

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.12 - phase: 0
(400 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
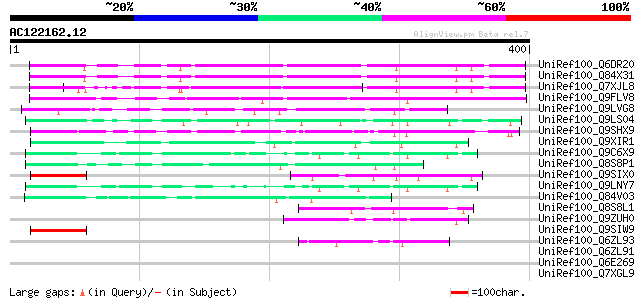
UniRef100_Q6DR20 Hypothetical protein [Arabidopsis thaliana] 160 5e-38
UniRef100_Q84X31 Hypothetical protein At2g17036/F6P23.4 [Arabido... 159 2e-37
UniRef100_Q7XJL8 At2g17030 protein [Arabidopsis thaliana] 134 6e-30
UniRef100_Q9FLV8 Gb|AAD22308.1 [Arabidopsis thaliana] 99 2e-19
UniRef100_Q9LVG8 Gb|AAF18598.1 [Arabidopsis thaliana] 84 9e-15
UniRef100_Q9LS04 Gb|AAC14516.2 [Arabidopsis thaliana] 81 5e-14
UniRef100_Q9SHX9 F1E22.13 [Arabidopsis thaliana] 74 6e-12
UniRef100_Q9XIR1 F13O11.14 protein [Arabidopsis thaliana] 67 1e-09
UniRef100_Q9C6X9 Hypothetical protein T7O23.22 [Arabidopsis thal... 64 6e-09
UniRef100_Q8S8P1 Hypothetical protein At2g26160 [Arabidopsis tha... 63 1e-08
UniRef100_Q9SIX0 Hypothetical protein At2g16290 [Arabidopsis tha... 58 5e-07
UniRef100_Q9LNY7 F9C16.31 [Arabidopsis thaliana] 55 3e-06
UniRef100_Q84V03 Hypothetical protein At2g16365/F16F14.28 [Arabi... 55 3e-06
UniRef100_Q8S8L1 Hypothetical protein At2g24255 [Arabidopsis tha... 49 3e-04
UniRef100_Q9ZUH0 Expressed protein [Arabidopsis thaliana] 49 3e-04
UniRef100_Q9SIW9 Hypothetical protein At2g16300 [Arabidopsis tha... 47 7e-04
UniRef100_Q6ZL93 Hypothetical protein OJ1065_B06.21 [Oryza sativa] 47 7e-04
UniRef100_Q6ZL91 F-box domain containing protein-like [Oryza sat... 47 0.001
UniRef100_Q6E269 Hypothetical protein [Arabidopsis thaliana] 47 0.001
UniRef100_Q7XGL9 Hypothetical protein [Oryza sativa] 46 0.002
>UniRef100_Q6DR20 Hypothetical protein [Arabidopsis thaliana]
Length = 404
Score = 160 bits (406), Expect = 5e-38
Identities = 130/417 (31%), Positives = 207/417 (49%), Gaps = 69/417 (16%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPN----DHNILPFQFPLLKYV 71
+DW+ LPKD++ LIS+ L+ DLI+FRSVCS+WRS+ P HN L +
Sbjct: 2 MDWATLPKDLLDLISKCLESSFDLIQFRSVCSSWRSAAGPKRLLWAHN--------LPFF 53
Query: 72 PTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQN- 130
P+ D +N I+ ++ +S L+KP + Q + + W++++ N
Sbjct: 54 PSDDKPFLSNVIL-----------RVAHQSILLIKPNEPQCEADLF----GWIVKVWDNI 98
Query: 131 -SSGKTQLLKPPFLSLTSTAF-SHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSY- 187
S K LLKP LS + F HLP + D +KF+ + L + + D H++
Sbjct: 99 YVSRKMTLLKP--LSSSRNYFPQHLPRIFDMSKFTVRELCREVKLYHPDYYCVPGHTALE 156
Query: 188 LYPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSN-LSTAHGDICLFKGRF 246
L K + + K ++L L Y K +F D+ WT +++ + + D+ +F GRF
Sbjct: 157 LELGKTVVKYLNDDKFVLLTILEY--GKLAVFRSWDREWTVINDYIPSRCQDLIMFDGRF 214
Query: 247 YVVDQSGQTVTVGPDS-STELAAQPLYRRCVRGENRKLLVENEGELLLLDI--------- 296
+ +D +G+TV V S LAA PL + G ++K L+E+ GE+ L+DI
Sbjct: 215 FAIDYNGRTVVVDYSSFKLTLAANPL----IGGGDKKFLIESCGEMFLVDIEFCLNEKPE 270
Query: 297 ---------HQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL---C 344
++T + KFFKL E EK+WV+++ LG ++ F+G +FSAS D+ C
Sbjct: 271 FTGGFYSYFNETTVSYKFKFFKLVEREKRWVEVEDLGDKMFFLGDDSTFSASTADIIPRC 330
Query: 345 LPKGNCVIFI---DTSVLSSDNMAGGNRVFHLDRGQLSHVSEYPECENLFL-PPEWI 397
+ G+ V F ++ V+ D G VF G+ V++ PE LF PP WI
Sbjct: 331 VGTGSFVFFYTHEESLVVMDDRNLG---VFDFRSGKTELVNKLPEYAKLFWPPPPWI 384
>UniRef100_Q84X31 Hypothetical protein At2g17036/F6P23.4 [Arabidopsis thaliana]
Length = 404
Score = 159 bits (401), Expect = 2e-37
Identities = 129/417 (30%), Positives = 206/417 (48%), Gaps = 69/417 (16%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPN----DHNILPFQFPLLKYV 71
+DW+ LPKD++ IS+ L+ DLI+FRSVCS+WRS+ P HN L +
Sbjct: 2 MDWATLPKDLLDXISKCLESSFDLIQFRSVCSSWRSAAGPKRLLWAHN--------LPFF 53
Query: 72 PTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQN- 130
P+ D +N I+ ++ +S L+KP + Q + + W++++ N
Sbjct: 54 PSDDKPFLSNVIL-----------RVAHQSILLIKPNEPQCEADLF----GWIVKVWDNI 98
Query: 131 -SSGKTQLLKPPFLSLTSTAF-SHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSY- 187
S K LLKP LS + F HLP + D +KF+ + L + + D H++
Sbjct: 99 YVSRKMTLLKP--LSSSRNYFPQHLPRIFDMSKFTVRELCREVKLYHPDYYCVPGHTALE 156
Query: 188 LYPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSN-LSTAHGDICLFKGRF 246
L K + + K ++L L Y K +F D+ WT +++ + + D+ +F GRF
Sbjct: 157 LELGKTVVKYLNDDKFVLLTILEY--GKLAVFRSWDREWTVINDYIPSRCQDLIMFDGRF 214
Query: 247 YVVDQSGQTVTVGPDS-STELAAQPLYRRCVRGENRKLLVENEGELLLLDI--------- 296
+ +D +G+TV V S LAA PL + G ++K L+E+ GE+ L+DI
Sbjct: 215 FAIDYNGRTVVVDYSSFKLTLAANPL----IGGGDKKFLIESCGEMFLVDIEFCLNEKPE 270
Query: 297 ---------HQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL---C 344
++T + KFFKL E EK+WV+++ LG ++ F+G +FSAS D+ C
Sbjct: 271 FTGGFYSYFNETTVSYKFKFFKLVEREKRWVEVEDLGDKMFFLGDDSTFSASTADIIPRC 330
Query: 345 LPKGNCVIFI---DTSVLSSDNMAGGNRVFHLDRGQLSHVSEYPECENLFL-PPEWI 397
+ G+ V F ++ V+ D G VF G+ V++ PE LF PP WI
Sbjct: 331 VGTGSFVFFYTHEESLVVMDDRNLG---VFDFRSGKTELVNKLPEYAKLFWPPPPWI 384
>UniRef100_Q7XJL8 At2g17030 protein [Arabidopsis thaliana]
Length = 1169
Score = 134 bits (336), Expect = 6e-30
Identities = 117/391 (29%), Positives = 187/391 (46%), Gaps = 69/391 (17%)
Query: 42 FRSVCSTWRSSPIPND----HNILPFQFPLLKYVPTPDSISNNNEIIDNIENTSTSFGHL 97
FRSVCS+WRS+ P HN L + P+ D +N I+ +
Sbjct: 768 FRSVCSSWRSAAGPKRLLWAHN--------LPFFPSDDKPFLSNVIL-----------RV 808
Query: 98 SKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNS--SGKTQLLKPPFLSLTSTAF-SHLP 154
+ +S L+KP + Q + + W++++ N S K LLKP LS + F HLP
Sbjct: 809 AHQSILLIKPNEPQCEADLF----GWIVKVWDNIYVSRKMTLLKP--LSSSRNYFPQHLP 862
Query: 155 NVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSY-LYPQKILAVTCPEKKPLVLGTLSYCS 213
+ D +KF+ + L + + D H++ L K + + K ++L L Y
Sbjct: 863 RIFDMSKFTVRELCREVKLYHPDYYCVPGHTALELELGKTVVKYLNDDKFVLLTILEY-- 920
Query: 214 SKRVLFHDRDKRWTPVSN-LSTAHGDICLFKGRFYVVDQSGQTVTVGPDS-STELAAQPL 271
K +F D+ WT +++ + + D+ +F GRF+ +D +G+TV V S LAA PL
Sbjct: 921 GKLAVFRSWDREWTVINDYIPSRCQDLIMFDGRFFAIDYNGRTVVVDYSSFKLTLAANPL 980
Query: 272 YRRCVRGENRKLLVENEGELLLLDI------------------HQTFFQFSIKFFKLDEM 313
+ G ++K L+E+ GE+ L+DI ++T + KFFKL E
Sbjct: 981 ----IGGGDKKFLIESCGEMFLVDIEFCLNEKPEFTGGFYSYFNETTVSYKFKFFKLVER 1036
Query: 314 EKKWVKLKKLGGRVLFVGSGCSFSASGWDL---CLPKGNCVIFI---DTSVLSSDNMAGG 367
EK+WV+++ LG ++ F+G +FSAS D+ C+ G+ V F ++ V+ D G
Sbjct: 1037 EKRWVEVEDLGDKMFFLGDDSTFSASTADIIPRCVGTGSFVFFYTHEESLVVMDDRNLG- 1095
Query: 368 NRVFHLDRGQLSHVSEYPECENLFL-PPEWI 397
VF G+ V++ PE LF PP WI
Sbjct: 1096 --VFDFRSGKTELVNKLPEYAKLFWPPPPWI 1124
Score = 115 bits (288), Expect = 2e-24
Identities = 85/265 (32%), Positives = 133/265 (50%), Gaps = 32/265 (12%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRS-----SPIPNDHNILPFQFPLLKY 70
VDWS LPKD++ LIS+ L+ DLI+FRSVCS+WRS SP+P H P+L
Sbjct: 2 VDWSTLPKDLLDLISKSLESSFDLIQFRSVCSSWRSAAEPKSPLPTHH------LPIL-- 53
Query: 71 VPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQ- 129
PD N + +++ F LS+RS L+KP + + ++ WLI++ +
Sbjct: 54 ---PD---NGGSL---FPDSAVGF-RLSQRSILLIKPHEPCIESDSF----GWLIKVEED 99
Query: 130 -NSSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYL 188
N K LL P ++ + P V+D +KF + L +F + + S YL
Sbjct: 100 LNVPRKVTLL-DPLCDTRNSIPENFPRVLDMSKFKVRELGREFKLHYFNTVGDIVESLYL 158
Query: 189 YPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYV 248
+ + C VL T+ + S K +F D+ WT ++++ + + D+ LF GRF+
Sbjct: 159 EKAVVKYLDCDGDYKFVLLTI-HVSGKLAVFRSWDRAWTVINDMPSPYDDVMLFDGRFFA 217
Query: 249 VDQSGQTVTVGPDS-STELAAQPLY 272
VD +G+TV V S L A P++
Sbjct: 218 VDNNGRTVVVDYSSLKLTLVASPVF 242
>UniRef100_Q9FLV8 Gb|AAD22308.1 [Arabidopsis thaliana]
Length = 373
Score = 99.4 bits (246), Expect = 2e-19
Identities = 99/399 (24%), Positives = 180/399 (44%), Gaps = 46/399 (11%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVPTPD 75
V WSELP +I+ LIS ++D DLI FRSVCS WRSS + ++ + PL P
Sbjct: 2 VKWSELPPEILHLISLKIDNPFDLIHFRSVCSFWRSSSLLKFRHMTSLRCPL---PLDPG 58
Query: 76 SISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKT 135
++ I LS R + L P +++ Q W+ ++ +G+
Sbjct: 59 GCGDDCHI-------------LSSRVYLLKCPNRDRPQY--------WMFKLQAKENGEV 97
Query: 136 QLLKPPFLSLTSTAFSHL-PNV-VDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQKI 193
+L FL S+A+ L P++ +D LA + + + F+ ++I
Sbjct: 98 -VLHSLFLRRNSSAYGCLYPSLSIDLLNCQIFELAQEHVACYSE-WFELFECISKCEERI 155
Query: 194 --LAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGD-ICLFKGRFYVVD 250
+ + + + ++LG LS+ + ++ D RWT + + + I FKG+FY +D
Sbjct: 156 GFMGLNREKNEYMILGKLSF--NGLAMYRSVDNRWTELEITPDSFFEGIVPFKGKFYAID 213
Query: 251 QSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVENEGELLLLDIHQTFFQFSI----- 305
++G+T V P + E+ R C + R LL+ + +L+ ++ + F I
Sbjct: 214 RTGKTTVVDP--TLEVNTFQRSRPCDKTRKRWLLMTGDKLILVEMCTKSRYDFHIPNIRE 271
Query: 306 -----KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDLCLPKGNCVIFIDTSVLS 360
+ +L+E W +++ + GRVLF+ C+FS ++ + N +IF+D S
Sbjct: 272 KKIWFEISELNEERNDWDQVEDMDGRVLFLEHYCTFSCLATEIPGFRANSIIFMDLWGGS 331
Query: 361 SDNMAGGNRVFHLDRGQLSHVSEYPECENLF-LPPEWIL 398
+ V+ + + + + E LF PP W++
Sbjct: 332 NSYELESILVYEFNEKGVRSLRDKLEYIELFPFPPGWVI 370
>UniRef100_Q9LVG8 Gb|AAF18598.1 [Arabidopsis thaliana]
Length = 374
Score = 83.6 bits (205), Expect = 9e-15
Identities = 95/359 (26%), Positives = 163/359 (44%), Gaps = 72/359 (20%)
Query: 10 TSSMASVDWSELPKDIIFLISQRLDVE----LDLIRFRSVCSTWRSS-PIPNDHNILPFQ 64
+SS+ WS+LP DI+ LIS RLD + + L+ RSVC+TWR S P+ N +N L +
Sbjct: 3 SSSLLPSQWSDLPLDILELISDRLDHDSSDTIHLLCLRSVCATWRLSLPLSNKNNRLS-K 61
Query: 65 FPLLKYVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWL 124
FP KY+P S S+ S+ F L + + + ++ P + R L
Sbjct: 62 FP--KYLPFWSSSSS-----------SSGFFTLKQSNVYKLEAP---------LNPRTCL 99
Query: 125 IRITQNSSGKTQLLKPPFLSLTSTAF--SHLPNVVDFTKFSPQHLATDFIIDKDDLTFQN 182
+++ + + G ++L S F + P+ +D +F + + + ++ + N
Sbjct: 100 VKLQETTPGIMRVLD--LFSNDRICFLPENFPSKIDLQEFHVRLVRRTYRME-----YAN 152
Query: 183 QHSSY------LYPQKILAVTCPEKKPLVL----GTLSYCSSKRVLFHDRDKRWTPVSNL 232
L+ K++ ++ E ++ G L + S D++W + N
Sbjct: 153 NGGGAVPCFWSLHSDKVVILSSGEDSAIIAIHSGGKLGFLKS------GNDEKWKILDNS 206
Query: 233 -STAHGDICLFK-GRFYVVDQSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVE-NEG 289
+ + DI L+K R VVD G+TV D A+ L G ++K LVE + G
Sbjct: 207 WNVIYEDIMLYKDNRCIVVDDKGKTVIYDVDFKVSDLAEGLVGG---GGHKKHLVECSGG 263
Query: 290 ELLLLD-----------IHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFS 337
E+ L+D I ++ +F + + L EK+W +++ LG LF+G CSFS
Sbjct: 264 EVFLVDKYVKTVWCKSDISKSVVEFRV--YNLKREEKRWEEVRDLGDVALFIGDDCSFS 320
>UniRef100_Q9LS04 Gb|AAC14516.2 [Arabidopsis thaliana]
Length = 377
Score = 81.3 bits (199), Expect = 5e-14
Identities = 106/416 (25%), Positives = 166/416 (39%), Gaps = 77/416 (18%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVP 72
M +WS+LP++++ LI+ R +D++R RS C +WRS+ + + QF +Y+P
Sbjct: 1 MEKTEWSDLPEELLDLIANRYSSNIDVLRIRSTCKSWRSAVAMSKERL---QFRFERYLP 57
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSS 132
T +N +I HLS +FF + P + WL+R Q S
Sbjct: 58 T-----SNKKI----------KAHLSPTTFFRITLPSS-------CPNKGWLVRTRQASK 95
Query: 133 --GKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIID-----------KDDLT 179
K LL P L+ + +D K + + I D
Sbjct: 96 MYRKITLLCP----LSGERITRSHQTLDLLKVGVSEIRQSYEIQIFDGLKDEKIPLDSEI 151
Query: 180 FQN--QHSSYLYPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRD--KRWTPVSNLSTA 235
F N ++S + V + + ++Y +K R+ + WT + N
Sbjct: 152 FSNYIKNSDKIPSGVFFRVAFLDN---IFFAVNYHDNKIWCCKTREGSRSWTKIKNQVED 208
Query: 236 HGDICLFKGRFYVVDQSG--------QTVTVGPDSSTELAAQPLYRRCVRGENRKLLVEN 287
DI L GR Y VD G Q V SST L Y C LVE
Sbjct: 209 FSDIILHMGRIYAVDLKGAIWWISLSQLTIVQQTSSTPLDYYK-YDSC----QDTRLVEY 263
Query: 288 EGELLL---LDIHQTFFQFSI--KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFS--ASG 340
G+L + L I + Q ++ K +K+DE KWV++ LG L V F+ AS
Sbjct: 264 CGDLCIVHELSITRNHIQRTVGFKVYKMDEDLAKWVEVSCLGDNTLIVACNSCFTVVASE 323
Query: 341 WDLCLPKGNCVIFIDTSVLSSDNMAGGNRVFHLDRGQLSHVSEY--PECENLFLPP 394
+ CL N + F V ++N+ +VF LD G ++ ++ C ++F P
Sbjct: 324 YHGCLK--NSIYFSYYDVKKAENI----KVFKLDDGSITQRTDISSQSCFHMFSLP 373
>UniRef100_Q9SHX9 F1E22.13 [Arabidopsis thaliana]
Length = 360
Score = 74.3 bits (181), Expect = 6e-12
Identities = 99/399 (24%), Positives = 166/399 (40%), Gaps = 66/399 (16%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQF-PLLKYVPTPD 75
DWS LP D++ +I+ RL ++L RFRS+C +WRSS +P PF+ PL+ P P
Sbjct: 3 DWSTLPVDLLNMIAGRLFSNIELKRFRSICRSWRSS-VPGAGKKNPFRTRPLILLNPNP- 60
Query: 76 SISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQN-SSGK 134
N + D+ LS+ +FF V Q WLI+ + SSGK
Sbjct: 61 ----NKPLTDHRRRGE----FLSRSAFFRVTLSSSPSQ--------GWLIKSDVDVSSGK 104
Query: 135 TQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQKIL 194
LL P L+ H VD ++F+ + + + ++ + + +++
Sbjct: 105 LHLLDP----LSRLPMEHSRKRVDLSEFTITEIREAYQVH----DWRTRKETRPIFKRVA 156
Query: 195 AVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYVVDQSGQ 254
V E VLG S+ ++++ D K W DI + KG+ Y +D G
Sbjct: 157 LVKDKEGDNQVLGIR---STGKMMYWD-IKTW-KAKEEGYEFSDIIVHKGQTYALDSIGI 211
Query: 255 TVTVGPDSSTELAAQPLYRRCVRGENRKLLVENEGELLLLD-----------IHQTFFQF 303
+ D + PL G+ R LVE GE +++ T +++
Sbjct: 212 VYWIRSDLKF-IRFGPLVGDWT-GDRR--LVECCGEFYIVERLVGESTWKRKADDTGYEY 267
Query: 304 S----IKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDLCLPKGNCVIFIDTSVL 359
+ K +K D+ + K +++K LG + + + FS + N + F D +++
Sbjct: 268 AKTVGFKVYKFDDEQGKMMEVKSLGDKAFVIATDTCFSVLAHEFYGCLENAIYFTDDTMI 327
Query: 360 SSDNMAGGNRVFHLDRGQLSHVSE--YP---ECENLFLP 393
+VF LD G S + YP C +F+P
Sbjct: 328 ---------KVFKLDNGNGSSIETTIYPYAQSCFQMFVP 357
>UniRef100_Q9XIR1 F13O11.14 protein [Arabidopsis thaliana]
Length = 383
Score = 66.6 bits (161), Expect = 1e-09
Identities = 87/358 (24%), Positives = 139/358 (38%), Gaps = 57/358 (15%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSS-PIPNDHNILPFQFPLLKYVPTPD 75
+WS+LP++++ LIS+ LD D++ RS+C +WRS+ P P+ + L + P P
Sbjct: 3 NWSQLPEELLNLISKNLDNCFDVVHARSICRSWRSAFPFPSSLSTLSYSLPTFAKFPL-- 60
Query: 76 SISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKT 135
S L K FL + +L I Q+ S
Sbjct: 61 --------------VSKDLCTLKKIQIFLFRARNPAADIPEY-----FLGGIDQDQSNDH 101
Query: 136 QLLKPPFLSLTSTAFSHL-PNVVDFTKFSPQHLATDFI-IDKDDLTFQNQHSSYLYPQKI 193
L P F P +V+ + L +I I D + N + +
Sbjct: 102 MELPSPLQCSVKVKFPQSDPFLVNMLDYQIIPLGFQYIMIGWDPESLANGYVGVAF---- 157
Query: 194 LAVTCPEKK---PLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAH-GDICLFKGRFYVV 249
P KK + L Y + VL + RW V S A + F+GRFYV
Sbjct: 158 ----LPVKKNGGDEFVVLLRYRNHLLVL-RSSEMRWMKVKKTSIASCKGLVSFRGRFYVT 212
Query: 250 DQSGQTVTVGPDSSTE---LAAQPLYRRCVRGENRKLLVENEGELLLLDIHQTF------ 300
+G P S + + ++PL ++ L+ EL L++ F
Sbjct: 213 FLNGDIYVFDPYSLEQTLLMPSEPL------RSSKYLIPNGSDELFLVEKFNPFPEADVL 266
Query: 301 --FQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL---CLPKGNCVIF 353
+F+ + +LDE +WV++ LG RVLF+G + S +L C GN ++F
Sbjct: 267 DLSRFACRVSRLDEEAGQWVEVIDLGDRVLFIGHFGNVCCSAKELPDGCGVSGNSILF 324
>UniRef100_Q9C6X9 Hypothetical protein T7O23.22 [Arabidopsis thaliana]
Length = 347
Score = 64.3 bits (155), Expect = 6e-09
Identities = 90/369 (24%), Positives = 146/369 (39%), Gaps = 77/369 (20%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVP 72
M V WS+L +D+I L++ L ++L+RFRS+C WRS+ V
Sbjct: 1 MGKVGWSDLHEDLIDLLANNLSSNINLLRFRSICKPWRST------------------VA 42
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSS 132
T + N+ E N+ +S +FF V P R + WLI+ Q S
Sbjct: 43 TKKRLHNHFE--RNLPTFKKKKTVVSPSTFFRVTLPSP-------CRNKGWLIKNRQVSE 93
Query: 133 GKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQK 192
L P T T +++ F + F D D + F + + +
Sbjct: 94 SSKNNLLSPLSGKTITPSDKTLDLLKVECFRDSSILQLF-ADSDRVVFLD--NVFFVVDF 150
Query: 193 ILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHG--DICLFKGRFYVVD 250
+ C C S + + WT ++N A G DI L KG+ Y +D
Sbjct: 151 KNEIWC-------------CKS-----GEETRHWTRINN-EEAKGFLDIILHKGKIYALD 191
Query: 251 QSGQTVTVGPDSSTELA------AQPLYRRCVRGENRKLLVENEGELLLLDIHQTFFQFS 304
+G + S +EL+ + P+ + K LVE GEL + +H+ + +F
Sbjct: 192 LTGAIWWI---SLSELSIYQYGPSTPVDFYEIDNCKEKRLVEYCGELCV--VHRFYKKFC 246
Query: 305 I-----------KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSF--SASGWDLCLPKGNCV 351
+ K +K+D+ +WV++ LG + L V + F AS + CL N +
Sbjct: 247 VKRVLTERTVCFKVYKMDKNLVEWVEVSSLGDKALIVATDNCFLVLASEYYGCLE--NAI 304
Query: 352 IFIDTSVLS 360
F D +S
Sbjct: 305 YFNDREDVS 313
>UniRef100_Q8S8P1 Hypothetical protein At2g26160 [Arabidopsis thaliana]
Length = 359
Score = 63.2 bits (152), Expect = 1e-08
Identities = 80/322 (24%), Positives = 124/322 (37%), Gaps = 53/322 (16%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVP 72
M +WSELP D+I L + R D++R RS+C WRS+ P F
Sbjct: 1 MEKPEWSELPGDLINLTANRFSSISDVLRVRSICKPWRSAA------ATPKSFQC----- 49
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQ-NS 131
++ ++N++I+ + LS +FF V P + WLIR Q +
Sbjct: 50 ---NLPSSNKMIETV---------LSPTTFFRVTGPSS-------CSYKGWLIRTKQVSE 90
Query: 132 SGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQ 191
S K LL P F L + + +D KF + + I D P
Sbjct: 91 SSKINLLSPFFRQLLTPS----QQTLDLLKFEVSEIRQSYEIHIFDKYLIQGVIGKEGPS 146
Query: 192 KILA-VTCPEKKPLVLG---TLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFY 247
IL+ V + +G + C S + + WT + N DI L KG+ Y
Sbjct: 147 HILSRVVFLDNLIFAVGQDDKIWCCKSG----EESSRIWTKIKNQVEDFLDIILHKGQVY 202
Query: 248 VVDQSGQTVTVGPDSSTELAAQPLYRRCVRGEN--RKLLVENEGELLLLD--------IH 297
+D +G + + L P G + K LVE G+L ++ I
Sbjct: 203 ALDLTGAIWWISLSPLSLLQFTPSIPMDYDGYDSCNKRLVEYCGDLCIIHQLRLKKAYIR 262
Query: 298 QTFFQFSIKFFKLDEMEKKWVK 319
++ K +K+DE KWV+
Sbjct: 263 RSQRTVGFKVYKMDEYVAKWVE 284
>UniRef100_Q9SIX0 Hypothetical protein At2g16290 [Arabidopsis thaliana]
Length = 415
Score = 57.8 bits (138), Expect = 5e-07
Identities = 47/165 (28%), Positives = 81/165 (48%), Gaps = 21/165 (12%)
Query: 217 VLFHDRDKRWTPVSNL--STAHGDICLFKGRFYVVDQSGQTVTVGPDSSTELAAQPLYRR 274
V++ R+ WT V + A+ D+ FKG+FY +D SG+ G EL+ +
Sbjct: 207 VMYRSRNMSWTQVVEHPETYAYQDLVAFKGKFYALDSSGR----GRVFVVELSLRVTEIP 262
Query: 275 CVRGE---NRKLLVENEGELLLLDI-------HQTFFQFSIKFFKLDEM--EKKWVKLKK 322
V+G +++ LV+ ELLL+ + + K F+LDE ++KWV++
Sbjct: 263 SVKGSQYCSKESLVQLGEELLLVQRFIPAGRRYDEYIYTWFKVFRLDEEGGKRKWVQVNY 322
Query: 323 LGGRVLFVGSGCSFSASGWDLCLPKGNCVIFI---DTSVLSSDNM 364
L RV+F+G+ S L NC++F+ D S+ +D++
Sbjct: 323 LNDRVIFLGARTKLCCSVHKLPGANENCIVFLASNDGSIFDNDSV 367
Score = 47.4 bits (111), Expect = 7e-04
Identities = 20/43 (46%), Positives = 28/43 (64%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHN 59
DWS LP D++ LI L+ +++ FRSVC +WRS P DH+
Sbjct: 3 DWSLLPNDLLELIVGHLETSFEIVLFRSVCRSWRSVVPPLDHS 45
>UniRef100_Q9LNY7 F9C16.31 [Arabidopsis thaliana]
Length = 311
Score = 55.1 bits (131), Expect = 3e-06
Identities = 90/369 (24%), Positives = 141/369 (37%), Gaps = 113/369 (30%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVP 72
M V WS+L +D+I L++ L ++L+RFRS+C WRS+ V
Sbjct: 1 MGKVGWSDLHEDLIDLLANNLSSNINLLRFRSICKPWRST------------------VA 42
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSS 132
T + N+ E N+ +S +FF V P R + WLI+ Q S
Sbjct: 43 TKKRLHNHFE--RNLPTFKKKKTVVSPSTFFRVTLPSP-------CRNKGWLIKNRQVSE 93
Query: 133 GKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQK 192
+ S L NV F++D F+N+
Sbjct: 94 S---------IFADSDRVVFLDNVF-------------FVVD-----FKNE--------- 117
Query: 193 ILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHG--DICLFKGRFYVVD 250
+ C C S H WT ++N A G DI L KG+ Y +D
Sbjct: 118 ---IWC-------------CKSGEETRH-----WTRINN-EEAKGFLDIILHKGKIYALD 155
Query: 251 QSGQTVTVGPDSSTELA------AQPLYRRCVRGENRKLLVENEGELLLLDIHQTFFQFS 304
+G + S +EL+ + P+ + K LVE GEL + +H+ + +F
Sbjct: 156 LTGAIWWI---SLSELSIYQYGPSTPVDFYEIDNCKEKRLVEYCGELCV--VHRFYKKFC 210
Query: 305 I-----------KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSF--SASGWDLCLPKGNCV 351
+ K +K+D+ +WV++ LG + L V + F AS + CL N +
Sbjct: 211 VKRVLTERTVCFKVYKMDKNLVEWVEVSSLGDKALIVATDNCFLVLASEYYGCLE--NAI 268
Query: 352 IFIDTSVLS 360
F D +S
Sbjct: 269 YFNDREDVS 277
>UniRef100_Q84V03 Hypothetical protein At2g16365/F16F14.28 [Arabidopsis thaliana]
Length = 332
Score = 55.1 bits (131), Expect = 3e-06
Identities = 75/297 (25%), Positives = 119/297 (39%), Gaps = 34/297 (11%)
Query: 12 SMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYV 71
++ DW LP D++ LI L+ ++ FRSVCS+WRS P DH+ +
Sbjct: 18 NLTMADWPLLPNDLLELIMGHLETSFEIFLFRSVCSSWRSVVPPLDHS-------RCLGI 70
Query: 72 PTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNS 131
T D IS N N T+ + L K +LVK L R + +
Sbjct: 71 KTHD-ISFNVGFSFNGRPTN-EYCTLKKIPIYLVKFWTPFGDDYLLAEMR-------ERN 121
Query: 132 SGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQ 191
G+ +LL P LS + N V F + + + + + + +P
Sbjct: 122 DGEPKLLLSP-LSSNGIKYGMGINKVLFNSLTSPIIPFGQYYEITYIEKRPSEYRFGFPY 180
Query: 192 KILAVTCPEKKPL------------VLGTLSYCSSKRVLFHDRDKRWTPVSN--LSTAHG 237
K+ V E+ VL C+ V++ R+ WT V A+
Sbjct: 181 KLEWVEITERVEFLKLDSEDSRDFAVLFAGRMCN--LVMYRSRNMSWTQVVEHPEKYAYQ 238
Query: 238 DICLFKGRFYVVDQSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVENEGELLLL 294
D+ FKG+FY VD SG+ + S E+ P + +++ LV++ ELLL+
Sbjct: 239 DLVAFKGKFYAVDSSGRGRVFVVELSFEVTEIPSVGGS-QQSSKESLVQSGEELLLV 294
>UniRef100_Q8S8L1 Hypothetical protein At2g24255 [Arabidopsis thaliana]
Length = 336
Score = 48.9 bits (115), Expect = 3e-04
Identities = 48/150 (32%), Positives = 70/150 (46%), Gaps = 25/150 (16%)
Query: 223 DKRWTPVSNLS-TAHGDICLFKGRFYVVDQSGQTVTVGPDS--STELAAQPLYRRCVRGE 279
+ RW + N+ T DI F+GRFY + SGQ V + P S T L + P R E
Sbjct: 161 EMRWKQLENVPYTICVDIVTFRGRFYALFLSGQIVVIDPYSLEVTLLLSSPQDPR----E 216
Query: 280 NRKLLVENEGELLLL------DIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGS- 332
R L+ + EL L+ D+ F + + + ++DE KWV++ LG VL +G+
Sbjct: 217 LRSLVPYGKDELFLVEVIIPNDVVIDFSRLARRVRRVDEKAGKWVEVGDLGDCVLLLGNL 276
Query: 333 GCSFSASGWDLCLPKG-----NCVIFIDTS 357
CS P+G N V+F D S
Sbjct: 277 SCSAKE------FPRGFGLTENAVLFGDQS 300
>UniRef100_Q9ZUH0 Expressed protein [Arabidopsis thaliana]
Length = 374
Score = 48.5 bits (114), Expect = 3e-04
Identities = 43/147 (29%), Positives = 68/147 (46%), Gaps = 15/147 (10%)
Query: 212 CSSKRVLFHDRDKRWTPVSNLSTAH-GDICLFKGRFYVVDQSGQTVTVGPDSSTELAAQP 270
C+ ++ + RW + STA ++ F+GRFY +G T + P S L A P
Sbjct: 185 CTDLFLVLRSTEMRWIRLEKPSTASCKELFTFRGRFYATFFNGDTFVIDPSS---LEATP 241
Query: 271 LYRRCVRGENRKLLVENEGELLLLDIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFV 330
L + LV + E L L + F + + +LDE +WV++ LG RVLF+
Sbjct: 242 LTPHI----DSNFLVPSGNEELFL-VKTDFLRCRVS--RLDEEAAEWVEVSDLGDRVLFL 294
Query: 331 GSGC-SFSASGWDL---CLPKGNCVIF 353
G +F S +L C G+ ++F
Sbjct: 295 GGHLGNFYCSAKELPHGCGLTGDSILF 321
Score = 42.4 bits (98), Expect = 0.023
Identities = 22/51 (43%), Positives = 35/51 (68%), Gaps = 3/51 (5%)
Query: 8 VATSSMASVDWSELPKDIIFLISQRL-DVELDLIRFRSVCSTWRSS-PIPN 56
++TSS+ DWS+LP++++ +IS L D D + RSVC +WRS+ P P+
Sbjct: 7 ISTSSIMP-DWSQLPEELLHIISTHLEDHYFDAVHARSVCRSWRSTFPFPS 56
>UniRef100_Q9SIW9 Hypothetical protein At2g16300 [Arabidopsis thaliana]
Length = 467
Score = 47.4 bits (111), Expect = 7e-04
Identities = 20/43 (46%), Positives = 28/43 (64%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHN 59
DWS LP D++ LI L+ +++ FRSVCS+WRS P D +
Sbjct: 3 DWSLLPNDLLELIVGHLETSFEIVLFRSVCSSWRSVVPPQDQS 45
Score = 45.8 bits (107), Expect = 0.002
Identities = 40/146 (27%), Positives = 65/146 (44%), Gaps = 16/146 (10%)
Query: 217 VLFHDRDKRWTPVSNLST--AHGDICLFKGRFYVVDQSGQTVTVGPDSSTELAAQPLYRR 274
V++ R+ WT V A+ D+ FKG+FY +D SG+ + S E+ P R
Sbjct: 203 VMYRSRNMSWTQVVEHPEKYAYQDLVAFKGKFYALDSSGRGRVFVVELSLEVMEIPSVRG 262
Query: 275 CVRGENRKLLVENEGELLLLDIHQTFFQFSIKFFKLDEMEKKWVKLKKLG---GRVLFVG 331
+ L++ E ELLL+ +F+ + DE W ++ +L GR
Sbjct: 263 SQQSSKENLVLSGE-ELLLVQ------RFTPEVRHYDEYLYTWFRVFRLDEEEGRETQTN 315
Query: 332 SGCSFSASGWDLCLPKGNCVIFIDTS 357
CS L K NC++FI+++
Sbjct: 316 LCCSVH----KLPCAKENCIVFIESN 337
Score = 40.4 bits (93), Expect = 0.089
Identities = 20/53 (37%), Positives = 29/53 (53%), Gaps = 6/53 (11%)
Query: 18 WSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKY 70
WS L D++ LI + +++ RSVCS+WRS P D+ FPL +Y
Sbjct: 402 WSLLLNDLLKLIVGHFETSFEIVNLRSVCSSWRSVVPPLDN------FPLFRY 448
>UniRef100_Q6ZL93 Hypothetical protein OJ1065_B06.21 [Oryza sativa]
Length = 366
Score = 47.4 bits (111), Expect = 7e-04
Identities = 41/130 (31%), Positives = 60/130 (45%), Gaps = 19/130 (14%)
Query: 223 DKRWTPVSNLSTAHGDICLFKGRFYVVD-----------QSGQTVTVGPDSSTELAAQPL 271
DK WT V +LS I F G FY V Q G+ +L Q
Sbjct: 190 DKCWTAV-DLSDHLRPIMSFAGPFYAVTTHQRCHHGGGGQPGEPDAAAGGGRRDLTLQHR 248
Query: 272 YRRCVRGENRKLLVENEGELLLLDIHQTFF--QFSIKFFKLDEMEKKWVKLKKLGGRVLF 329
+ R + + LV+N GELLL +H+T + + +++D +K V ++ LGGR +F
Sbjct: 249 FSRMLGSAH---LVDNNGELLL--VHRTLSGDKRLYQAYRVDLDGRKMVPVRGLGGRAVF 303
Query: 330 VGSGCSFSAS 339
+G CS S S
Sbjct: 304 IGHDCSLSVS 313
>UniRef100_Q6ZL91 F-box domain containing protein-like [Oryza sativa]
Length = 377
Score = 47.0 bits (110), Expect = 0.001
Identities = 45/125 (36%), Positives = 60/125 (48%), Gaps = 11/125 (8%)
Query: 223 DKRWTPVSNLSTAHGDICLFKGRFY-VVDQSGQTVTVGPDSST----ELAAQPLYRRCVR 277
DK WT V NLS GRFY V + V V +S T E A L R R
Sbjct: 193 DKCWTAV-NLSDCLRPSMSLAGRFYGVTSDAIMVVEVSRESQTPQLVEAADLTLQHRFSR 251
Query: 278 GENRKLLVENE-GELLLLDIHQTFF--QFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGC 334
LV+N GELLL +H+T + + +++D +K V ++ LGGR +F+G C
Sbjct: 252 MLGSAHLVDNNIGELLL--VHRTLSGNKRLYQAYRVDLDGRKTVPVRGLGGRAVFIGHDC 309
Query: 335 SFSAS 339
S S S
Sbjct: 310 SLSVS 314
>UniRef100_Q6E269 Hypothetical protein [Arabidopsis thaliana]
Length = 336
Score = 47.0 bits (110), Expect = 0.001
Identities = 48/150 (32%), Positives = 68/150 (45%), Gaps = 25/150 (16%)
Query: 223 DKRWTPVSNLS-TAHGDICLFKGRFYVVDQSGQTVTVGPDS--STELAAQPLYRRCVRGE 279
+ RW + N+ T DI F+GRFY SGQ V + P S T L + P R E
Sbjct: 161 EMRWKQLENVPYTICVDIVTFRGRFYAPFLSGQIVVIDPYSLEVTLLLSSPQDPR----E 216
Query: 280 NRKLLVENEGELLLL------DIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGS- 332
R L + EL L+ D+ F + + + ++DE KWV++ LG VL +G+
Sbjct: 217 LRSLAPYGKDELFLVEVIIPNDVVIDFSRLARRVRRVDEKAGKWVEVGDLGDCVLLLGNL 276
Query: 333 GCSFSASGWDLCLPKG-----NCVIFIDTS 357
CS P+G N V+F D S
Sbjct: 277 SCSAKE------FPRGFGLTENAVLFGDQS 300
>UniRef100_Q7XGL9 Hypothetical protein [Oryza sativa]
Length = 450
Score = 46.2 bits (108), Expect = 0.002
Identities = 25/59 (42%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVPTP 74
+DWS LP D++ L S RL LD +RFR+VC WR++ + PF P L P P
Sbjct: 1 MDWSGLPGDLLRLFSGRLHDPLDFLRFRAVCRAWRAATAAAAASPPPF-LPWLLARPAP 58
Score = 39.3 bits (90), Expect = 0.20
Identities = 26/83 (31%), Positives = 40/83 (47%), Gaps = 13/83 (15%)
Query: 275 CVRGENRKLLVENEGELL----LLDIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFV 330
CV R+ E GELL L++H+ ++ + +WV+++ +G R+LFV
Sbjct: 236 CVIRYFRRKNTEQAGELLEDCRALEVHR---------LEIAGEKSRWVQMRSIGDRMLFV 286
Query: 331 GSGCSFSASGWDLCLPKGNCVIF 353
G FS D +GNCV F
Sbjct: 287 GLYQGFSLRAADFAGLEGNCVYF 309
Score = 35.4 bits (80), Expect = 2.9
Identities = 27/97 (27%), Positives = 41/97 (41%), Gaps = 17/97 (17%)
Query: 261 DSSTELAAQPLYRRCVRGENRKLLVENEGELL----LLDIHQTFFQFSIKFFKLDEMEKK 316
D E P C +N E GELL L++H+ ++ + +
Sbjct: 327 DGQIEELPGPSMHACTWSKNS----EQAGELLEDCCALEVHR---------LEIAGEKSR 373
Query: 317 WVKLKKLGGRVLFVGSGCSFSASGWDLCLPKGNCVIF 353
WV+++ + R+LFVG FS D +GNCV F
Sbjct: 374 WVQMRSIRDRMLFVGLYQGFSLRAADFAGLEGNCVYF 410
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 707,564,050
Number of Sequences: 2790947
Number of extensions: 30459548
Number of successful extensions: 67063
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 49
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 66916
Number of HSP's gapped (non-prelim): 175
length of query: 400
length of database: 848,049,833
effective HSP length: 129
effective length of query: 271
effective length of database: 488,017,670
effective search space: 132252788570
effective search space used: 132252788570
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC122162.12


