

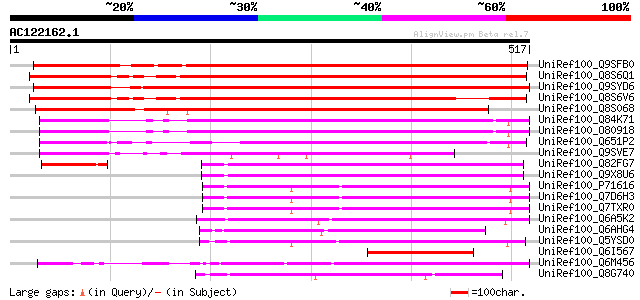
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.1 + phase: 0 /pseudo/partial
(517 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SFB0 Putative integral membrane protein [Arabidopsis... 582 e-164
UniRef100_Q8S6Q1 Putative membrane protein [Oryza sativa] 535 e-150
UniRef100_Q9SYD6 F11M15.20 protein [Arabidopsis thaliana] 529 e-149
UniRef100_Q8S6V6 Putative membrane protein [Oryza sativa] 464 e-129
UniRef100_Q8S068 P0678F11.10 protein [Oryza sativa] 452 e-125
UniRef100_Q84K71 Hypothetical protein At2g38330 [Arabidopsis tha... 337 6e-91
UniRef100_O80918 Hypothetical protein At2g38330 [Arabidopsis tha... 337 6e-91
UniRef100_Q651P2 Hypothetical protein OSJNBa0038K02.39 [Oryza sa... 308 2e-82
UniRef100_Q9SVE7 Hypothetical protein F22I13.150 [Arabidopsis th... 224 6e-57
UniRef100_Q82FG7 Putative DNA-damage-inducible protein F [Strept... 150 1e-34
UniRef100_Q9X8U6 Putative membrane protein [Streptomyces coelico... 142 2e-32
UniRef100_P71616 POSSIBLE DNA-DAMAGE-INDUCIBLE PROTEIN F DINF [M... 130 9e-29
UniRef100_Q7D6H3 DNA-damage-inducible protein F, putative [Mycob... 130 9e-29
UniRef100_Q7TXR0 POSSIBLE DNA-DAMAGE-INDUCIBLE PROTEIN F DINF [M... 129 2e-28
UniRef100_Q6A5K2 Conserved membrane protein, MatE domain [Propio... 128 5e-28
UniRef100_Q6AHG4 DNA-damage-inducible protein F [Leifsonia xyli] 120 7e-26
UniRef100_Q5YSD0 Putative DNA-damage-inducible protein F [Nocard... 120 7e-26
UniRef100_Q6I567 Hypothetical protein OSJNBb0014K18.14 [Oryza sa... 118 5e-25
UniRef100_Q6M456 PUTATIVE DNA-DAMAGE-INDUCIBLE MEMBRANE PROTEIN ... 114 5e-24
UniRef100_Q8G740 Hypothetical protein [Bifidobacterium longum] 110 1e-22
>UniRef100_Q9SFB0 Putative integral membrane protein [Arabidopsis thaliana]
Length = 526
Score = 582 bits (1499), Expect = e-164
Identities = 312/494 (63%), Positives = 379/494 (76%), Gaps = 17/494 (3%)
Query: 24 VFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRI 83
VF D+ +EILGIAFP+ALA+AADPIASLIDTAF+G LG V+LAA GVSIA+FNQASRI
Sbjct: 27 VFSRDTTGREILGIAFPAALALAADPIASLIDTAFVGRLGAVQLAAVGVSIAIFNQASRI 86
Query: 84 TIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQD-IEAGATKQ 142
TIFPLVS+TTSFVAEEDTM+++ +A K+N V + L+QD +E G +
Sbjct: 87 TIFPLVSLTTSFVAEEDTMEKMKEEAN----------KANLVHAETILVQDSLEKGISSP 136
Query: 143 DSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLI 202
S N + +NS NKS K +KR I +ASTA++ G +LGL+QA LI
Sbjct: 137 TS---NDTNQPQQPPAPDTKSNSGNKSNKK---EKRTIRTASTAMILGLILGLVQAIFLI 190
Query: 203 FAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIV 262
F++K LLG MG+K +SPML PA KYL +RALGAPA+LLSLAMQGIFRGFKDT TPL+ V
Sbjct: 191 FSSKLLLGVMGVKPNSPMLSPAHKYLSIRALGAPALLLSLAMQGIFRGFKDTKTPLFATV 250
Query: 263 SGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIF 322
+N+ +DP+ IF +LGI GAAI+HV+SQY M +L L KKV+L+PP+ DLQ
Sbjct: 251 VADVINIVLDPIFIFVLRLGIIGAAIAHVISQYFMTLILFVFLAKKVNLIPPNFGDLQFG 310
Query: 323 RFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQ 382
RFLKNG LLLAR IAVTFC TL+A++AARLG PMAAFQ CLQVW+TSSLL DGLAVA Q
Sbjct: 311 RFLKNGLLLLARTIAVTFCQTLAAAMAARLGTTPMAAFQICLQVWLTSSLLNDGLAVAGQ 370
Query: 383 AILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRL 442
AILACSFAEKDYNKVT A+R LQM FVLG+GLS+ VG GLYFGAGVFSK+ AVIHL+ +
Sbjct: 371 AILACSFAEKDYNKVTAVASRVLQMGFVLGLGLSVFVGLGLYFGAGVFSKDPAVIHLMAI 430
Query: 443 GLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWI 502
G+PF+AATQPINSLAFV DGVN+GASDFAY+AYS+V V+ S+ ++ ++ K+ GFIGIWI
Sbjct: 431 GIPFIAATQPINSLAFVLDGVNFGASDFAYTAYSMVGVAAISIAAVIYMAKTNGFIGIWI 490
Query: 503 ALTIYMSLRMFAGV 516
ALTIYM+LR G+
Sbjct: 491 ALTIYMALRAITGI 504
>UniRef100_Q8S6Q1 Putative membrane protein [Oryza sativa]
Length = 501
Score = 535 bits (1377), Expect = e-150
Identities = 286/496 (57%), Positives = 367/496 (73%), Gaps = 22/496 (4%)
Query: 20 ECSLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQ 79
+ L F+ D L +EI+GIA P ALA+ ADP+ASL+DTAFIGH+GPVELAA GVSIAVFNQ
Sbjct: 20 DARLAFRWDELGREIMGIAVPGALALMADPVASLVDTAFIGHIGPVELAAVGVSIAVFNQ 79
Query: 80 ASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGA 139
SRI IFPLVS+TTSFVAEED T + +++ +G+ NE D +++
Sbjct: 80 VSRIAIFPLVSVTTSFVAEEDA-----TSSDREKYEINGE---NEFNVSDSEMEE----- 126
Query: 140 TKQDSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAA 199
L + ++A++ SKSS T+SS + K K+++I S STALL G VLGL+QA
Sbjct: 127 ------LVSHEEASAAPSKSSFETDSS---DVKIEHKRKNIPSVSTALLLGGVLGLLQAL 177
Query: 200 TLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLY 259
L+ AKPLLG MG+K S ML+PA+KYL +R+LGAPAVLLSLAMQG+FRG KDT TPLY
Sbjct: 178 LLVICAKPLLGYMGVKQGSAMLMPALKYLVVRSLGAPAVLLSLAMQGVFRGLKDTKTPLY 237
Query: 260 VIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDL 319
V+G A N+ +DP+ +F F+ G+ GAAI+HV+SQY +A++LL+ L VDLLPPS K +
Sbjct: 238 ATVAGDATNIVLDPIFMFVFQYGVSGAAIAHVISQYFIASILLWRLRLHVDLLPPSFKHM 297
Query: 320 QIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAV 379
Q RFLKNG LLLARVIA T CVTLSAS+AARLG +PMAAFQ CLQ+W+ SSLLADGLA
Sbjct: 298 QFSRFLKNGFLLLARVIAATCCVTLSASMAARLGSVPMAAFQICLQIWLASSLLADGLAF 357
Query: 380 AIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHL 439
A QAILA +FA +D++K A+R LQ+ VLG+ LS+ +G GL G+ +F+ + V+H
Sbjct: 358 AGQAILASAFARQDHSKAAATASRILQLGLVLGLLLSIFLGIGLRLGSRLFTDDQDVLHH 417
Query: 440 IRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIG 499
I LG+PFV+ TQPIN+LAFVFDG+NYGASDF Y+AYS+++V+I S+ + L GF+G
Sbjct: 418 IYLGIPFVSLTQPINALAFVFDGINYGASDFGYAAYSMILVAIVSIIFIVTLASYNGFVG 477
Query: 500 IWIALTIYMSLRMFAG 515
IWIALT+YMSLRM AG
Sbjct: 478 IWIALTVYMSLRMLAG 493
>UniRef100_Q9SYD6 F11M15.20 protein [Arabidopsis thaliana]
Length = 501
Score = 529 bits (1363), Expect = e-149
Identities = 291/495 (58%), Positives = 352/495 (70%), Gaps = 20/495 (4%)
Query: 24 VFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRI 83
V K D L EI IA P+ALA+ ADPIASL+DTAFIG +GPVELAA GVSIA+FNQ SRI
Sbjct: 25 VLKFDELGLEIARIALPAALALTADPIASLVDTAFIGQIGPVELAAVGVSIALFNQVSRI 84
Query: 84 TIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQ- 142
IFPLVSITTSFVAEED S + DH + IE G
Sbjct: 85 AIFPLVSITTSFVAEEDA------------------CSSQQDTVRDHK-ECIEIGINNPT 125
Query: 143 DSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLI 202
+ T++ + + + T+SS S SKP KKR+I SAS+AL+ G VLGL QA LI
Sbjct: 126 EETIELIPEKHKDSLSDEFKTSSSIFSISKPPAKKRNIPSASSALIIGGVLGLFQAVFLI 185
Query: 203 FAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIV 262
AAKPLL MG+K+DSPM+ P+ +YL LR+LGAPAVLLSLA QG+FRGFKDTTTPL+ V
Sbjct: 186 SAAKPLLSFMGVKHDSPMMRPSQRYLSLRSLGAPAVLLSLAAQGVFRGFKDTTTPLFATV 245
Query: 263 SGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIF 322
G N+ +DP+ IF F+LG+ GAA +HV+SQY+M +LL+ LM +VD+ S K LQ
Sbjct: 246 IGDVTNIILDPIFIFVFRLGVTGAATAHVISQYLMCGILLWKLMGQVDIFNMSTKHLQFC 305
Query: 323 RFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQ 382
RF+KNG LLL RVIAVTFCVTLSASLAAR G MAAFQ CLQVW+ +SLLADG AVA Q
Sbjct: 306 RFMKNGFLLLMRVIAVTFCVTLSASLAAREGSTSMAAFQVCLQVWLATSLLADGYAVAGQ 365
Query: 383 AILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRL 442
AILA +FA+KDY + A+R LQ+ VLG L++++G GL+FGA VF+K+ V+HLI +
Sbjct: 366 AILASAFAKKDYKRAAATASRVLQLGLVLGFVLAVILGAGLHFGARVFTKDDKVLHLISI 425
Query: 443 GLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWI 502
GLPFVA TQPIN+LAFVFDGVN+GASDF Y+A SLVMV+I S+ L FL + GFIG+W
Sbjct: 426 GLPFVAGTQPINALAFVFDGVNFGASDFGYAAASLVMVAIVSILCLLFLSSTHGFIGLWF 485
Query: 503 ALTIYMSLRMFAGVW 517
LTIYMSLR G W
Sbjct: 486 GLTIYMSLRAAVGFW 500
>UniRef100_Q8S6V6 Putative membrane protein [Oryza sativa]
Length = 469
Score = 464 bits (1193), Expect = e-129
Identities = 261/496 (52%), Positives = 337/496 (67%), Gaps = 54/496 (10%)
Query: 20 ECSLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQ 79
+ L F+ D L +EI+GIA P ALA+ ADP+ASL+DTAFIGH+GPVELAA GVSIAVFNQ
Sbjct: 20 DARLAFRWDELGREIMGIAVPGALALMADPVASLVDTAFIGHIGPVELAAVGVSIAVFNQ 79
Query: 80 ASRITIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGA 139
SRI IFPLVS+TTSFVAEED T + +++ +G+ NE D +++
Sbjct: 80 VSRIAIFPLVSVTTSFVAEEDA-----TSSDREKYEINGE---NEFNVSDSEMEE----- 126
Query: 140 TKQDSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAA 199
L + ++A++ SKSS T+SS + K K+++I S STALL G VLGL+QA
Sbjct: 127 ------LVSHEEASAAPSKSSFETDSS---DVKIEHKRKNIPSVSTALLLGGVLGLLQAL 177
Query: 200 TLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLY 259
L+ AKPLLG MG+K S ML+PA+KYL +R+LGAPAVLLSLAMQG+FRG KDT TPLY
Sbjct: 178 LLVICAKPLLGYMGVKQGSAMLMPALKYLVVRSLGAPAVLLSLAMQGVFRGLKDTKTPLY 237
Query: 260 VIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDL 319
V+G A N+ +DP+ +F F+ G+ GAAI+HV+SQY +A++LL+ L VDLLPPS K +
Sbjct: 238 ATVAGDATNIVLDPIFMFVFQYGVSGAAIAHVISQYFIASILLWRLRLHVDLLPPSFKHM 297
Query: 320 QIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAV 379
Q RFLKNG LLLARVIA T CVTLSAS+AARLG +PMAAFQ CLQ+W+ SSLLADGLA
Sbjct: 298 QFSRFLKNGFLLLARVIAATCCVTLSASMAARLGSVPMAAFQICLQIWLASSLLADGLAF 357
Query: 380 AIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHL 439
A QAILA +FA +D++K A+R LQ+ VLG+ LS+ +G GL G+ +F+ + V+H
Sbjct: 358 AGQAILASAFARQDHSKAAATASRILQLGLVLGLLLSIFLGIGLRLGSRLFTDDQDVLHH 417
Query: 440 IRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIG 499
I LG+P ++V+I S+ + L GF+G
Sbjct: 418 IYLGIP--------------------------------ILVAIVSIIFIVTLASYNGFVG 445
Query: 500 IWIALTIYMSLRMFAG 515
IWIALT+YMSLRM AG
Sbjct: 446 IWIALTVYMSLRMLAG 461
>UniRef100_Q8S068 P0678F11.10 protein [Oryza sativa]
Length = 558
Score = 452 bits (1163), Expect = e-125
Identities = 241/459 (52%), Positives = 326/459 (70%), Gaps = 15/459 (3%)
Query: 26 KMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITI 85
++D L E+L IA P++LA+ ADP+ASLIDTAFIG +G VE+AA GV+IAVFNQ ++ I
Sbjct: 104 RLDELGAEVLRIAVPASLALTADPLASLIDTAFIGRIGSVEIAAVGVAIAVFNQVMKVCI 163
Query: 86 FPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDST 145
+PLVS+TTSFVAEED + A+ ++ AK + A A +
Sbjct: 164 YPLVSVTTSFVAEEDAILSKGAAGADDDNDDGHDAKGHGA--------SAAAVADPEKQQ 215
Query: 146 LKNGDDANSN---ISKSSIVTNSSNKSESKPIR----KKRHIASASTALLFGTVLGLIQA 198
+ D A +N +S +++ T K+ + + ++R + S ++AL+ G LGL+QA
Sbjct: 216 VVGVDSAETNGAEVSTAAVRTTDDKKAAAAGVGVGKCRRRFVPSVTSALIVGAFLGLLQA 275
Query: 199 ATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPL 258
L+ A KPLL MG+K SPM++PA++YL +R+LGAPAVLLSLAMQG+FRGFKDT TPL
Sbjct: 276 VFLVAAGKPLLRIMGVKPGSPMMIPALRYLVVRSLGAPAVLLSLAMQGVFRGFKDTKTPL 335
Query: 259 YVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKD 318
Y V+G N+A+DP+LIF + G+ GAAI+HV+SQY++ ++L L++KVD++P S+K
Sbjct: 336 YATVTGDLANIALDPILIFTCRFGVVGAAIAHVISQYLITLIMLCKLVRKVDVIPSSLKS 395
Query: 319 LQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLA 378
L+ RFL G LLLARV+AVTFCVTL+ASLAAR G MAAFQ C QVW+ SSLLADGLA
Sbjct: 396 LKFRRFLGCGFLLLARVVAVTFCVTLAASLAARHGATAMAAFQICAQVWLASSLLADGLA 455
Query: 379 VAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIH 438
VA QA+LA +FA+KD+ KV R LQ++ VLGVGL+ + G++FGAGVF+ + AVI
Sbjct: 456 VAGQALLASAFAKKDHYKVAVTTARVLQLAVVLGVGLTAFLAAGMWFGAGVFTSDAAVIS 515
Query: 439 LIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSL 477
I G+PFVA TQ IN+LAFVFDGVN+GASD+A++AYS+
Sbjct: 516 TIHRGVPFVAGTQTINTLAFVFDGVNFGASDYAFAAYSM 554
>UniRef100_Q84K71 Hypothetical protein At2g38330 [Arabidopsis thaliana]
Length = 521
Score = 337 bits (863), Expect = 6e-91
Identities = 194/490 (39%), Positives = 286/490 (57%), Gaps = 66/490 (13%)
Query: 30 LAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLV 89
+ EI+ IA P+ALA+AADPI SL+DTAF+GH+G ELAA GVS++VFN S++ PL+
Sbjct: 76 IGMEIMSIALPAALALAADPITSLVDTAFVGHIGSAELAAVGVSVSVFNLVSKLFNVPLL 135
Query: 90 SITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLKNG 149
++TTSFVAEE +A A K D
Sbjct: 136 NVTTSFVAEE------------------------------------QAIAAKDD------ 153
Query: 150 DDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLL 209
N +I S K+ + S ST+L+ +G+ +A L + L+
Sbjct: 154 ---NDSIETS-----------------KKVLPSVSTSLVLAAGVGIAEAIALSLGSDFLM 193
Query: 210 GAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNV 269
M + +DSPM +PA ++LRLRA GAP ++++LA QG FRGFKDTTTPLY +V+G LN
Sbjct: 194 DVMAIPFDSPMRIPAEQFLRLRAYGAPPIVVALAAQGAFRGFKDTTTPLYAVVAGNVLNA 253
Query: 270 AMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFRFLKNGG 329
+DP+LIF GI GAA + V+S+Y++A +LL+ L + V LL P +K + ++LK+GG
Sbjct: 254 VLDPILIFVLGFGISGAAAATVISEYLIAFILLWKLNENVVLLSPQIKVGRANQYLKSGG 313
Query: 330 LLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSF 389
LL+ R +A+ TL+ SLAA+ GP MA Q L++W+ SLL D LA+A Q++LA ++
Sbjct: 314 LLIGRTVALLVPFTLATSLAAQNGPTQMAGHQIVLEIWLAVSLLTDALAIAAQSLLATTY 373
Query: 390 AEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAA 449
++ +Y + LQ+ G GL+ V+ + +F+ + V+ + G FVA
Sbjct: 374 SQGEYKQAREVLFGVLQVGLATGTGLAAVLFITFEPFSSLFTTDSEVLKIALSGTLFVAG 433
Query: 450 TQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSK--GFIGIWIALTIY 507
+QP+N+LAFV DG+ YG SDF ++AYS+V+V ++SLF L + G GIW L ++
Sbjct: 434 SQPVNALAFVLDGLYYGVSDFGFAAYSMVIVGF--ISSLFMLVAAPTFGLAGIWTGLFLF 491
Query: 508 MSLRMFAGVW 517
M+LR+ AG W
Sbjct: 492 MALRLVAGAW 501
>UniRef100_O80918 Hypothetical protein At2g38330 [Arabidopsis thaliana]
Length = 539
Score = 337 bits (863), Expect = 6e-91
Identities = 194/490 (39%), Positives = 286/490 (57%), Gaps = 66/490 (13%)
Query: 30 LAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLV 89
+ EI+ IA P+ALA+AADPI SL+DTAF+GH+G ELAA GVS++VFN S++ PL+
Sbjct: 76 IGMEIMSIALPAALALAADPITSLVDTAFVGHIGSAELAAVGVSVSVFNLVSKLFNVPLL 135
Query: 90 SITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLKNG 149
++TTSFVAEE +A A K D
Sbjct: 136 NVTTSFVAEE------------------------------------QAIAAKDD------ 153
Query: 150 DDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLL 209
N +I S K+ + S ST+L+ +G+ +A L + L+
Sbjct: 154 ---NDSIETS-----------------KKVLPSVSTSLVLAAGVGIAEAIALSLGSDFLM 193
Query: 210 GAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNV 269
M + +DSPM +PA ++LRLRA GAP ++++LA QG FRGFKDTTTPLY +V+G LN
Sbjct: 194 DVMAIPFDSPMRIPAEQFLRLRAYGAPPIVVALAAQGAFRGFKDTTTPLYAVVAGNVLNA 253
Query: 270 AMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFRFLKNGG 329
+DP+LIF GI GAA + V+S+Y++A +LL+ L + V LL P +K + ++LK+GG
Sbjct: 254 VLDPILIFVLGFGISGAAAATVISEYLIAFILLWKLNENVVLLSPQIKVGRANQYLKSGG 313
Query: 330 LLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSF 389
LL+ R +A+ TL+ SLAA+ GP MA Q L++W+ SLL D LA+A Q++LA ++
Sbjct: 314 LLIGRTVALLVPFTLATSLAAQNGPTQMAGHQIVLEIWLAVSLLTDALAIAAQSLLATTY 373
Query: 390 AEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAA 449
++ +Y + LQ+ G GL+ V+ + +F+ + V+ + G FVA
Sbjct: 374 SQGEYKQAREVLFGVLQVGLATGTGLAAVLFITFEPFSSLFTTDSEVLKIALSGTLFVAG 433
Query: 450 TQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSK--GFIGIWIALTIY 507
+QP+N+LAFV DG+ YG SDF ++AYS+V+V ++SLF L + G GIW L ++
Sbjct: 434 SQPVNALAFVLDGLYYGVSDFGFAAYSMVIVGF--ISSLFMLVAAPTFGLAGIWTGLFLF 491
Query: 508 MSLRMFAGVW 517
M+LR+ AG W
Sbjct: 492 MALRLVAGAW 501
>UniRef100_Q651P2 Hypothetical protein OSJNBa0038K02.39 [Oryza sativa]
Length = 533
Score = 308 bits (790), Expect = 2e-82
Identities = 189/488 (38%), Positives = 274/488 (55%), Gaps = 87/488 (17%)
Query: 30 LAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLV 89
+ KE+L +A P+ + A DP+A L++TA+IG LGPVELA+A V ++VFN S++ PL+
Sbjct: 119 IKKELLNLALPAIVGQAIDPVAQLLETAYIGRLGPVELASAAVGVSVFNIISKLFNIPLL 178
Query: 90 SITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLKNG 149
SITTSFVA ED + QF G S E+G K+
Sbjct: 179 SITTSFVA-EDV-----ARHDSDQFTSEGNMSS-------------ESGGRKR------- 212
Query: 150 DDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLL 209
+ S S+A+L +G+I+A+ LI
Sbjct: 213 ------------------------------LPSISSAILLAAAIGVIEASALI------- 235
Query: 210 GAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNV 269
L ALGAPAV++SLA+QGIFRG KDT TPL G V
Sbjct: 236 --------------------LGALGAPAVVVSLAIQGIFRGLKDTKTPLLYSGLGNISAV 275
Query: 270 AMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFRFLKNGG 329
+ P L++ LG+ GAA++ + SQY+ LLL+ L K+ LLPP ++DL ++K+GG
Sbjct: 276 LLLPFLVYSLNLGLNGAALATIASQYLGMFLLLWSLSKRAVLLPPKIEDLDFVGYIKSGG 335
Query: 330 LLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSF 389
+LL R ++V +TL ++AAR G I MAA Q CLQVW+ SLL+D LAV+ QA++A SF
Sbjct: 336 MLLGRTLSVLITMTLGTAMAARQGTIAMAAHQICLQVWLAVSLLSDALAVSAQALIASSF 395
Query: 390 AEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAA 449
A+ DY KV L++ ++G L+L++ A +FSK+ V+ ++ G+ FV+A
Sbjct: 396 AKLDYEKVKEVTYYVLKIGLLVGAALALLLFASFGRIAELFSKDPMVLQIVGSGVLFVSA 455
Query: 450 TQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSK--GFIGIWIALTIY 507
+QPIN+LAF+FDG+++G SDF+YSA S M+++ +++SLF LY K G G+W L ++
Sbjct: 456 SQPINALAFIFDGLHFGVSDFSYSASS--MITVGAISSLFLLYAPKVFGLPGVWAGLALF 513
Query: 508 MSLRMFAG 515
M LRM AG
Sbjct: 514 MGLRMTAG 521
>UniRef100_Q9SVE7 Hypothetical protein F22I13.150 [Arabidopsis thaliana]
Length = 479
Score = 224 bits (570), Expect = 6e-57
Identities = 154/463 (33%), Positives = 241/463 (51%), Gaps = 101/463 (21%)
Query: 30 LAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLV 89
+ +E++ ++ P+ A DP+ L++TA+IG LG VEL +AGVS+A+FN S++ PL+
Sbjct: 33 IKRELVMLSLPAIAGQAIDPLTLLMETAYIGRLGSVELGSAGVSMAIFNTISKLFNIPLL 92
Query: 90 SITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLKNG 149
S+ TSFVAE+ I AA QD+ +
Sbjct: 93 SVATSFVAED-----IAKIAA----------------------QDLAS------------ 113
Query: 150 DDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLL 209
+D+ S+I S+ + +++ ++S STAL+ +G+ +A L A+ P L
Sbjct: 114 EDSQSDIP-------------SQGLPERKQLSSVSTALVLAIGIGIFEALALSLASGPFL 160
Query: 210 GAMGLKYDSP------MLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVS 263
MG++ S M +PA ++L LRALGAPA ++SLA+QGIFRGFKDT TP+Y +V
Sbjct: 161 RLMGIQSVSSVQRMSEMFIPARQFLVLRALGAPAYVVSLALQGIFRGFKDTKTPVYCLVL 220
Query: 264 GYA----------LNVAMDPLLIFYFKLGIRGAAISHVLSQ-----------YIMATLLL 302
+ L V + PL I+ F++G+ GAAIS V+SQ Y +A L+L
Sbjct: 221 SFPNFHNSGIGNFLAVFLFPLFIYKFRMGVAGAAISSVISQMVLNPFPLIHRYTVAILML 280
Query: 303 FILMKKVDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQT 362
+L K+V LLPP + L+ +LK+GG +L R ++V +T++ S+AAR G MAA Q
Sbjct: 281 ILLNKRVILLPPKIGSLKFGDYLKSGGFVLGRTLSVLVTMTVATSMAARQGVFAMAAHQI 340
Query: 363 CLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKV----------------------TTA 400
C+QVW+ SLL D LA + QA++A S +++D+ V
Sbjct: 341 CMQVWLAVSLLTDALASSGQALIASSASKRDFEGVKEFIFTFWGCYLISCYIYIYRERCN 400
Query: 401 ATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLG 443
+Q+ V G+ L++V+G A +N+ +H + G
Sbjct: 401 VFGVVQIGVVTGIALAIVLGMSFSSIADGGGRNIISVHAVCTG 443
>UniRef100_Q82FG7 Putative DNA-damage-inducible protein F [Streptomyces avermitilis]
Length = 448
Score = 150 bits (378), Expect = 1e-34
Identities = 103/325 (31%), Positives = 167/325 (50%), Gaps = 7/325 (2%)
Query: 192 VLGLIQAATLIFAAKPLLGAMGLKYDSPMLVP-AVKYLRLRALGAPAVLLSLAMQGIFRG 250
+LG + A + A ++ G S P A+ YLR+ ALG PA+L+ LA G+ RG
Sbjct: 108 LLGAVVIAVFLPTAPAVVDLFGA---SETAAPYAITYLRISALGIPAMLVVLAATGVLRG 164
Query: 251 FKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVD 310
+DT TPLYV V+G+ N ++ +L++ LGI G+A V++QY MA L+++++
Sbjct: 165 LQDTKTPLYVAVAGFVANAVLNVVLVYGAGLGIAGSAWGTVIAQYGMAVAYLYVVVRGAR 224
Query: 311 LL-PPSMKDLQIFRFLKNGGL-LLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWM 368
L P D+ R G LL R +++ + ++ ++AARLG +AA Q L +W
Sbjct: 225 KLGAPLRPDIAGIRACAQAGAPLLVRTLSLRAVLMIATAVAARLGDADIAAHQIILSLWS 284
Query: 369 TSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAG 428
+ D +A+A QAI+ D A R ++ GV L L+V
Sbjct: 285 LLAFALDAIAIAGQAIIGRYLGAGDTEGARAACRRMVEWGIAAGVVLGLLVVVARPLFLP 344
Query: 429 VFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVM-VSIASVTS 487
+F+ + AV L VA +QPI + FV DGV GA D Y A+++++ +++ + +
Sbjct: 345 LFTGDSAVKDAALPALLLVALSQPICGVVFVLDGVLMGAGDGLYLAWAMLLTLAVFTPVA 404
Query: 488 LFFLYKSKGFIGIWIALTIYMSLRM 512
L G +W A+T+ M++RM
Sbjct: 405 LLVPMLGGGLTALWGAMTLMMTVRM 429
Score = 47.8 bits (112), Expect = 8e-04
Identities = 25/66 (37%), Positives = 41/66 (61%), Gaps = 1/66 (1%)
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSI 91
+EI+ +A P+ A+ A+P+ + D+A +GHLG +LA GV+ A+ A + +F L
Sbjct: 20 REIVALAVPAFGALVAEPLFVMADSAIVGHLGTAQLAGLGVASALLTTAVSVFVF-LAYA 78
Query: 92 TTSFVA 97
TT+ VA
Sbjct: 79 TTAAVA 84
>UniRef100_Q9X8U6 Putative membrane protein [Streptomyces coelicolor]
Length = 448
Score = 142 bits (358), Expect = 2e-32
Identities = 100/325 (30%), Positives = 167/325 (50%), Gaps = 7/325 (2%)
Query: 192 VLGLIQAATLIFAAKPLLGAMGLKYDSPMLVP-AVKYLRLRALGAPAVLLSLAMQGIFRG 250
+LG A + AA L+ G S P A YLR+ +LG PA+L+ LA G+ RG
Sbjct: 108 LLGAAVVAVFLPAAPSLVELFGA---SDTAAPYATTYLRISSLGIPAMLVVLASTGVLRG 164
Query: 251 FKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVD 310
++T TPLYV V+G+ N ++ LL++ LGI G+A V++Q MA + L+++++
Sbjct: 165 LQNTRTPLYVAVAGFIANAVLNVLLVYGAGLGIAGSAWGTVIAQCGMAAVYLWVVVRGAR 224
Query: 311 LLPPSMK-DLQIFRFLKNGGL-LLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWM 368
S++ DL R G+ LL R +++ + ++ ++AARLG +AA Q L +W
Sbjct: 225 QHGASLRPDLVGIRASAQAGMPLLVRTLSLRAILMIATAVAARLGDADIAAHQIVLSLWS 284
Query: 369 TSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAG 428
+ D +A+A QAI+ D A R ++ +GV L L+V
Sbjct: 285 LLAFALDAIAIAGQAIIGRYLGADDAQGAREACRRMVEWGVAVGVVLGLLVVLSRPVFLP 344
Query: 429 VFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVM-VSIASVTS 487
+F+ + V L VA QP+ + FV DGV GA D Y A+++++ +++ + +
Sbjct: 345 LFTGDSMVTDAALPALVIVAVAQPVCGVVFVLDGVLMGAGDGPYLAWAMLLTLAVFTPAA 404
Query: 488 LFFLYKSKGFIGIWIALTIYMSLRM 512
L G +W A+T+ M++R+
Sbjct: 405 LLVPTLGGGLTALWAAMTLMMAMRL 429
Score = 46.2 bits (108), Expect = 0.002
Identities = 23/66 (34%), Positives = 41/66 (61%), Gaps = 1/66 (1%)
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSI 91
+EI+ +A P+ A+ A+P+ + D+A +GHLG +LA G++ A+ A + +F L
Sbjct: 20 REIVALAVPAFGALVAEPLFVMADSAIVGHLGTAQLAGLGIASALLTTAVSVFVF-LAYA 78
Query: 92 TTSFVA 97
TT+ V+
Sbjct: 79 TTAAVS 84
>UniRef100_P71616 POSSIBLE DNA-DAMAGE-INDUCIBLE PROTEIN F DINF [Mycobacterium
tuberculosis]
Length = 439
Score = 130 bits (327), Expect = 9e-29
Identities = 92/331 (27%), Positives = 159/331 (47%), Gaps = 10/331 (3%)
Query: 193 LGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFK 252
LG + + A PL+ A+ + A+ +LR+ LG PA+L+SLA G RG +
Sbjct: 100 LGALVVVVVEATATPLVSAIAS--GDGITAAALPWLRIAILGTPAILVSLAGNGWLRGVQ 157
Query: 253 DTTTPLYVIVSGYALNVAMDPLLIFYF----KLGIRGAAISHVLSQYIMATLLLFILMKK 308
DT PL +V+G+ + + PLL++ + + G+ G+A+++++ Q++ A L L+ +
Sbjct: 158 DTVRPLRYVVAGFGSSALLCPLLVYGWLGLPRWGLTGSAVANLVGQWLAALLFAGALLAE 217
Query: 309 VDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWM 368
L P L + L+ R +A C +A++AAR G +AA Q LQ+W
Sbjct: 218 RVSLRPDRAVLGAQLMMARD--LIVRTLAFQVCYVSAAAVAARFGAAALAAHQVVLQLWG 275
Query: 369 TSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAG 428
+L+ D LA+A Q+++ + D A R S + L+ +G G G
Sbjct: 276 LLALVLDSLAIAAQSLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGSSVLPG 335
Query: 429 VFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSL 488
+F+ + +V+ I + F+ P + F DGV GA D A+ + V ++ L
Sbjct: 336 LFTDDRSVLAAIGVPWWFMVVQLPFAGIVFAVDGVLLGAGDAAFMRTATVASALVGFLPL 395
Query: 489 FFLYKSKGF--IGIWIALTIYMSLRMFAGVW 517
+L + G+ GIW L ++ LR+ W
Sbjct: 396 VWLSLAYGWGLAGIWSGLGTFIVLRLIFVGW 426
>UniRef100_Q7D6H3 DNA-damage-inducible protein F, putative [Mycobacterium
tuberculosis]
Length = 436
Score = 130 bits (327), Expect = 9e-29
Identities = 92/331 (27%), Positives = 159/331 (47%), Gaps = 10/331 (3%)
Query: 193 LGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFK 252
LG + + A PL+ A+ + A+ +LR+ LG PA+L+SLA G RG +
Sbjct: 97 LGALVVVVVEATATPLVSAIAS--GDGITAAALPWLRIAILGTPAILVSLAGNGWLRGVQ 154
Query: 253 DTTTPLYVIVSGYALNVAMDPLLIFYF----KLGIRGAAISHVLSQYIMATLLLFILMKK 308
DT PL +V+G+ + + PLL++ + + G+ G+A+++++ Q++ A L L+ +
Sbjct: 155 DTVRPLRYVVAGFGSSALLCPLLVYGWLGLPRWGLTGSAVANLVGQWLAALLFAGALLAE 214
Query: 309 VDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWM 368
L P L + L+ R +A C +A++AAR G +AA Q LQ+W
Sbjct: 215 RVSLRPDRAVLGAQLMMARD--LIVRTLAFQVCYVSAAAVAARFGAAALAAHQVVLQLWG 272
Query: 369 TSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAG 428
+L+ D LA+A Q+++ + D A R S + L+ +G G G
Sbjct: 273 LLALVLDSLAIAAQSLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGSSVLPG 332
Query: 429 VFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSL 488
+F+ + +V+ I + F+ P + F DGV GA D A+ + V ++ L
Sbjct: 333 LFTDDRSVLAAIGVPWWFMVVQLPFAGIVFAVDGVLLGAGDAAFMRTATVASALVGFLPL 392
Query: 489 FFLYKSKGF--IGIWIALTIYMSLRMFAGVW 517
+L + G+ GIW L ++ LR+ W
Sbjct: 393 VWLSLAYGWGLAGIWSGLGTFIVLRLIFVGW 423
>UniRef100_Q7TXR0 POSSIBLE DNA-DAMAGE-INDUCIBLE PROTEIN F DINF [Mycobacterium bovis]
Length = 439
Score = 129 bits (325), Expect = 2e-28
Identities = 92/331 (27%), Positives = 159/331 (47%), Gaps = 10/331 (3%)
Query: 193 LGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFK 252
LG + + A PL+ A+ + A+ +LR+ LG PA+L+SLA G RG +
Sbjct: 100 LGALVVVVVEATATPLVSAIAS--GDGITAAALPWLRIAILGTPAILVSLAGNGWLRGVQ 157
Query: 253 DTTTPLYVIVSGYALNVAMDPLLIFYF----KLGIRGAAISHVLSQYIMATLLLFILMKK 308
DT PL +V+G+ + + PLL++ + + G+ G+A+++++ Q++ A L L+ +
Sbjct: 158 DTVRPLRYVVAGFGSSALLCPLLVYGWLGLPRWGLTGSAVANLVGQWLAALLFAGALLAE 217
Query: 309 VDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWM 368
L P L + L+ R +A C +A++AAR G +AA Q LQ+W
Sbjct: 218 RVSLRPDRAVLGAQLMMARD--LIVRTLAFQVCYVSAAAVAARFGAAALAAHQVVLQLWG 275
Query: 369 TSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAG 428
+L+ D LA+A Q+++ + D A R S + L+ +G G G
Sbjct: 276 LLALVLDSLAIAAQSLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGSSVLPG 335
Query: 429 VFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSL 488
+F+ + +V+ I + F+ P + F DGV GA D A+ + V ++ L
Sbjct: 336 LFTDDRSVLAAIGVLWWFMVVQLPFAGIVFAVDGVLLGAGDAAFMRTATVASALVGFLPL 395
Query: 489 FFLYKSKGF--IGIWIALTIYMSLRMFAGVW 517
+L + G+ GIW L ++ LR+ W
Sbjct: 396 VWLSLAYGWGLAGIWSGLGTFIVLRLIFVGW 426
>UniRef100_Q6A5K2 Conserved membrane protein, MatE domain [Propionibacterium acnes]
Length = 448
Score = 128 bits (321), Expect = 5e-28
Identities = 92/341 (26%), Positives = 166/341 (47%), Gaps = 14/341 (4%)
Query: 187 LLFGTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQG 246
L ++GL+ A L+ + G G + A +YLR+ G PA+L ++A+ G
Sbjct: 100 LWLSVIIGLLVAIMLVAIPTTVAGWFGAS--GAVAEQAGRYLRITGFGVPAMLATMAITG 157
Query: 247 IFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILM 306
+ RGF+DT TPL V V ++ N+ ++ + GI+G+AI ++ Q MA L+++L
Sbjct: 158 VLRGFQDTRTPLVVTVVTFSANLVLNLWFVLGMGWGIQGSAIGTLVCQIAMAVALVWVLR 217
Query: 307 ---KKVDL-LPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQT 362
+ +DL L P + I L++G LL R +A+ + ++ +AAR G I MA++Q
Sbjct: 218 IRTRGLDLSLVPHLSG--IASSLRDGIPLLIRTLALRAALYVTTWVAARSGAITMASYQV 275
Query: 363 CLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGG 422
+ +W + D L +A QA+ S D + + + V GV + +V+
Sbjct: 276 TMTMWNLLLMAMDALGIAGQALTGASLGAGDVRRTRSLTATMTRWGLVAGVVIGIVLAAF 335
Query: 423 LYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSI 482
+++ + AV + GL VAA Q + AFV DGV GA D + + + V++ +
Sbjct: 336 HQVVPALYTDDPAVHRAVAAGLLVVAAQQIVAGPAFVLDGVLIGAGDGRWLSGAQVVMLL 395
Query: 483 ASVTSLFFLY------KSKGFIGIWIALTIYMSLRMFAGVW 517
+ + ++ + + +W+A + +M +R W
Sbjct: 396 VYLPMAWAVHLLAPSDPAAAVVWLWVAFSGFMIVRCAILAW 436
Score = 45.4 bits (106), Expect = 0.004
Identities = 21/63 (33%), Positives = 38/63 (59%)
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSI 91
++IL +A P+ L++ A+P+ + D+A +GHLG ELA GV+ A + + +F +
Sbjct: 17 RQILNLAVPAFLSLIAEPLFLVADSAVVGHLGTAELAGLGVASAALTTFTGLFVFLAYAT 76
Query: 92 TTS 94
T +
Sbjct: 77 TAT 79
>UniRef100_Q6AHG4 DNA-damage-inducible protein F [Leifsonia xyli]
Length = 461
Score = 120 bits (302), Expect = 7e-26
Identities = 89/290 (30%), Positives = 137/290 (46%), Gaps = 11/290 (3%)
Query: 190 GTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFR 249
G LG + AA + A P+L ++ + + A YL + G PA+L A G+ R
Sbjct: 82 GLGLGAVLAAGRL--ASPVLVSL-FAASAAVSEQAEVYLSISMAGLPAMLFVFAATGLLR 138
Query: 250 GFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKV 309
G +DT TPL V G+A N+A++ + I LGI G+A+ V++Q+ M + ++ +
Sbjct: 139 GLQDTRTPLAVAGGGFAANIALNAVFIGVLGLGIAGSALGTVVAQWAMVAVYAVVVARHA 198
Query: 310 -----DLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCL 364
LLP + + R GG L R ++ + L+ + A RLGP +AAFQ +
Sbjct: 199 RRAGAGLLP---RHTGLGRTAVAGGWLFLRTASLRGAMLLAIAAATRLGPDDLAAFQVAM 255
Query: 365 QVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLY 424
V+ T + D LA+A QA++ +V R +Q G L V
Sbjct: 256 TVFATLAFALDTLAIAAQALVGKGLGAGKLPEVRAVLRRCVQWGVGSGAVLGAVTVALSP 315
Query: 425 FGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSA 474
AG+F+++ AV L+ L V + P+ FV DGV GA D Y A
Sbjct: 316 VAAGLFTRDAAVTALLPAALAIVGLSAPLGGYVFVLDGVLIGAGDTRYLA 365
Score = 38.1 bits (87), Expect = 0.62
Identities = 19/53 (35%), Positives = 29/53 (53%)
Query: 40 PSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSIT 92
P+ A+ A+P+ L D+A +GHLG LA G++ AV + +F S T
Sbjct: 2 PALGALVAEPLFLLADSAVVGHLGVAPLAGLGIAAAVLQTIVGLMVFLAYSTT 54
>UniRef100_Q5YSD0 Putative DNA-damage-inducible protein F [Nocardia farcinica]
Length = 449
Score = 120 bits (302), Expect = 7e-26
Identities = 90/331 (27%), Positives = 157/331 (47%), Gaps = 15/331 (4%)
Query: 190 GTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFR 249
G +L ++QAA A P+ A+ D + A+ +LR+ G P +L+++A G R
Sbjct: 102 GLILVVMQAA-----AGPVTAAISGGGD--IAAEALPWLRIALFGVPLILIAMAGNGWLR 154
Query: 250 GFKDTTTPLYVIVSGYALNVAMDPLLIFYF----KLGIRGAAISHVLSQYIMATLLLFIL 305
G +DT PL +V G L+ + P+L+ +L + G+A+++V Q + A L + +
Sbjct: 155 GVQDTRRPLVFVVCGLGLSAVLCPVLVHGLLGAPRLELAGSAVANVAGQTVTAALFVTAV 214
Query: 306 MKKVDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQ 365
+++ L P ++ L G L+ R ++ C +A++AAR G +AA Q LQ
Sbjct: 215 VRERVPLTPHWSVMRAQLVL--GRDLILRSLSFQACFVSAAAVAARFGAASVAAHQLVLQ 272
Query: 366 VWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYF 425
+W +L D LA+A Q ++ + D A R + S + + L+ V G
Sbjct: 273 LWSFLALTLDALAIAAQTLVGAALGGGDATGARRLAGRITRWSELFALALAAVFAAGYTV 332
Query: 426 GAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASV 485
+F+ + AV+ ++ F A P+ + F DGV GA D A+ + + ++
Sbjct: 333 IPALFTTDAAVLERTQVAWWFFVALIPVAGVVFALDGVLLGAGDAAFLRTATLAAALLGF 392
Query: 486 TSLFFLYKS--KGFIGIWIALTIYMSLRMFA 514
+L + G GIW L +M LR+ A
Sbjct: 393 LPAIWLSLAFDWGIAGIWSGLAAFMVLRLAA 423
Score = 37.0 bits (84), Expect = 1.4
Identities = 22/73 (30%), Positives = 31/73 (42%)
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSI 91
+ ILG+A P+ + A+P+ L D A +G LG + LA V + Q S F
Sbjct: 11 RRILGLALPTLGVLVAEPLYLLFDLAVVGRLGALALAGLAVGGLILAQVSTQLTFLSYGT 70
Query: 92 TTSFVAEEDTMDR 104
T DR
Sbjct: 71 TARSARRHGAGDR 83
>UniRef100_Q6I567 Hypothetical protein OSJNBb0014K18.14 [Oryza sativa]
Length = 117
Score = 118 bits (295), Expect = 5e-25
Identities = 58/106 (54%), Positives = 76/106 (70%)
Query: 357 MAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLS 416
MAAFQ C QVW+ +SLLAD L +A QA+ A FA+KD+ K+ R LQ++ VLGVGL+
Sbjct: 1 MAAFQICAQVWLATSLLADDLTIAGQALFASVFAKKDHYKMAVTTARVLQLAVVLGVGLT 60
Query: 417 LVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDG 462
+ G++FG+GVF+ + AVI I G+PFVA Q IN+LAFVFDG
Sbjct: 61 AFLATGMWFGSGVFTSDTAVISTIHKGVPFVAGMQTINTLAFVFDG 106
>UniRef100_Q6M456 PUTATIVE DNA-DAMAGE-INDUCIBLE MEMBRANE PROTEIN [Corynebacterium
glutamicum]
Length = 437
Score = 114 bits (286), Expect = 5e-24
Identities = 119/493 (24%), Positives = 204/493 (41%), Gaps = 81/493 (16%)
Query: 28 DSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFP 87
D AK+I G+AFP+ +AA P+ L+DTA +G LG G +A A+ I
Sbjct: 14 DVSAKQIFGLAFPALGVLAAMPLYLLLDTAVVGTLG-------GFELAALGAATTIQ--- 63
Query: 88 LVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLK 147
+TT L + G T + S +
Sbjct: 64 -AQVTTQ-------------------------------------LTFLSYGTTARSSRIF 85
Query: 148 NGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKP 207
D I++ + + AL G LG++ TL+ P
Sbjct: 86 GMGDRRGAIAEG--------------------VQATWVALFVG--LGIL---TLMLIGAP 120
Query: 208 LLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYAL 267
A+ L D + A +LR+ A P +L+ +A G RG ++T PLY ++G
Sbjct: 121 TF-ALWLSGDEALAQEAGHWLRVAAFAVPLILMIMAGNGWLRGIQNTKLPLYFTLAGVIP 179
Query: 268 NVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVD-LLPPSMKDLQIFRFLK 326
+ P IF K G+ G+A ++++++ I A+L L L+K + PS ++ L
Sbjct: 180 GAILIP--IFVAKFGLVGSAWANLIAEAITASLFLGALIKHHEGSWKPSWTVMK--NQLV 235
Query: 327 NGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILA 386
G L+ R ++ +A++AAR G +AA Q LQ+W +L+ D LA+A Q +
Sbjct: 236 LGRDLIMRSMSFQVAFLSAAAVAARFGTASLAAHQVLLQLWNFITLVLDSLAIAAQTLTG 295
Query: 387 CSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPF 446
+ + ++ S + GL LV + +F+++ V+ I
Sbjct: 296 AALGAGTAKVARRVGNQVIKYSLIFAGGLGLVFVVLHSWIPRIFTQDADVLDAIASPWWI 355
Query: 447 VAATQPINSLAFVFDGVNYGASDFAY-SAYSLVMVSIASVTSLFFLYK-SKGFIGIWIAL 504
+ A + + F DGV GA+D + S++ V + + ++ Y G G+W L
Sbjct: 356 MVAMIILGGIVFAIDGVLLGAADAVFLRNASILAVVVGFLPGVWISYALDAGLTGVWCGL 415
Query: 505 TIYMSLRMFAGVW 517
++ +R+FA +W
Sbjct: 416 LAFILIRLFAVIW 428
>UniRef100_Q8G740 Hypothetical protein [Bifidobacterium longum]
Length = 531
Score = 110 bits (275), Expect = 1e-22
Identities = 86/310 (27%), Positives = 138/310 (43%), Gaps = 12/310 (3%)
Query: 186 ALLFGTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQ 245
AL GTVLGL L AA+PL A+G + + +L AV Y R LGAP +L+ A
Sbjct: 185 ALSIGTVLGL----GLFAAAEPLCRALGGQGE--VLEQAVTYTRAIVLGAPGMLMVYAAN 238
Query: 246 GIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLF-- 303
GIFRG + L V G +N +D L + GI G+ ++ +++Q+ M L+
Sbjct: 239 GIFRGLQKVRITLIAAVGGAVVNTVLDVLFVIVLNWGIAGSGVATLVAQWFMGLFLVIPA 298
Query: 304 ILMKKVDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTC 363
IL + D + I +G L R +A+ + + + AAR+G +A FQ
Sbjct: 299 ILWSRADGASLRPRLAGIAAAGGDGLPLFIRTLAIRAAMVTTVACAARMGTAVLAGFQAV 358
Query: 364 LQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLG--VGLSLVVGG 421
W + + D + +A Q ++A + + T + V G +G + V G
Sbjct: 359 NSSWNFAMNMLDSVGIAGQTLVATALGAGSVQQARRLTRATGRAGLVTGAVIGTAFAVVG 418
Query: 422 GLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVS 481
F FS + LI +G+ + P+ DG+ GA D+ Y A + + +
Sbjct: 419 --LFAGHFFSPTPHIQTLIAVGMVTMGIFFPLQGWMMAIDGILIGARDYRYLAVTCTLTA 476
Query: 482 IASVTSLFFL 491
+ VT + L
Sbjct: 477 VVYVTLILIL 486
Score = 40.0 bits (92), Expect = 0.16
Identities = 24/69 (34%), Positives = 36/69 (51%), Gaps = 1/69 (1%)
Query: 29 SLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPL 88
S + I+ +A P+ + A+P LIDTA +GH+G LA + + A + IF L
Sbjct: 96 STNRRIMALALPTFGQLIAEPTFILIDTAIVGHIGDAALAGLSIGSTIILTAVGLCIF-L 154
Query: 89 VSITTSFVA 97
TT+ VA
Sbjct: 155 AYSTTAQVA 163
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.327 0.139 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 720,341,786
Number of Sequences: 2790947
Number of extensions: 27738388
Number of successful extensions: 124490
Number of sequences better than 10.0: 1102
Number of HSP's better than 10.0 without gapping: 428
Number of HSP's successfully gapped in prelim test: 675
Number of HSP's that attempted gapping in prelim test: 122882
Number of HSP's gapped (non-prelim): 1903
length of query: 517
length of database: 848,049,833
effective HSP length: 132
effective length of query: 385
effective length of database: 479,644,829
effective search space: 184663259165
effective search space used: 184663259165
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC122162.1


