L E Q W+++ N + K+ L+K
Sbjct: 56 DDKPFLSNVILRVA--HQSILLIKPNEPQCEADLFGWIVKVWDNIYVSRKM--TLLKPLS 111
Query: 117 PQQEETLAH-PRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTCHGKDP 175
+ H PR+FD +K +V L + L ++ H L V D
Sbjct: 112 SSRNYFPQHLPRIFDMSKFTVRELCREVKLYHPDYYCVPGHTALELELGKTVVKYLNDDK 171
Query: 176 LILGSLGYCSYRPL-LFRDRDEHWKPI-PKMSSLYGDTCVFKGQLYAVHQSGQTVTIGPD 233
+L L Y L +FR D W I + S D +F G+ +A+ +G+TV +
Sbjct: 172 FVL--LTILEYGKLAVFRSWDREWTVINDYIPSRCQDLIMFDGRFFAIDYNGRTVVVDYS 229
Query: 234 S-SVELAAQPLGHDSPGGNKMLVESEGRLLLLGIEESSN---YFSIDFFELDEK-----K 284
S + LAA PL G K L+ES G + L+ IE N F+ F+ + K
Sbjct: 230 SFKLTLAANPL--IGGGDKKFLIESCGEMFLVDIEFCLNEKPEFTGGFYSYFNETTVSYK 287
Query: 285 KKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSASASDL---CIQKGNCAIFI--DE 339
K+ +L+ E+EK+WV++ + GD++FF+G +FSAS +D+ C+ G+ F +E
Sbjct: 288 FKFFKLV---EREKRWVEVEDLGDKMFFLGDDSTFSASTADIIPRCVGTGSFVFFYTHEE 344
Query: 340 SVLHNNNMVRGKRVFHLDQDRL-SRGSKYLNLFL-PPEWI 377
S++ ++ G F + L ++ +Y LF PP WI
Sbjct: 345 SLVVMDDRNLGVFDFRSGKTELVNKLPEYAKLFWPPPPWI 384
>UniRef100_Q7XJL8 At2g17030 protein [Arabidopsis thaliana]
Length = 1169
Score = 103 bits (256), Expect = 1e-20
Identities = 84/270 (31%), Positives = 126/270 (46%), Gaps = 40/270 (14%)
Query: 5 DWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRS-----SSIPNHHNILPFKFPLLKCE 59
DWS LPKDLL LIS+ +++ DLI+FRS+CS+WRS S +P HH P+L
Sbjct: 3 DWSTLPKDLLDLISKSLESSFDLIQFRSVCSSWRSAAEPKSPLPTHH------LPILPDN 56
Query: 60 DNSIKYNDNEISSSSNHHPSQLEEKQTLI--PRRP--------WLIRFTQ--NSHGKTKL 107
S+ + D+ + +L ++ L+ P P WLI+ + N K L
Sbjct: 57 GGSL-FPDSAVG-------FRLSQRSILLIKPHEPCIESDSFGWLIKVEEDLNVPRKVTL 108
Query: 108 FHPLIKYPRPQQEETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLA 167
PL E PRV D +K V LG +F L N +Y+ V
Sbjct: 109 LDPLCDTRNSIPEN---FPRVLDMSKFKVRELGREFKLHYFNTVGDIVESLYLEKAVVKY 165
Query: 168 VTCHGKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQT 227
+ C G +L ++ + S + +FR D W I M S Y D +F G+ +AV +G+T
Sbjct: 166 LDCDGDYKFVLLTI-HVSGKLAVFRSWDRAWTVINDMPSPYDDVMLFDGRFFAVDNNGRT 224
Query: 228 VTIGPDS-SVELAAQPLGHDSPGGNKMLVE 256
V + S + L A P+ GG+K +++
Sbjct: 225 VVVDYSSLKLTLVASPVF----GGDKNVLK 250
Score = 91.7 bits (226), Expect = 3e-17
Identities = 109/375 (29%), Positives = 171/375 (45%), Gaps = 45/375 (12%)
Query: 30 FRSICSNWRSSSIPNH----HNILPFKFPLLKCEDNSIKYNDNEISSSSNHHPSQL---- 81
FRS+CS+WRS++ P HN LPF FP + + N I + H S L
Sbjct: 768 FRSVCSSWRSAAGPKRLLWAHN-LPF-FP-----SDDKPFLSNVILRVA--HQSILLIKP 818
Query: 82 EEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPRPQQEETLAH-PRVFDFNKLSVLHLG 140
E Q W+++ N + K+ L+K + H PR+FD +K +V L
Sbjct: 819 NEPQCEADLFGWIVKVWDNIYVSRKM--TLLKPLSSSRNYFPQHLPRIFDMSKFTVRELC 876
Query: 141 TDFILDKVNFTSRNRHGIYMLPKTVLAVTCHGKDPLILGSLGYCSYRPL-LFRDRDEHWK 199
+ L ++ H L V D +L L Y L +FR D W
Sbjct: 877 REVKLYHPDYYCVPGHTALELELGKTVVKYLNDDKFVL--LTILEYGKLAVFRSWDREWT 934
Query: 200 PIPK-MSSLYGDTCVFKGQLYAVHQSGQTVTIGPDS-SVELAAQPLGHDSPGGNKMLVES 257
I + S D +F G+ +A+ +G+TV + S + LAA PL G K L+ES
Sbjct: 935 VINDYIPSRCQDLIMFDGRFFAIDYNGRTVVVDYSSFKLTLAANPL--IGGGDKKFLIES 992
Query: 258 EGRLLLLGIEESSNY---FSIDFFELDEK-----KKKWVRLMDFDEKEKKWVKLRNFGDR 309
G + L+ IE N F+ F+ + K K+ +L+ E+EK+WV++ + GD+
Sbjct: 993 CGEMFLVDIEFCLNEKPEFTGGFYSYFNETTVSYKFKFFKLV---EREKRWVEVEDLGDK 1049
Query: 310 VFFIGRGCSFSASASDL---CIQKGNCAIFI--DESVLHNNNMVRGKRVFHLDQDRL-SR 363
+FF+G +FSAS +D+ C+ G+ F +ES++ ++ G F + L ++
Sbjct: 1050 MFFLGDDSTFSASTADIIPRCVGTGSFVFFYTHEESLVVMDDRNLGVFDFRSGKTELVNK 1109
Query: 364 GSKYLNLFL-PPEWI 377
+Y LF PP WI
Sbjct: 1110 LPEYAKLFWPPPPWI 1124
>UniRef100_Q9FLV8 Gb|AAD22308.1 [Arabidopsis thaliana]
Length = 373
Score = 93.2 bits (230), Expect = 1e-17
Identities = 93/386 (24%), Positives = 166/386 (42%), Gaps = 32/386 (8%)
Query: 6 WSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSIKY 65
WS+LP ++L LIS +ID DLI FRS+CS WRSSS+ ++ + PL D
Sbjct: 4 WSELPPEILHLISLKIDNPFDLIHFRSVCSFWRSSSLLKFRHMTSLRCPLPL--DPGGCG 61
Query: 66 NDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKYPR-------PQ 118
+D I SS + L + + W+ + +G+ L ++ P
Sbjct: 62 DDCHILSSRVY----LLKCPNRDRPQYWMFKLQAKENGEVVLHSLFLRRNSSAYGCLYPS 117
Query: 119 QEETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTCHGKDPLIL 178
L + ++F+ + V F L + R G L + + +IL
Sbjct: 118 LSIDLLNCQIFELAQEHVACYSEWFELFECISKCEERIGFMGLNR-------EKNEYMIL 170
Query: 179 GSLGYCSYRPLLFRDRDEHWKPIP-KMSSLYGDTCVFKGQLYAVHQSGQTVTIGPDSSVE 237
G L + ++R D W + S + FKG+ YA+ ++G+T + P V
Sbjct: 171 GKLSFNGLA--MYRSVDNRWTELEITPDSFFEGIVPFKGKFYAIDRTGKTTVVDPTLEVN 228
Query: 238 LAAQPLGHDSPGGNKMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKK-WVRLMDFDEK 296
+ D +L+ + +L+L+ + S Y DF + ++KK W + + +E+
Sbjct: 229 TFQRSRPCDKTRKRWLLMTGD-KLILVEMCTKSRY---DFHIPNIREKKIWFEISELNEE 284
Query: 297 EKKWVKLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFHL 356
W ++ + RV F+ C+FS A+++ + N IF+D N+ + V+
Sbjct: 285 RNDWDQVEDMDGRVLFLEHYCTFSCLATEIPGFRANSIIFMDLWGGSNSYELESILVYEF 344
Query: 357 DQD---RLSRGSKYLNLF-LPPEWIL 378
++ L +Y+ LF PP W++
Sbjct: 345 NEKGVRSLRDKLEYIELFPFPPGWVI 370
>UniRef100_Q9SHX9 F1E22.13 [Arabidopsis thaliana]
Length = 360
Score = 80.1 bits (196), Expect = 9e-14
Identities = 94/362 (25%), Positives = 155/362 (41%), Gaps = 38/362 (10%)
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKF-PLLKCEDNS 62
ADWS LP DLL +I+ R+ + I+L RFRSIC +WR SS+P PF+ PL+ N
Sbjct: 2 ADWSTLPVDLLNMIAGRLFSNIELKRFRSICRSWR-SSVPGAGKKNPFRTRPLILLNPNP 60
Query: 63 IK-YNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQN-SHGKTKLFHPLIKYPRPQQE 120
K D+ + + P + WLI+ + S GK L PL + P
Sbjct: 61 NKPLTDHRRRGEFLSRSAFFRVTLSSSPSQGWLIKSDVDVSSGKLHLLDPLSRLPMEHSR 120
Query: 121 ETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTCHGKDPLILG- 179
+ + D ++ ++ + + + R R + K V V D +LG
Sbjct: 121 KRV------DLSEFTITEIREAYQV----HDWRTRKETRPIFKRVALVKDKEGDNQVLGI 170
Query: 180 -SLGYCSYRPL-LFRDRDEHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQTVTIGPDSSVE 237
S G Y + ++ ++E ++ + D V KGQ YA+ G I D
Sbjct: 171 RSTGKMMYWDIKTWKAKEEGYE--------FSDIIVHKGQTYALDSIGIVYWIRSDLKF- 221
Query: 238 LAAQPLGHDSPGGNKMLVESEGRLLLLG--IEESSNYFSIDFFELDEKKKKWVRLMDFDE 295
+ PL D G++ LVE G ++ + ES+ D + K ++ FD+
Sbjct: 222 IRFGPLVGDWT-GDRRLVECCGEFYIVERLVGESTWKRKADDTGYEYAKTVGFKVYKFDD 280
Query: 296 KEKKWVKLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFIDESVLHNNNMVRGKRVFH 355
++ K +++++ GD+ F I FS A + N F D++++ +VF
Sbjct: 281 EQGKMMEVKSLGDKAFVIATDTCFSVLAHEFYGCLENAIYFTDDTMI---------KVFK 331
Query: 356 LD 357
LD
Sbjct: 332 LD 333
>UniRef100_Q9XIR1 F13O11.14 protein [Arabidopsis thaliana]
Length = 383
Score = 70.5 bits (171), Expect = 7e-11
Identities = 91/349 (26%), Positives = 153/349 (43%), Gaps = 38/349 (10%)
Query: 5 DWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRS-----SSIPNHHNILP--FKFPLLK 57
+WSQLP++LL LIS+ +D D++ RSIC +WRS SS+ LP KFPL+
Sbjct: 3 NWSQLPEELLNLISKNLDNCFDVVHARSICRSWRSAFPFPSSLSTLSYSLPTFAKFPLVS 62
Query: 58 CEDNSIKYNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPL---IKY 114
+ ++K + + N E I + S+ +L PL +K
Sbjct: 63 KDLCTLKKIQIFLFRARNPAADIPEYFLGGIDQ--------DQSNDHMELPSPLQCSVKV 114
Query: 115 PRPQQEETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTCHGKD 174
PQ + L + + D+ ++ LG +I+ + + + Y + L V +G D
Sbjct: 115 KFPQSDPFLVN--MLDY---QIIPLGFQYIM--IGWDPESLANGY-VGVAFLPVKKNGGD 166
Query: 175 PLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYGDTCV-FKGQLYAVHQSGQTVTIGPD 233
++ L Y ++ L+ R + W + K S V F+G+ Y +G P
Sbjct: 167 EFVV-LLRYRNHL-LVLRSSEMRWMKVKKTSIASCKGLVSFRGRFYVTFLNGDIYVFDPY 224
Query: 234 SSVELAAQPLGHDSPGGNKMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDF 293
S + P + +K L+ + L L +E+ + + D +L + RL
Sbjct: 225 SLEQTLLMP--SEPLRSSKYLIPNGSDELFL-VEKFNPFPEADVLDLSRFACRVSRL--- 278
Query: 294 DEKEKKWVKLRNFGDRVFFIGRGCSFSASASDL---CIQKGNCAIFIDE 339
DE+ +WV++ + GDRV FIG + SA +L C GN +F +E
Sbjct: 279 DEEAGQWVEVIDLGDRVLFIGHFGNVCCSAKELPDGCGVSGNSILFTNE 327
>UniRef100_Q9LVG8 Gb|AAF18598.1 [Arabidopsis thaliana]
Length = 374
Score = 68.6 bits (166), Expect = 3e-10
Identities = 90/346 (26%), Positives = 140/346 (40%), Gaps = 49/346 (14%)
Query: 4 ADWSQLPKDLLTLISQRIDTE----IDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCE 59
+ WS LP D+L LIS R+D + I L+ RS+C+ WR S ++ N KFP
Sbjct: 9 SQWSDLPLDILELISDRLDHDSSDTIHLLCLRSVCATWRLSLPLSNKNNRLSKFP----- 63
Query: 60 DNSIKYNDNEISSSSNH-----HPSQLEEKQTLIPRRPWLIRFTQNSHGKTKLFHPLIKY 114
KY SSSS+ S + + + + R L++ + + G ++ L
Sbjct: 64 ----KYLPFWSSSSSSSGFFTLKQSNVYKLEAPLNPRTCLVKLQETTPGIMRVLD-LFSN 118
Query: 115 PR----PQQEETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTC 170
R P+ P D + V + + ++ N + L + +
Sbjct: 119 DRICFLPEN-----FPSKIDLQEFHVRLVRRTYRMEYANNGGGAVPCFWSLHSDKVVILS 173
Query: 171 HGKDPLIL-----GSLGYCSYRPLLFRDRDEHWKPIPKM-SSLYGDTCVFK-GQLYAVHQ 223
G+D I+ G LG+ L DE WK + + +Y D ++K + V
Sbjct: 174 SGEDSAIIAIHSGGKLGF------LKSGNDEKWKILDNSWNVIYEDIMLYKDNRCIVVDD 227
Query: 224 SGQTVTIGPDSSVELAAQPLGHDSPGGNKMLVESEGRLLLLGIEESSNYFSIDFFELD-E 282
G+TV D V A+ L G K LVE G + L Y + + D
Sbjct: 228 KGKTVIYDVDFKVSDLAEGLVGGG-GHKKHLVECSGGEVFL----VDKYVKTVWCKSDIS 282
Query: 283 KKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSAS--ASDL 326
K R+ + +EK+W ++R+ GD FIG CSFS A DL
Sbjct: 283 KSVVEFRVYNLKREEKRWEEVRDLGDVALFIGDDCSFSVQNPAGDL 328
>UniRef100_Q84SZ0 Hypothetical protein OSJNBa0087C10.17 [Oryza sativa]
Length = 531
Score = 59.3 bits (142), Expect = 2e-07
Identities = 107/424 (25%), Positives = 167/424 (39%), Gaps = 67/424 (15%)
Query: 1 MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFK---FPLLK 57
M EA+ LP+DLL I R++ DL+R S+CS+W S+ H++ +K P L
Sbjct: 115 MVEAELPHLPEDLLVQILSRLEIP-DLLRASSVCSSWHSA-YTTLHSLGQYKRHQTPCLF 172
Query: 58 CEDNSIKYNDNEISSSSNHHPSQLEEKQTLIPRR-------PWLIRFTQNSHGKTKLFHP 110
S N I S + ++ I R WL+ T + + L +P
Sbjct: 173 YTSESAGKNVGCIYSLAEQRTYKITLPDPPIRDRYLIGSSDGWLV--TIDDKCEMHLLNP 230
Query: 111 LIKYPRPQQEETLAHPRVFDFNKLSVLHLGTDFIL----------DKVNFTSRNRHGIYM 160
+ + E +A P V +++ + + I+ D V F+SR+ G +
Sbjct: 231 VTR-------EQMALPPVITMEQVNPTYDESGAIVKYENRSQFWHDGVMFSSRSM-GSII 282
Query: 161 LPKT----------VLAVTCHGKDPLIL--GSLGYCSYRPLLFRDRDEHWKPIPKMSSLY 208
P+ V + T GK ++L G S+ R D+ W +P+ Y
Sbjct: 283 SPRWQQLFLTGRAFVFSETSTGKLLVVLIRNPFGQLSFA----RVGDDEWDYLPE----Y 334
Query: 209 G--DTCVFK-GQLYAVHQSGQTVTIGPDSSVELAAQPLGH-----DSPGGNKMLVESEGR 260
G + C +K G LYAV G+ I + + +G D +L G
Sbjct: 335 GRYEDCTYKDGLLYAVTTLGEIHAIDLSGPIAMVKVVMGKVMDIGDGDRNTYILHAPWGD 394
Query: 261 LLLLGIEESSNYF--SIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCS 318
+L + E +Y S D ++ K + + D E+K VK+ D V F+G +
Sbjct: 395 VLQIWKTEEDDYIHPSEDDYDAILKNTASIEVYKSDLVEEKLVKINRLQDHVLFVGHNQT 454
Query: 319 FSASASDLCIQKGNCAIFIDES---VLHNNNMVRGKRVFHL-DQDRLSRGSKYLNLFLP- 373
A + K N A F D+S + N R VF+L D R GS L P
Sbjct: 455 LCLRAEEFPSLKANHAYFTDDSQNWITEFKNNRRDIGVFNLEDNSRDELGSPQLWSNWPS 514
Query: 374 PEWI 377
P WI
Sbjct: 515 PVWI 518
>UniRef100_Q9LS04 Gb|AAC14516.2 [Arabidopsis thaliana]
Length = 377
Score = 58.5 bits (140), Expect = 3e-07
Identities = 88/394 (22%), Positives = 149/394 (37%), Gaps = 68/394 (17%)
Query: 1 MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFP-LLKCE 59
M + +WS LP++LL LI+ R + ID++R RS C +WR S++ L F+F L
Sbjct: 1 MEKTEWSDLPEELLDLIANRYSSNIDVLRIRSTCKSWR-SAVAMSKERLQFRFERYLPTS 59
Query: 60 DNSIKYNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNS--HGKTKLFHPLIKYPRP 117
+ IK + + + PS P + WL+R Q S + K L PL
Sbjct: 60 NKKIKAHLSPTTFFRITLPSS-------CPNKGWLVRTRQASKMYRKITLLCPLSGERIT 112
Query: 118 QQEETL-------------AHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIY----M 160
+ +TL ++FD K + L ++ + + + + G++
Sbjct: 113 RSHQTLDLLKVGVSEIRQSYEIQIFDGLKDEKIPLDSEIFSNYIKNSDKIPSGVFFRVAF 172
Query: 161 LPKTVLAVTCHGKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYGDTCVFKGQLYA 220
L AV H + C R+ W I + D + G++YA
Sbjct: 173 LDNIFFAVNYHD------NKIWCCK-----TREGSRSWTKIKNQVEDFSDIILHMGRIYA 221
Query: 221 VHQSGQTVTIGPDSSV---ELAAQPLG---HDSPGGNKMLVESEGRLLL---LGIEESSN 271
V G I + ++ PL +DS + LVE G L + L I +
Sbjct: 222 VDLKGAIWWISLSQLTIVQQTSSTPLDYYKYDSCQDTR-LVEYCGDLCIVHELSITRNHI 280
Query: 272 YFSIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSASASDL--CIQ 329
++ F ++ DE KWV++ GD + F+ AS+ C++
Sbjct: 281 QRTVGF-----------KVYKMDEDLAKWVEVSCLGDNTLIVACNSCFTVVASEYHGCLK 329
Query: 330 KGNCAIFIDESVLHNNNMVRGKRVFHLDQDRLSR 363
+ D N +VF LD +++
Sbjct: 330 NSIYFSYYDVKKAEN------IKVFKLDDGSITQ 357
>UniRef100_Q84V03 Hypothetical protein At2g16365/F16F14.28 [Arabidopsis thaliana]
Length = 332
Score = 56.2 bits (134), Expect = 1e-06
Identities = 74/294 (25%), Positives = 120/294 (40%), Gaps = 54/294 (18%)
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSI 63
ADW LP DLL LI ++T ++ FRS+CS+WRS P H+ L + + I
Sbjct: 22 ADWPLLPNDLLELIMGHLETSFEIFLFRSVCSSWRSVVPPLDHS------RCLGIKTHDI 75
Query: 64 KYNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRF-------------TQNSHGKTK-LFH 109
+N + S N P+ E TL +L++F + + G+ K L
Sbjct: 76 SFN---VGFSFNGRPT--NEYCTLKKIPIYLVKFWTPFGDDYLLAEMRERNDGEPKLLLS 130
Query: 110 PL----IKYPRPQQE---ETLAHPRV---------FDFNKLSVLHLGTDFILDKVNFTSR 153
PL IKY + +L P + + + S G + L+ V T R
Sbjct: 131 PLSSNGIKYGMGINKVLFNSLTSPIIPFGQYYEITYIEKRPSEYRFGFPYKLEWVEITER 190
Query: 154 NRHGIYMLPKTVLAVTCHGKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYG--DT 211
+ + + +D +L + C+ +++R R+ W + + Y D
Sbjct: 191 --------VEFLKLDSEDSRDFAVLFAGRMCNL--VMYRSRNMSWTQVVEHPEKYAYQDL 240
Query: 212 CVFKGQLYAVHQSGQTVTIGPDSSVELAAQP-LGHDSPGGNKMLVESEGRLLLL 264
FKG+ YAV SG+ + S E+ P +G + LV+S LLL+
Sbjct: 241 VAFKGKFYAVDSSGRGRVFVVELSFEVTEIPSVGGSQQSSKESLVQSGEELLLV 294
>UniRef100_Q8S8P1 Hypothetical protein At2g26160 [Arabidopsis thaliana]
Length = 359
Score = 55.8 bits (133), Expect = 2e-06
Identities = 76/310 (24%), Positives = 124/310 (39%), Gaps = 49/310 (15%)
Query: 1 MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCE- 59
M + +WS+LP DL+ L + R + D++R RSIC WRS++ P F +C
Sbjct: 1 MEKPEWSELPGDLINLTANRFSSISDVLRVRSICKPWRSAA------ATPKSF---QCNL 51
Query: 60 DNSIKYNDNEISSSSNHH---PSQLEEKQTLIPRRPWLIRFTQNSH-GKTKLFHPLIKYP 115
+S K + +S ++ PS K WLIR Q S K L P +
Sbjct: 52 PSSNKMIETVLSPTTFFRVTGPSSCSYK-------GWLIRTKQVSESSKINLLSPFFRQL 104
Query: 116 RPQQEET--LAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRH---GIYMLPKTVLAVTC 170
++T L V + + +H+ +++ V H + L + AV
Sbjct: 105 LTPSQQTLDLLKFEVSEIRQSYEIHIFDKYLIQGVIGKEGPSHILSRVVFLDNLIFAV-- 162
Query: 171 HGKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQT--V 228
G+D I +C + W I + D + KGQ+YA+ +G +
Sbjct: 163 -GQDDKI-----WCCKSG---EESSRIWTKIKNQVEDFLDIILHKGQVYALDLTGAIWWI 213
Query: 229 TIGPDSSVELAAQ-PLGHDS-PGGNKMLVESEGRLLLLG--------IEESSNYFSIDFF 278
++ P S ++ P+ +D NK LVE G L ++ I S +
Sbjct: 214 SLSPLSLLQFTPSIPMDYDGYDSCNKRLVEYCGDLCIIHQLRLKKAYIRRSQRTVGFKVY 273
Query: 279 ELDEKKKKWV 288
++DE KWV
Sbjct: 274 KMDEYVAKWV 283
>UniRef100_Q9C6X9 Hypothetical protein T7O23.22 [Arabidopsis thaliana]
Length = 347
Score = 52.4 bits (124), Expect = 2e-05
Identities = 77/343 (22%), Positives = 123/343 (35%), Gaps = 66/343 (19%)
Query: 1 MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNH--HNILPFKFPLLKC 58
M + WS L +DL+ L++ + + I+L+RFRSIC WRS+ HN P K
Sbjct: 1 MGKVGWSDLHEDLIDLLANNLSSNINLLRFRSICKPWRSTVATKKRLHNHFERNLPTFKK 60
Query: 59 EDNSIKYNDNEISSSSNHHPSQLEEKQTLIP--RRPWLIRFTQNSH-GKTKLFHPLIKYP 115
+ + PS P + WLI+ Q S K L PL
Sbjct: 61 KKTVVS-------------PSTFFRVTLPSPCRNKGWLIKNRQVSESSKNNLLSPLSGKT 107
Query: 116 RPQQEETLAHPRVFDFNKLSVLHLGTD----FILDKVNFTSRNRHGIYMLPKTVLAVTCH 171
++TL +V F S+L L D LD V F ++ I+
Sbjct: 108 ITPSDKTLDLLKVECFRDSSILQLFADSDRVVFLDNVFFVVDFKNEIWCCKS-------- 159
Query: 172 GKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSL-YGDTCVFKGQLYAVHQSGQTVTI 230
+ HW I + + D + KG++YA+ +G I
Sbjct: 160 --------------------GEETRHWTRINNEEAKGFLDIILHKGKIYALDLTGAIWWI 199
Query: 231 GPDSSVELAAQPLGHDSPGG--------NKMLVESEGRLLLLGIEESSNYFSIDFFELDE 282
S EL+ G +P K LVE G L ++ Y +
Sbjct: 200 ---SLSELSIYQYGPSTPVDFYEIDNCKEKRLVEYCGELCVV----HRFYKKFCVKRVLT 252
Query: 283 KKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSASASD 325
++ ++ D+ +WV++ + GD+ + F AS+
Sbjct: 253 ERTVCFKVYKMDKNLVEWVEVSSLGDKALIVATDNCFLVLASE 295
>UniRef100_Q9SHY1 F1E22.11 [Arabidopsis thaliana]
Length = 526
Score = 51.6 bits (122), Expect = 4e-05
Identities = 78/362 (21%), Positives = 131/362 (35%), Gaps = 55/362 (15%)
Query: 5 DWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSS-----SIPNHHNIL----PFKFPL 55
DWS LP++LL I+ R + ++ RF SIC +W SS P H L P
Sbjct: 3 DWSTLPEELLHFIAARSFSLVEYKRFSSICVSWHSSVSGVKKNPFHRRPLIDFNPIAPSE 62
Query: 56 LKCEDNSIKYNDNEISSSSNHHPSQLEEKQTLIPRRPWLIRFTQNSH-GKTKLFHPLIKY 114
ED+ N S + L P + W+I+ +++ G+ L +PL ++
Sbjct: 63 TLLEDHVFSCNPGAFLSRAAFFRVTLSSS----PSKGWIIKSDMDTNSGRFHLLNPLSRF 118
Query: 115 PRPQQEETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTV---LAVTCH 171
P E+ LD ++FT Y + K L +
Sbjct: 119 PLRISSES----------------------LDLLDFTVSEIQESYAVLKDAKGRLPNPGY 156
Query: 172 GKDPLILGSLGYCSYRPLLFRDRD---EHW-----KPIPKMSSLYGDTCVFKGQLYAVHQ 223
+ L+ G + +L RD +W +M + D V KG Y +
Sbjct: 157 QRSALVKVKEGDDHHHGILGIGRDGTINYWNGNVLNGFKQMGHHFSDIIVHKGVTYVLDS 216
Query: 224 SGQTVTIGPDSSVELAAQPLGHDSPGGNK----MLVESEGRLLLLG--IEESSNYFSIDF 277
G I D + L + G + V+ G L ++ +E+S
Sbjct: 217 KGIVWCINSDLEMSRYETSLDENMTNGCRRYYMRFVDCCGELYVIKRLPKENSRKRKSTL 276
Query: 278 FELDEKKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFI 337
F+ + ++ D++ KWV+++ GD F + FS A + N FI
Sbjct: 277 FQF--SRTAGFKVYKIDKELAKWVEVKTLGDNAFVMATDTCFSVLAHEYYGCLPNSIYFI 334
Query: 338 DE 339
++
Sbjct: 335 ED 336
>UniRef100_Q9SIX0 Hypothetical protein At2g16290 [Arabidopsis thaliana]
Length = 415
Score = 50.8 bits (120), Expect = 6e-05
Identities = 43/170 (25%), Positives = 72/170 (42%), Gaps = 18/170 (10%)
Query: 189 LLFRDRDEHWKPIPKMSSLYG--DTCVFKGQLYAVHQSGQTVTIGPDSSVELAAQPLGHD 246
+++R R+ W + + Y D FKG+ YA+ SG+ + S+ + P
Sbjct: 207 VMYRSRNMSWTQVVEHPETYAYQDLVAFKGKFYALDSSGRGRVFVVELSLRVTEIPSVKG 266
Query: 247 SPGGNKMLVESEGRLLLLG---IEESSNYFSIDFFELDEKKKKWVRLMDFDEK--EKKWV 301
S +K + G LLL I Y DE W ++ DE+ ++KWV
Sbjct: 267 SQYCSKESLVQLGEELLLVQRFIPAGRRY--------DEYIYTWFKVFRLDEEGGKRKWV 318
Query: 302 KLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIFI---DESVLHNNNMV 348
++ DRV F+G S L NC +F+ D S+ N++++
Sbjct: 319 QVNYLNDRVIFLGARTKLCCSVHKLPGANENCIVFLASNDGSIFDNDSVL 368
Score = 49.7 bits (117), Expect = 1e-04
Identities = 21/44 (47%), Positives = 29/44 (65%)
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHN 47
ADWS LP DLL LI ++T +++ FRS+C +WRS P H+
Sbjct: 2 ADWSLLPNDLLELIVGHLETSFEIVLFRSVCRSWRSVVPPLDHS 45
>UniRef100_Q9SIW9 Hypothetical protein At2g16300 [Arabidopsis thaliana]
Length = 467
Score = 49.7 bits (117), Expect = 1e-04
Identities = 20/36 (55%), Positives = 27/36 (74%)
Query: 4 ADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRS 39
ADWS LP DLL LI ++T +++ FRS+CS+WRS
Sbjct: 2 ADWSLLPNDLLELIVGHLETSFEIVLFRSVCSSWRS 37
Score = 40.0 bits (92), Expect = 0.11
Identities = 19/48 (39%), Positives = 28/48 (57%), Gaps = 1/48 (2%)
Query: 6 WSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKF 53
WS L DLL LI +T +++ RS+CS+WR S +P N F++
Sbjct: 402 WSLLLNDLLKLIVGHFETSFEIVNLRSVCSSWR-SVVPPLDNFPLFRY 448
Score = 39.7 bits (91), Expect = 0.14
Identities = 36/154 (23%), Positives = 60/154 (38%), Gaps = 21/154 (13%)
Query: 189 LLFRDRDEHWKPIPKMSSLYG--DTCVFKGQLYAVHQSGQTVTIGPDSSVELAAQPLGHD 246
+++R R+ W + + Y D FKG+ YA+ SG+ + S+E+ P
Sbjct: 203 VMYRSRNMSWTQVVEHPEKYAYQDLVAFKGKFYALDSSGRGRVFVVELSLEVMEIPSVRG 262
Query: 247 SPGGNKMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNF 306
S +K + G LLL F+ + DE W R+ DE+E + +
Sbjct: 263 SQQSSKENLVLSGEELLL-----VQRFTPEVRHYDEYLYTWFRVFRLDEEEGRETQ---- 313
Query: 307 GDRVFFIGRGCSFSASASDLCIQKGNCAIFIDES 340
+ S L K NC +FI+ +
Sbjct: 314 ----------TNLCCSVHKLPCAKENCIVFIESN 337
>UniRef100_Q6ESP4 Hypothetical protein P0684A08.38 [Oryza sativa]
Length = 377
Score = 47.8 bits (112), Expect = 5e-04
Identities = 81/363 (22%), Positives = 131/363 (35%), Gaps = 78/363 (21%)
Query: 3 EADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNS 62
E W+QLP +LL LI +++ D +RFR++CS WR ++ + +P + P +
Sbjct: 21 EDRWTQLPPELLPLICKKLPDSADFVRFRTVCSAWRDAAPLSD---VPPQLPWV------ 71
Query: 63 IKYNDNEISSSSNHHPSQLEEKQTLIPR----RPWLIR--------FTQNSHGKTKLFHP 110
++ + + ++ +T R R WL+ T T L++P
Sbjct: 72 VERRGSAFQARAHFRFYSPSSGRTYGVRGYGGRSWLVMGGACQEHLVTTVDLSTTALYNP 131
Query: 111 LIKYPRPQQEETLAHPRVFDFNKLSVLHLGTDFILDKVNFTSRNRHGIYMLPKTVLAVTC 170
L E P + + V+H V R R G P V A T
Sbjct: 132 L------TGERLALPPAPYPQWRHGVVH---------VVAAGRRRGG---APLVVNASTR 173
Query: 171 HGKDPLILGSLGYCSYRPLLFRDRDEHWKPIPKMSSLYGDTCVFKGQLYAVHQSGQTVTI 230
G GYC R D W + + G G+ Y + +T+ I
Sbjct: 174 TG-------HFGYC-------RQGDTKWTLVDGRQDM-GHRAYHGGRFYVNTNAQETLVI 218
Query: 231 GPDS-SVELAAQPLGHDSPGG-----NKMLVESEGRLLLL-------GIEESSNYFSIDF 277
+ +VE P + G LVES G+L+ + S+ + ++
Sbjct: 219 DASTGAVESVLPPPPRSADAGAGVSCGDYLVESRGKLIRAVLFPRDGVVTTSAEDYYLNV 278
Query: 278 FELDEKKKKWVRLMDFDEKEKKWVKLRNFGDRV-FFIGRGCSFSASASDLCIQKGNCAIF 336
++L E K W K+ + GD V FF G FS + K +C F
Sbjct: 279 YQLQEDGK----------AAAAWAKVESVGDSVLFFDKHGHGFSLEPNGAAELKRDCVYF 328
Query: 337 IDE 339
+ E
Sbjct: 329 MHE 331
>UniRef100_Q9LNY7 F9C16.31 [Arabidopsis thaliana]
Length = 311
Score = 45.8 bits (107), Expect = 0.002
Identities = 18/40 (45%), Positives = 28/40 (70%)
Query: 1 MAEADWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSS 40
M + WS L +DL+ L++ + + I+L+RFRSIC WRS+
Sbjct: 1 MGKVGWSDLHEDLIDLLANNLSSNINLLRFRSICKPWRST 40
>UniRef100_Q650W5 Hypothetical protein OSJNBa0070E11.6 [Oryza sativa]
Length = 861
Score = 45.8 bits (107), Expect = 0.002
Identities = 25/74 (33%), Positives = 38/74 (50%), Gaps = 3/74 (4%)
Query: 6 WSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSSIPNHHNILPFKFPLLKCEDNSIKY 65
W+ LP DLL IS+R+ D RF ++C +WR + P+ LP+ LL D +
Sbjct: 39 WADLPFDLLADISRRLHATADFARFHAVCKSWRRTLPPHPPTFLPW---LLSPGDATGHR 95
Query: 66 NDNEISSSSNHHPS 79
+ S S+ HP+
Sbjct: 96 TARCVFSKSSRHPA 109
>UniRef100_Q7XGL9 Hypothetical protein [Oryza sativa]
Length = 450
Score = 45.4 bits (106), Expect = 0.003
Identities = 18/37 (48%), Positives = 26/37 (69%)
Query: 5 DWSQLPKDLLTLISQRIDTEIDLIRFRSICSNWRSSS 41
DWS LP DLL L S R+ +D +RFR++C WR+++
Sbjct: 2 DWSGLPGDLLRLFSGRLHDPLDFLRFRAVCRAWRAAT 38
Score = 41.6 bits (96), Expect = 0.037
Identities = 18/57 (31%), Positives = 33/57 (57%)
Query: 280 LDEKKKKWVRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIF 336
L++ + V ++ ++ +WV++R+ GDR+ F+G FS A+D +GNC F
Sbjct: 253 LEDCRALEVHRLEIAGEKSRWVQMRSIGDRMLFVGLYQGFSLRAADFAGLEGNCVYF 309
Score = 38.1 bits (87), Expect = 0.41
Identities = 16/49 (32%), Positives = 28/49 (56%)
Query: 288 VRLMDFDEKEKKWVKLRNFGDRVFFIGRGCSFSASASDLCIQKGNCAIF 336
V ++ ++ +WV++R+ DR+ F+G FS A+D +GNC F
Sbjct: 362 VHRLEIAGEKSRWVQMRSIRDRMLFVGLYQGFSLRAADFAGLEGNCVYF 410
>UniRef100_Q8S8L1 Hypothetical protein At2g24255 [Arabidopsis thaliana]
Length = 336
Score = 43.9 bits (102), Expect = 0.007
Identities = 43/165 (26%), Positives = 68/165 (41%), Gaps = 18/165 (10%)
Query: 189 LLFRDRDEHWKPIPKMS-SLYGDTCVFKGQLYAVHQSGQTVTIGPDSSVELAAQPLGHDS 247
L+ + WK + + ++ D F+G+ YA+ SGQ V I P S+E+
Sbjct: 155 LVLTSAEMRWKQLENVPYTICVDIVTFRGRFYALFLSGQIVVIDP-YSLEVTLLLSSPQD 213
Query: 248 PGGNKMLVESEGRLLLLGIEESSNYFSIDFFELDEKKKKWVRLMDFDEKEKKWVKLRNFG 307
P + LV L L N IDF L + ++ DEK KWV++ + G
Sbjct: 214 PRELRSLVPYGKDELFLVEVIIPNDVVIDFSRLARRVRR------VDEKAGKWVEVGDLG 267
Query: 308 DRVFFIG----------RGCSFSASASDLCIQKGNCAIFIDESVL 342
D V +G RG + +A Q GN ++ +S +
Sbjct: 268 DCVLLLGNLSCSAKEFPRGFGLTENAVLFGDQSGNAFLYYKQSAV 312
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 695,843,453
Number of Sequences: 2790947
Number of extensions: 30692494
Number of successful extensions: 80905
Number of sequences better than 10.0: 97
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 80775
Number of HSP's gapped (non-prelim): 147
length of query: 380
length of database: 848,049,833
effective HSP length: 129
effective length of query: 251
effective length of database: 488,017,670
effective search space: 122492435170
effective search space used: 122492435170
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC122162.9


