

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.9 + phase: 0
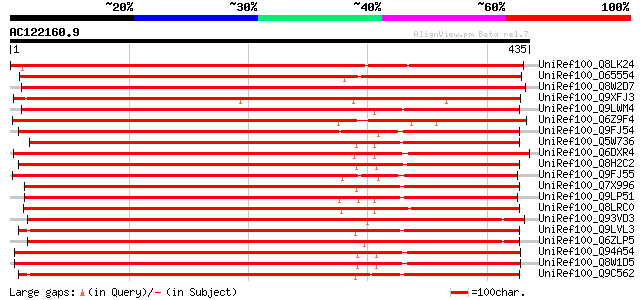
(435 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LK24 SOS2-like protein kinase [Glycine max] 710 0.0
UniRef100_O65554 Hypothetical protein F6I18.130 [Arabidopsis tha... 648 0.0
UniRef100_Q8W2D7 CBL-interacting protein kinase [Brassica napus] 646 0.0
UniRef100_Q9XFJ3 Putative serine/threonine protein kinase [Mesem... 555 e-157
UniRef100_Q9LWM4 Putative CBL-interacting protein kinase 2 [Oryz... 538 e-151
UniRef100_Q6Z9F4 Putative CBL-interacting protein kinase [Oryza ... 521 e-146
UniRef100_Q9FJ54 Serine/threonine protein kinase [Arabidopsis th... 517 e-145
UniRef100_Q5W736 Putative serine/threonine protein kinase [Oryza... 514 e-144
UniRef100_Q6DXR4 Putative serine-threonine kinase [Gossypium hir... 512 e-144
UniRef100_Q8H2C2 Serine/threonine kinase [Persea americana] 508 e-142
UniRef100_Q9FJ55 Serine/threonine protein kinase [Arabidopsis th... 503 e-141
UniRef100_Q7X996 Putative Serine/threonine Kinase [Oryza sativa] 501 e-140
UniRef100_Q9LP51 Hypothetical protein F28N24.9 [Arabidopsis thal... 486 e-136
UniRef100_Q8LRC0 Putative serine/threonine protein kinase [Oryza... 480 e-134
UniRef100_Q93VD3 At1g30270/F12P21_6 [Arabidopsis thaliana] 474 e-132
UniRef100_Q9LVL3 Serine/threonine protein kinase [Arabidopsis th... 474 e-132
UniRef100_Q6ZLP5 Putative CBL-interacting protein kinase 23 [Ory... 473 e-132
UniRef100_Q94A54 AT5g25110/T11H3_120 [Arabidopsis thaliana] 473 e-132
UniRef100_Q8W1D5 CBL-interacting protein kinase CIPK25 [Arabidop... 473 e-132
UniRef100_Q9C562 SOS2-like protein kinase PKS2 [Arabidopsis thal... 473 e-132
>UniRef100_Q8LK24 SOS2-like protein kinase [Glycine max]
Length = 438
Score = 710 bits (1832), Expect = 0.0
Identities = 355/433 (81%), Positives = 394/433 (90%), Gaps = 6/433 (1%)
Query: 1 MCEKENPNS---HSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKV 57
M EK N ++ LL GKYELGR+LGHGTFAKVYHA++L TGK+VAMKVVGKEKV+KV
Sbjct: 1 MGEKSNVGGDAINTTLLHGKYELGRLLGHGTFAKVYHARHLKTGKSVAMKVVGKEKVVKV 60
Query: 58 GMIEQIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVA 117
GM+EQIKREIS M MVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKI +GRL+E++A
Sbjct: 61 GMMEQIKREISAMNMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIARGRLREEMA 120
Query: 118 RVYFQQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTT 177
R+YFQQLISAVDFCHSRGVYHRDLKPENLLLD+DGNLKV+DFGL TFSEHLR DGLLHTT
Sbjct: 121 RLYFQQLISAVDFCHSRGVYHRDLKPENLLLDDDGNLKVTDFGLSTFSEHLRHDGLLHTT 180
Query: 178 CGTPAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKC 237
CGTPAYV+PEVI K+GYDGAKADIWSCGVILYVLLAGFLPFQDDNLVA+YKKIYRGDFKC
Sbjct: 181 CGTPAYVAPEVIGKRGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVALYKKIYRGDFKC 240
Query: 238 PPWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEF 297
PPWFS EARRLITKLLDPNPNTRI+ISKIMDSSWFKKPVPK++ +K+E+ + E+K+ +
Sbjct: 241 PPWFSSEARRLITKLLDPNPNTRITISKIMDSSWFKKPVPKNLMGKKREELDLEEKIKQH 300
Query: 298 MECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKA 357
++ STTMNAFHIISLSEGFDLSPLFEEKKREE+ E+RFAT S VIS+LE +AKA
Sbjct: 301 E--QEVSTTMNAFHIISLSEGFDLSPLFEEKKREEK-ELRFATTRPASSVISRLEDLAKA 357
Query: 358 VKFDVKSSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKE 417
VKFDVK S+TKVRLQGQE+GRKGKLAIAAD+YAVTPSFLVVEVKKD GDTLEYNQFCSKE
Sbjct: 358 VKFDVKKSETKVRLQGQEKGRKGKLAIAADLYAVTPSFLVVEVKKDNGDTLEYNQFCSKE 417
Query: 418 LRPALKDIFWTSA 430
LRPALKDI W ++
Sbjct: 418 LRPALKDIVWRTS 430
>UniRef100_O65554 Hypothetical protein F6I18.130 [Arabidopsis thaliana]
Length = 441
Score = 648 bits (1671), Expect = 0.0
Identities = 318/426 (74%), Positives = 369/426 (85%), Gaps = 7/426 (1%)
Query: 9 SHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREIS 68
S + LL G+YELGR+LGHGTFAKVYHA+N+ TGK+VAMKVVGKEKV+KVGM++QIKREIS
Sbjct: 15 SSTGLLHGRYELGRLLGHGTFAKVYHARNIQTGKSVAMKVVGKEKVVKVGMVDQIKREIS 74
Query: 69 VMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAV 128
VM+MVKHPNIV+LHEVMASKSKIY AMELVRGGELF K+ KGRL+EDVARVYFQQLISAV
Sbjct: 75 VMRMVKHPNIVELHEVMASKSKIYFAMELVRGGELFAKVAKGRLREDVARVYFQQLISAV 134
Query: 129 DFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEV 188
DFCHSRGVYHRDLKPENLLLDE+GNLKV+DFGL F+EHL+QDGLLHTTCGTPAYV+PEV
Sbjct: 135 DFCHSRGVYHRDLKPENLLLDEEGNLKVTDFGLSAFTEHLKQDGLLHTTCGTPAYVAPEV 194
Query: 189 IAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRL 248
I KKGYDGAKAD+WSCGVIL+VLLAG+LPFQDDNLV MY+KIYRGDFKCP W S +ARRL
Sbjct: 195 ILKKGYDGAKADLWSCGVILFVLLAGYLPFQDDNLVNMYRKIYRGDFKCPGWLSSDARRL 254
Query: 249 ITKLLDPNPNTRISISKIMDSSWFKKPVPKS----IAMRKKEKEEEEDKVFEFMECEKSS 304
+TKLLDPNPNTRI+I K+MDS WFKK +S +A EE+ D F + ++ +
Sbjct: 255 VTKLLDPNPNTRITIEKVMDSPWFKKQATRSRNEPVAATITTTEEDVD--FLVHKSKEET 312
Query: 305 TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAK-AVKFDVK 363
T+NAFHII+LSEGFDLSPLFEEKK+EE+ EMRFAT S VIS LE+ A+ KFDV+
Sbjct: 313 ETLNAFHIIALSEGFDLSPLFEEKKKEEKREMRFATSRPASSVISSLEEAARVGNKFDVR 372
Query: 364 SSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKELRPALK 423
S+++VR++G++ GRKGKLA+ A+I+AV PSF+VVEVKKD GDTLEYN FCS LRPALK
Sbjct: 373 KSESRVRIEGKQNGRKGKLAVEAEIFAVAPSFVVVEVKKDHGDTLEYNNFCSTALRPALK 432
Query: 424 DIFWTS 429
DIFWTS
Sbjct: 433 DIFWTS 438
>UniRef100_Q8W2D7 CBL-interacting protein kinase [Brassica napus]
Length = 441
Score = 646 bits (1666), Expect = 0.0
Identities = 317/424 (74%), Positives = 365/424 (85%), Gaps = 2/424 (0%)
Query: 11 SALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVM 70
+ LL G+YELGR+LGHGTFAKVYHA+N+ TGK+VAMKVVGKEKV+KVGM++QIKREISVM
Sbjct: 17 TGLLHGRYELGRLLGHGTFAKVYHARNVTTGKSVAMKVVGKEKVVKVGMVDQIKREISVM 76
Query: 71 KMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAVDF 130
+MVKHPNIV+LHEVMASK+KIY AMELVRGGELF K+ KGRL+EDVARVYFQQLISAVDF
Sbjct: 77 RMVKHPNIVELHEVMASKTKIYFAMELVRGGELFAKVAKGRLREDVARVYFQQLISAVDF 136
Query: 131 CHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIA 190
CHSRGVYHRDLKPENLLLDE+GNLKV+DFGL F+EHL+QDGLLHTTCGTPAYV+PEVI
Sbjct: 137 CHSRGVYHRDLKPENLLLDEEGNLKVTDFGLSAFTEHLKQDGLLHTTCGTPAYVAPEVIL 196
Query: 191 KKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLIT 250
KKGYDGAKAD+WSCGVIL+VLLAG+LPFQDDNLV MY+KIYRGDFKCP W S +ARRL+T
Sbjct: 197 KKGYDGAKADLWSCGVILFVLLAGYLPFQDDNLVNMYRKIYRGDFKCPGWLSSDARRLVT 256
Query: 251 KLLDPNPNTRISISKIMDSSWFKKPVPKS-IAMRKKEKEEEEDKVFEFMECEKSSTTMNA 309
KLLDPNPNTRI+I K+MDS WFKK +S E EED + ++ + T+NA
Sbjct: 257 KLLDPNPNTRITIDKVMDSHWFKKSSTRSRNEPVAATPEAEEDVDVSVHKSKEETETLNA 316
Query: 310 FHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAK-AVKFDVKSSDTK 368
FHII+LSEGFDLSPLFEEKK+EE+ EMRFAT S VIS LE+ AK KFDV+ S+ +
Sbjct: 317 FHIIALSEGFDLSPLFEEKKKEEKMEMRFATSRPASSVISSLEEAAKVGNKFDVRKSECR 376
Query: 369 VRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKELRPALKDIFWT 428
VR++G++ GRKGKLA+ A+I+AV PSF+VVEVKKD GDTLEYN FCS LRPALKDIFWT
Sbjct: 377 VRIEGKQNGRKGKLAVEAEIFAVAPSFVVVEVKKDHGDTLEYNNFCSTALRPALKDIFWT 436
Query: 429 SADP 432
S P
Sbjct: 437 STTP 440
>UniRef100_Q9XFJ3 Putative serine/threonine protein kinase [Mesembryanthemum
crystallinum]
Length = 462
Score = 555 bits (1430), Expect = e-157
Identities = 290/440 (65%), Positives = 343/440 (77%), Gaps = 16/440 (3%)
Query: 4 KENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQI 63
+E H AL GKYELGRVLG GTFA+VYHA+NL TG ++A+KVV K+KVIKVGM+EQ+
Sbjct: 20 RETEMKHPAL-QGKYELGRVLGQGTFARVYHARNLVTGNSLAIKVVAKDKVIKVGMMEQM 78
Query: 64 KREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQ 122
KREISVMK VKHPNIVQLHEVMASKSKIY+AM+LVRGGELF+K+ K G+L EDVAR YF+
Sbjct: 79 KREISVMKRVKHPNIVQLHEVMASKSKIYLAMDLVRGGELFSKVSKGGKLDEDVARSYFK 138
Query: 123 QLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGL-CTFSEHLRQDGLLHTTCGTP 181
QLISAVDFCHSRGVYHRDLKPENLLLDE GNLK++DFGL +F LL T+CGTP
Sbjct: 139 QLISAVDFCHSRGVYHRDLKPENLLLDEHGNLKITDFGLSASFENEDHHQTLLRTSCGTP 198
Query: 182 AYVSPEVIAKK---GYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCP 238
YV+PEVI++K GY GAK+DIWSCG+IL+VLLAGFLPF DDN++ MY+K+++GDF CP
Sbjct: 199 HYVAPEVISRKGQQGYHGAKSDIWSCGIILFVLLAGFLPFHDDNIIRMYRKVFKGDFTCP 258
Query: 239 PWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKE-------KEEEE 291
W S +AR LIT+LLD NP TRI++S+IM S W+ A+ E
Sbjct: 259 SWLSSQARDLITRLLDINPATRITVSEIMQSDWYCMDTSSMTAVTTDEFDLDHNNNNNNN 318
Query: 292 DKVFEFMECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKL 351
+K +NAFHIISLS+GFDLSPLFEEK REER++MRFAT + S V+S+L
Sbjct: 319 SNYNNIHHHKKEPEMLNAFHIISLSQGFDLSPLFEEKHREERQDMRFATTKSASTVVSRL 378
Query: 352 EQV-AKAVKFDVKS--SDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTL 408
E+V AKA KF VK D+ VRLQGQERGRKGKLAIAA+I +V+PS LVVEVKKD GDTL
Sbjct: 379 EEVAAKAGKFIVKKFCGDSVVRLQGQERGRKGKLAIAAEIMSVSPSLLVVEVKKDNGDTL 438
Query: 409 EYNQFCSKELRPALKDIFWT 428
EYNQFC+K+LRPALKDI WT
Sbjct: 439 EYNQFCTKQLRPALKDIVWT 458
>UniRef100_Q9LWM4 Putative CBL-interacting protein kinase 2 [Oryza sativa]
Length = 461
Score = 538 bits (1385), Expect = e-151
Identities = 267/431 (61%), Positives = 333/431 (76%), Gaps = 15/431 (3%)
Query: 11 SALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVM 70
+++L +YELGR+LG GTFAKVYHA+NL + ++VA+KV+ KEKV++VGMI+QIKREIS+M
Sbjct: 5 ASILMNRYELGRMLGQGTFAKVYHARNLASNQSVAIKVIDKEKVLRVGMIDQIKREISIM 64
Query: 71 KMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAVDF 130
++V+HPNIVQLHEVMASKSKIY AME VRGGELF+++ +GRLKED AR YFQQLI AVDF
Sbjct: 65 RLVRHPNIVQLHEVMASKSKIYFAMEYVRGGELFSRVARGRLKEDAARKYFQQLIGAVDF 124
Query: 131 CHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIA 190
CHSRGVYHRDLKPENLL+DE+GNLKVSDFGL F E +QDGLLHTTCGTPAYV+PE+I
Sbjct: 125 CHSRGVYHRDLKPENLLVDENGNLKVSDFGLSAFKECQKQDGLLHTTCGTPAYVAPEIIN 184
Query: 191 KKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLIT 250
K+GYDGAKADIWSCGVIL+VLLAG+LPF D NL+ MY+KI +GD K P WF+ + RRL++
Sbjct: 185 KRGYDGAKADIWSCGVILFVLLAGYLPFHDSNLMEMYRKISKGDVKFPQWFTTDVRRLLS 244
Query: 251 KLLDPNPNTRISISKIMDSSWFKKPV-PKSIAMRKKEKEEEEDKVFEFMECEKSS----- 304
+LLDPNPN RI++ K+++ WFKK P + + E +D F K +
Sbjct: 245 RLLDPNPNIRITVEKLVEHPWFKKGYKPAVMLSQPNESNNLKDVHTAFSADHKDNEGKAK 304
Query: 305 --------TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAK 356
++NAF IISLS+GFDLS LFE K E++ + RF T S ++SKLEQ+A+
Sbjct: 305 EPASSLKPVSLNAFDIISLSKGFDLSGLFENDK-EQKADSRFMTQKPASAIVSKLEQIAE 363
Query: 357 AVKFDVKSSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSK 416
F VK D V+LQG + GRKG+LAI A+I+ VTPSF VVEVKK GDTLEY +FC+K
Sbjct: 364 TESFKVKKQDGLVKLQGSKEGRKGQLAIDAEIFEVTPSFFVVEVKKSAGDTLEYEKFCNK 423
Query: 417 ELRPALKDIFW 427
LRP+L+DI W
Sbjct: 424 GLRPSLRDICW 434
>UniRef100_Q6Z9F4 Putative CBL-interacting protein kinase [Oryza sativa]
Length = 451
Score = 521 bits (1342), Expect = e-146
Identities = 270/447 (60%), Positives = 331/447 (73%), Gaps = 25/447 (5%)
Query: 3 EKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQ 62
E E +L G+YE+GRVLGHG F +V+ A++L TG++VA+KVV KEKV++ GM+EQ
Sbjct: 8 EGEGKKGGGTVLQGRYEMGRVLGHGNFGRVHVARDLRTGRSVAVKVVAKEKVVRAGMMEQ 67
Query: 63 IKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYF 121
IKREI+VMK V HPNIV+LHEVMA++SKIY+A+ELVRGGELF +IV+ GR++ED AR YF
Sbjct: 68 IKREIAVMKRVSHPNIVELHEVMATRSKIYLALELVRGGELFGRIVRLGRVREDAARHYF 127
Query: 122 QQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTP 181
+QL+SAVDFCHSRGVYHRDLKPENLLLDE GNLKV DFGL ++H R DGLLHT CGTP
Sbjct: 128 RQLVSAVDFCHSRGVYHRDLKPENLLLDEAGNLKVVDFGLSALADHARADGLLHTLCGTP 187
Query: 182 AYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWF 241
Y +PEV+ KGYDGAKAD+WSCGVILYVLLAG LPF DDN+V +Y+K RGD++CP W
Sbjct: 188 GYAAPEVLRDKGYDGAKADLWSCGVILYVLLAGSLPFPDDNIVTLYRKAQRGDYRCPAWL 247
Query: 242 SLEARRLITKLLDPNPNTRISISKIMDSSWFKK---------PVPKSIAMRKKEKEEEED 292
S +ARRLI +LLDPNP TRIS+++++++ WFKK +P + A KEE E
Sbjct: 248 STDARRLIPRLLDPNPTTRISVAQLVETPWFKKTSISRPVSIELPPAFADPAPAKEEAE- 306
Query: 293 KVFEFMECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREEREE--MRFATGGTPSRVISK 350
+ T+NAFH+ISLSEGFDLSPLFE + R + M FAT S VIS+
Sbjct: 307 --------KDEPETLNAFHLISLSEGFDLSPLFEGDSAKGRRDGGMLFATREPASGVISR 358
Query: 351 LEQVAK---AVKFDVKSSDTKVRLQGQER-GRKGKLAIAADIYAVTPSFLVVEVKKDKGD 406
LE VA KS VRL+G ER G KG+LA+AADI++V PS LVV+VKKD GD
Sbjct: 359 LEGVAARGGGRMRVTKSGARGVRLEGAERGGAKGRLAVAADIFSVAPSVLVVDVKKDGGD 418
Query: 407 TLEYNQFCSKELRPALKDIFWTSADPP 433
TLEY FCS+ELRPAL+DI W +A P
Sbjct: 419 TLEYRSFCSEELRPALQDIVWGAAADP 445
>UniRef100_Q9FJ54 Serine/threonine protein kinase [Arabidopsis thaliana]
Length = 439
Score = 517 bits (1332), Expect = e-145
Identities = 255/424 (60%), Positives = 329/424 (77%), Gaps = 8/424 (1%)
Query: 8 NSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREI 67
+ + +L KYELGR+LG GTFAKVYHA+N+ TG++VA+KV+ K+KV KVG+I+QIKREI
Sbjct: 2 DKNGIVLMRKYELGRLLGQGTFAKVYHARNIKTGESVAIKVIDKQKVAKVGLIDQIKREI 61
Query: 68 SVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISA 127
SVM++V+HP++V LHEVMASK+KIY AME V+GGELF+K+ KG+LKE++AR YFQQLI A
Sbjct: 62 SVMRLVRHPHVVFLHEVMASKTKIYFAMEYVKGGELFDKVSKGKLKENIARKYFQQLIGA 121
Query: 128 VDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPE 187
+D+CHSRGVYHRDLKPENLLLDE+G+LK+SDFGL E +QDGLLHTTCGTPAYV+PE
Sbjct: 122 IDYCHSRGVYHRDLKPENLLLDENGDLKISDFGLSALRESKQQDGLLHTTCGTPAYVAPE 181
Query: 188 VIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARR 247
VI KKGYDGAKAD+WSCGV+LYVLLAGFLPF + NLV MY+KI +G+FKCP WF E ++
Sbjct: 182 VIGKKGYDGAKADVWSCGVVLYVLLAGFLPFHEQNLVEMYRKITKGEFKCPNWFPPEVKK 241
Query: 248 LITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFMECEKSSTTM 307
L++++LDPNPN+RI I KIM++SWF+K K I K + + D + + S M
Sbjct: 242 LLSRILDPNPNSRIKIEKIMENSWFQKGF-KKIETPKSPESHQIDSLISDVHAAFSVKPM 300
Query: 308 --NAFHII-SLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVA-KAVKFDVK 363
NAF +I SLS+GFDLS LFE +EER E +F T ++SK E++A + +F++
Sbjct: 301 SYNAFDLISSLSQGFDLSGLFE---KEERSESKFTTKKDAKEIVSKFEEIATSSERFNLT 357
Query: 364 SSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKELRPALK 423
SD V+++ + GRKG LAI +I+ VT SF +VE KK GDT+EY QFC +ELRP+LK
Sbjct: 358 KSDVGVKMEDKREGRKGHLAIDVEIFEVTNSFHMVEFKKSGGDTMEYKQFCDRELRPSLK 417
Query: 424 DIFW 427
DI W
Sbjct: 418 DIVW 421
>UniRef100_Q5W736 Putative serine/threonine protein kinase [Oryza sativa]
Length = 457
Score = 514 bits (1325), Expect = e-144
Identities = 251/429 (58%), Positives = 327/429 (75%), Gaps = 20/429 (4%)
Query: 17 KYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKHP 76
+YE+G++LG GTFAKVYH +N+ T ++VA+KV+ K+K+ KVG+++QIKREISVMK+V+HP
Sbjct: 14 RYEIGKLLGQGTFAKVYHGRNIVTSQSVAIKVIDKDKIFKVGLMDQIKREISVMKLVRHP 73
Query: 77 NIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAVDFCHSRGV 136
NIVQL+EVMA+KSKIY +E V+GGELFNK+ KGRLKED AR YFQQL+SAVDFCHSRGV
Sbjct: 74 NIVQLYEVMATKSKIYFVLEYVKGGELFNKVAKGRLKEDAARKYFQQLVSAVDFCHSRGV 133
Query: 137 YHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGYDG 196
YHRDLKPENLL+DE+GNLK++DFGL +E RQDGLLHTTCGTPAYV+PEVI++KGYDG
Sbjct: 134 YHRDLKPENLLVDENGNLKITDFGLSALAESRRQDGLLHTTCGTPAYVAPEVISRKGYDG 193
Query: 197 AKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDPN 256
K D WSCGVIL+VL+AG+LPFQD NL+ MY+KI + +FKCP WFS + R+L++++LDPN
Sbjct: 194 VKVDTWSCGVILFVLMAGYLPFQDSNLMEMYRKIGKAEFKCPAWFSSDVRKLVSRILDPN 253
Query: 257 PNTRISISKIMDSSWFKKPVPKSIAMRKKEKEE------EEDKVFEFMECEKSS------ 304
P +R+ I+KIM++ WFKK + + ++ E E + + VF M S
Sbjct: 254 PRSRMPITKIMETYWFKKGLDSKLILKNVETNEPVTALADVNVVFSSMGSSSSKKTEEKQ 313
Query: 305 -----TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKAVK 359
T +NAF IISLSEGFDLS LFEE ++++E RF + + S +ISKLE VA K
Sbjct: 314 DAGKLTNLNAFDIISLSEGFDLSGLFEE--TDKKKEARFTSSQSASAIISKLEDVASCSK 371
Query: 360 FDVKSSDTKV-RLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKEL 418
VK + V +++G GRKG LAI A+I+ VTPSF +VE+KK+ GDTLEY +++
Sbjct: 372 LTVKKKEGGVLKMEGASEGRKGVLAIDAEIFEVTPSFHLVEIKKNNGDTLEYQHLWKEDM 431
Query: 419 RPALKDIFW 427
+PALKDI W
Sbjct: 432 KPALKDIVW 440
>UniRef100_Q6DXR4 Putative serine-threonine kinase [Gossypium hirsutum]
Length = 478
Score = 512 bits (1319), Expect = e-144
Identities = 258/454 (56%), Positives = 331/454 (72%), Gaps = 26/454 (5%)
Query: 4 KENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQI 63
K++ N+ LL G+YE+G++LGHGTFAKVYHA+N+ +G NVA+KV+ KEK++K G+I I
Sbjct: 14 KKDANNGQTLLLGRYEVGKLLGHGTFAKVYHARNVKSGDNVAIKVIDKEKILKSGLIAHI 73
Query: 64 KREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQ 123
KREIS+++ V+HPNIVQL EVMA+KSKIY ME VRGGELFNK+ KGRLKEDVAR YFQQ
Sbjct: 74 KREISILRRVRHPNIVQLFEVMATKSKIYFVMEYVRGGELFNKVAKGRLKEDVARKYFQQ 133
Query: 124 LISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAY 183
LISAV FCH+RGVYHRDLKPENLLLDE+G+LKVSDFGL S+ +RQDGL HT CGTPA+
Sbjct: 134 LISAVHFCHARGVYHRDLKPENLLLDENGDLKVSDFGLSAVSDQIRQDGLFHTFCGTPAF 193
Query: 184 VSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSL 243
V+PEV+A+KGYD AK DIWSCGVIL+VL+AG+LPFQD N++AMYKKIY+G+F+CP WFS
Sbjct: 194 VAPEVLARKGYDAAKVDIWSCGVILFVLMAGYLPFQDQNIMAMYKKIYKGEFRCPRWFSP 253
Query: 244 EARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEK----EEEEDKVFEFME 299
E RL+TKLLD NP TRI+I +IM+ WFKK + +K E++++ V +
Sbjct: 254 ELIRLLTKLLDTNPETRITIPEIMEKRWFKKGFKHIKFYIEDDKLCSVEDDDNDVGSCSD 313
Query: 300 CEKSS------------------TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATG 341
S ++NAF +IS S GF+LS LFEE + E RF +G
Sbjct: 314 QSSMSESETELETRKRVGTLPRPASLNAFDLISFSPGFNLSGLFEEGE----EGSRFVSG 369
Query: 342 GTPSRVISKLEQVAKAVKFDVKSSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVK 401
S +ISKLE++AK V F V+ D +V L+G G KG L+IAA+I+ +TPS +VVEVK
Sbjct: 370 APVSTIISKLEEIAKVVSFTVRKKDCRVSLEGSREGAKGPLSIAAEIFELTPSLVVVEVK 429
Query: 402 KDKGDTLEYNQFCSKELRPALKDIFWTSADPPAA 435
K G+ EY FC++EL+P LK++ + AA
Sbjct: 430 KKGGERGEYEDFCNRELKPGLKELMVEESQSSAA 463
>UniRef100_Q8H2C2 Serine/threonine kinase [Persea americana]
Length = 453
Score = 508 bits (1307), Expect = e-142
Identities = 253/433 (58%), Positives = 328/433 (75%), Gaps = 15/433 (3%)
Query: 8 NSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREI 67
++ +L ++ELGR+LG GTFAKVYHA+NL + ++VA+KV+ KEKV+KVGMI+QIKREI
Sbjct: 2 DNKGGILMQRFELGRLLGQGTFAKVYHARNLKSSQSVAIKVIDKEKVLKVGMIDQIKREI 61
Query: 68 SVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISA 127
S M++ HPN+VQL+EVMA+K+KIY +E V+GGELFNK+ KG+LKEDVA YFQQL+SA
Sbjct: 62 STMRLASHPNVVQLYEVMATKTKIYFVLEYVKGGELFNKVAKGKLKEDVAWKYFQQLVSA 121
Query: 128 VDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPE 187
VDFCHSRGVYHRDLKPENLL+DE+ NLKV+DFGL +E RQDGLLHTTCGTPAYV+PE
Sbjct: 122 VDFCHSRGVYHRDLKPENLLVDENENLKVTDFGLGALAESKRQDGLLHTTCGTPAYVAPE 181
Query: 188 VIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARR 247
VI++KGYDGAKADIWSCGVIL+VLLAG+LPF D NL+ +Y+K+ + ++KCP WF E RR
Sbjct: 182 VISRKGYDGAKADIWSCGVILFVLLAGYLPFHDSNLMELYRKVGKAEYKCPNWFPPEVRR 241
Query: 248 LITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEE----EEDKVFEFMECEKS 303
L+ ++LDPNPNTRISI+KI D+SWFK+ A + + +E + D F + +
Sbjct: 242 LLARILDPNPNTRISIAKIKDNSWFKRGFEAKFAKGETKSKELAPLDTDAAFASSDTGVA 301
Query: 304 STT--------MNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVA 355
+ +NAF IISLS GFDLS LFE+ R+E RFA+ S +ISKLE +A
Sbjct: 302 ARKPELVKPQHLNAFDIISLSAGFDLSGLFEDTNC--RKEARFASKQPASAIISKLEDIA 359
Query: 356 KAVKFDVKSSDTKV-RLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFC 414
K ++ VK D + +L+G + GRKG L+I A+I+ V PSF +VEVKK GDTLEY +
Sbjct: 360 KRLRLKVKKKDEGILKLEGSKEGRKGVLSIDAEIFVVNPSFHLVEVKKSSGDTLEYQKLW 419
Query: 415 SKELRPALKDIFW 427
++++RPAL DI W
Sbjct: 420 NQDMRPALTDIVW 432
>UniRef100_Q9FJ55 Serine/threonine protein kinase [Arabidopsis thaliana]
Length = 483
Score = 503 bits (1295), Expect = e-141
Identities = 254/454 (55%), Positives = 330/454 (71%), Gaps = 37/454 (8%)
Query: 3 EKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQ 62
+K++ ++H AL+ GKYE+GR+LGHGTFAKVY A+N +G++VA+KV+ KEKV+K G+I
Sbjct: 13 KKQDQSNHQALILGKYEMGRLLGHGTFAKVYLARNAQSGESVAIKVIDKEKVLKSGLIAH 72
Query: 63 IKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQ 122
IKREIS+++ V+HPNIVQL EVMA+KSKIY ME V+GGELFNK+ KGRLKE++AR YFQ
Sbjct: 73 IKREISILRRVRHPNIVQLFEVMATKSKIYFVMEYVKGGELFNKVAKGRLKEEMARKYFQ 132
Query: 123 QLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPA 182
QLISAV FCH RGVYHRDLKPENLLLDE+GNLKVSDFGL S+ +RQDGL HT CGTPA
Sbjct: 133 QLISAVSFCHFRGVYHRDLKPENLLLDENGNLKVSDFGLSAVSDQIRQDGLFHTFCGTPA 192
Query: 183 YVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFS 242
YV+PEV+A+KGYDGAK DIWSCGVIL+VL+AGFLPF D N++AMYKKIYRGDF+CP WF
Sbjct: 193 YVAPEVLARKGYDGAKVDIWSCGVILFVLMAGFLPFHDRNVMAMYKKIYRGDFRCPRWFP 252
Query: 243 LEARRLITKLLDPNPNTRISISKIMDSSWFKKPVP------------------------K 278
+E RL+ ++L+ P R ++ IM++SWFKK +
Sbjct: 253 VEINRLLIRMLETKPERRFTMPDIMETSWFKKGFKHIKFYVEDDHQLCNVADDDEIESIE 312
Query: 279 SIAMRKKEKEEEEDKVFEFMECEKSSTTM------NAFHIISLSEGFDLSPLFEEKKREE 332
S++ R E ED FE + + +M NAF +IS S GFDLS LFE ++
Sbjct: 313 SVSGRSSTVSEPED--FESFDGRRRGGSMPRPASLNAFDLISFSPGFDLSGLFE----DD 366
Query: 333 REEMRFATGGTPSRVISKLEQVAKAVKFDVKSSDTKVRLQGQERG-RKGKLAIAADIYAV 391
E RF +G ++ISKLE++A+ V F V+ D KV L+G G KG L+IAA+I+ +
Sbjct: 367 GEGSRFVSGAPVGQIISKLEEIARIVSFTVRKKDCKVSLEGSREGSMKGPLSIAAEIFEL 426
Query: 392 TPSFLVVEVKKDKGDTLEYNQFCSKELRPALKDI 425
TP+ +VVEVKK GD +EY++FC+KEL+P L+++
Sbjct: 427 TPALVVVEVKKKGGDKMEYDEFCNKELKPKLQNL 460
>UniRef100_Q7X996 Putative Serine/threonine Kinase [Oryza sativa]
Length = 443
Score = 501 bits (1290), Expect = e-140
Identities = 245/424 (57%), Positives = 324/424 (75%), Gaps = 12/424 (2%)
Query: 13 LLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKM 72
+L KYE+G++LG GTFAKVYHA+N T ++VA+K++ KEKV+K G+++QIKREISVMK+
Sbjct: 8 MLMKKYEMGKLLGQGTFAKVYHARNTETSESVAIKMIDKEKVLKGGLMDQIKREISVMKL 67
Query: 73 VKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAVDFCH 132
V+HPNIVQL+EVMA+K+KIY +E V+GGELFNK+ +GRLKED AR YFQQLI AVDFCH
Sbjct: 68 VRHPNIVQLYEVMATKTKIYFVLEHVKGGELFNKVQRGRLKEDAARKYFQQLICAVDFCH 127
Query: 133 SRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKK 192
SRGVYHRDLKPENLLLDE+ NLKVSDFGL ++ RQDGLLHTTCGTPAYV+PEVI ++
Sbjct: 128 SRGVYHRDLKPENLLLDENSNLKVSDFGLSALADCKRQDGLLHTTCGTPAYVAPEVINRR 187
Query: 193 GYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKL 252
GYDGAKADIWSCGVIL+VLLAG+LPF D NL+ MYKKI + +FKCP WF+ + RRL+ ++
Sbjct: 188 GYDGAKADIWSCGVILFVLLAGYLPFHDKNLMDMYKKIGKAEFKCPSWFNTDVRRLLLRI 247
Query: 253 LDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEE--------EEDKVFEFMECEKSS 304
LDPNP+TRIS+ KIM++ WF+K + + + ++ + D EK
Sbjct: 248 LDPNPSTRISMDKIMENPWFRKGLDAKLLRYNLQPKDAIPVDMSTDFDSFNSAPTLEKKP 307
Query: 305 TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKAVKFDVKS 364
+ +NAF IISLS G DLS +FEE +++E +F + T S +ISK+E +AK ++ +
Sbjct: 308 SNLNAFDIISLSTGLDLSGMFEE---SDKKESKFTSTSTASTIISKIEDIAKGLRLKLTK 364
Query: 365 SD-TKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKELRPALK 423
D ++++G + GRKG + I A+I+ VTP+F +VE+KK GDTLEY + ++E+RPALK
Sbjct: 365 KDGGLLKMEGSKPGRKGVMGIDAEIFEVTPNFHLVELKKTNGDTLEYRKVLNQEMRPALK 424
Query: 424 DIFW 427
DI W
Sbjct: 425 DIVW 428
>UniRef100_Q9LP51 Hypothetical protein F28N24.9 [Arabidopsis thaliana]
Length = 520
Score = 486 bits (1251), Expect = e-136
Identities = 248/437 (56%), Positives = 312/437 (70%), Gaps = 27/437 (6%)
Query: 13 LLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKM 72
+L GKYELG++LGHGTFAKVY A+N+ +G VA+KV+ KEK++K G++ IKREIS+++
Sbjct: 69 ILMGKYELGKLLGHGTFAKVYLAQNIKSGDKVAIKVIDKEKIMKSGLVAHIKREISILRR 128
Query: 73 VKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISAVDFCH 132
V+HP IV L EVMA+KSKIY ME V GGELFN + KGRL E+ AR YFQQLIS+V FCH
Sbjct: 129 VRHPYIVHLFEVMATKSKIYFVMEYVGGGELFNTVAKGRLPEETARRYFQQLISSVSFCH 188
Query: 133 SRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKK 192
RGVYHRDLKPENLLLD GNLKVSDFGL +E LRQDGL HT CGTPAY++PEV+ +K
Sbjct: 189 GRGVYHRDLKPENLLLDNKGNLKVSDFGLSAVAEQLRQDGLCHTFCGTPAYIAPEVLTRK 248
Query: 193 GYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKL 252
GYD AKAD+WSCGVIL+VL+AG +PF D N++ MYKKIY+G+F+CP WFS + RL+T+L
Sbjct: 249 GYDAAKADVWSCGVILFVLMAGHIPFYDKNIMVMYKKIYKGEFRCPRWFSSDLVRLLTRL 308
Query: 253 LDPNPNTRISISKIMDSSWFKKP-------VPKSIAMRKKEKEEEE-------------D 292
LD NP+TRI+I +IM + WFKK + R+ E EEEE D
Sbjct: 309 LDTNPDTRITIPEIMKNRWFKKGFKHVKFYIEDDKLCREDEDEEEEASSSGRSSTVSESD 368
Query: 293 KVFEFMECEKSS----TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVI 348
F+ S +++NAF IIS S GFDLS LFEE E E RF +G S++I
Sbjct: 369 AEFDVKRMGIGSMPRPSSLNAFDIISFSSGFDLSGLFEE---EGGEGTRFVSGAPVSKII 425
Query: 349 SKLEQVAKAVKFDVKSSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTL 408
SKLE++AK V F V+ + +RL+G G KG L IAA+I+ +TPS +VVEVKK GD
Sbjct: 426 SKLEEIAKIVSFTVRKKEWSLRLEGCREGAKGPLTIAAEIFELTPSLVVVEVKKKGGDRE 485
Query: 409 EYNQFCSKELRPALKDI 425
EY +FC+KELRP L+ +
Sbjct: 486 EYEEFCNKELRPELEKL 502
>UniRef100_Q8LRC0 Putative serine/threonine protein kinase [Oryza sativa]
Length = 502
Score = 480 bits (1235), Expect = e-134
Identities = 246/430 (57%), Positives = 315/430 (73%), Gaps = 16/430 (3%)
Query: 12 ALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMK 71
++L G+YE+G+ LG GTFAKVY+A+NL TG+ VA+K++ K+KV+KVG++EQIKREIS+M+
Sbjct: 6 SILMGRYEVGKQLGQGTFAKVYYARNLTTGQAVAIKMINKDKVMKVGLMEQIKREISIMR 65
Query: 72 MVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDF 130
+VKHPN++QL EVMASKSKIY +E +GGELFNKI K G+L ED AR YF QLI+AVD+
Sbjct: 66 LVKHPNVLQLFEVMASKSKIYFVLEYAKGGELFNKIAKEGKLSEDSARRYFHQLINAVDY 125
Query: 131 CHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIA 190
CHSRGVYHRDLKPENLLLDE+ NLKVSDFGL +E RQDGLLHTTCGTPAYV+PEV++
Sbjct: 126 CHSRGVYHRDLKPENLLLDENENLKVSDFGLSALAESKRQDGLLHTTCGTPAYVAPEVLS 185
Query: 191 KKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLIT 250
+KGYDGAKAD+WSCGVIL+VL+AG+LPF D NL+ MY+KI R DF+CP +FS E + LI
Sbjct: 186 RKGYDGAKADVWSCGVILFVLVAGYLPFHDPNLIEMYRKICRADFRCPRYFSAELKDLIH 245
Query: 251 KLLDPNPNTRISISKIMDSSWFKKPV----PKSIAMRKKEKEEEEDKVFEFMECEKSS-- 304
K+LD +P+TRISI +I S+W++KPV S A EC S
Sbjct: 246 KILDSDPSTRISIPRIKRSTWYRKPVEINAKNSEAATTNSISSGVATTSGSAECSTSEEN 305
Query: 305 ------TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKAV 358
+NAF IISLS GF+LS FE+ + E RF T + V+ KL+++AK +
Sbjct: 306 QGSLSLPNLNAFDIISLSTGFNLSGFFEDTHGHQEE--RFTTRQPVTTVLGKLKELAKRL 363
Query: 359 KFDVKSSDTKV-RLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKE 417
K VK D V RL + G+KG L + A+I+ VTPSFL+VE+KK GDT+EY + ++
Sbjct: 364 KLKVKKKDNGVLRLAAPKEGKKGFLELDAEIFEVTPSFLLVELKKTNGDTMEYRKLVKED 423
Query: 418 LRPALKDIFW 427
+RPALKDI W
Sbjct: 424 IRPALKDIVW 433
>UniRef100_Q93VD3 At1g30270/F12P21_6 [Arabidopsis thaliana]
Length = 482
Score = 474 bits (1220), Expect = e-132
Identities = 232/429 (54%), Positives = 313/429 (72%), Gaps = 14/429 (3%)
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
GKYELGR LG GTFAKV A+N+ G NVA+KV+ KEKV+K MI QIKREIS MK++KH
Sbjct: 29 GKYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMIAQIKREISTMKLIKH 88
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFCHSR 134
PN++++ EVMASK+KIY +E V GGELF+KI GRLKED AR YFQQLI+AVD+CHSR
Sbjct: 89 PNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDEARKYFQQLINAVDYCHSR 148
Query: 135 GVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGY 194
GVYHRDLKPENLLLD +G LKVSDFGL + +R+DGLLHTTCGTP YV+PEVI KGY
Sbjct: 149 GVYHRDLKPENLLLDANGALKVSDFGLSALPQQVREDGLLHTTCGTPNYVAPEVINNKGY 208
Query: 195 DGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLD 254
DGAKAD+WSCGVIL+VL+AG+LPF+D NL ++YKKI++ +F CPPWFS A++LI ++LD
Sbjct: 209 DGAKADLWSCGVILFVLMAGYLPFEDSNLTSLYKKIFKAEFTCPPWFSASAKKLIKRILD 268
Query: 255 PNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFM------------ECEK 302
PNP TRI+ ++++++ WFKK ++ D +F+ E K
Sbjct: 269 PNPATRITFAEVIENEWFKKGYKAPKFENADVSLDDVDAIFDDSGESKNLVVERREEGLK 328
Query: 303 SSTTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKAVKFDV 362
+ TMNAF +IS S+G +L LFE++ + + RF + + + +++K+E A + FDV
Sbjct: 329 TPVTMNAFELISTSQGLNLGSLFEKQMGLVKRKTRFTSKSSANEIVTKIEAAAAPMGFDV 388
Query: 363 KSSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKELRPAL 422
K+++ K++L G++ GRKG+LA+A +++ V PS +VE++K GDTLE+++F K L L
Sbjct: 389 KTNNYKMKLTGEKSGRKGQLAVATEVFQVAPSLYMVEMRKSGGDTLEFHKF-YKNLTTGL 447
Query: 423 KDIFWTSAD 431
KDI W + D
Sbjct: 448 KDIVWKTID 456
>UniRef100_Q9LVL3 Serine/threonine protein kinase [Arabidopsis thaliana]
Length = 464
Score = 474 bits (1219), Expect = e-132
Identities = 239/432 (55%), Positives = 321/432 (73%), Gaps = 16/432 (3%)
Query: 8 NSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREI 67
N S L D KY++GR+LG GTFAKVY+ +++ T ++VA+K++ KEKV+KVG+IEQIKREI
Sbjct: 3 NKPSVLTD-KYDVGRLLGQGTFAKVYYGRSILTNQSVAIKMIDKEKVMKVGLIEQIKREI 61
Query: 68 SVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISA 127
SVM++ +HPN+V+L+EVMA+K++IY ME +GGELFNK+ KG+L++DVA YF QLI+A
Sbjct: 62 SVMRIARHPNVVELYEVMATKTRIYFVMEYCKGGELFNKVAKGKLRDDVAWKYFYQLINA 121
Query: 128 VDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPE 187
VDFCHSR VYHRD+KPENLLLD++ NLKVSDFGL ++ RQDGLLHTTCGTPAYV+PE
Sbjct: 122 VDFCHSREVYHRDIKPENLLLDDNENLKVSDFGLSALADCKRQDGLLHTTCGTPAYVAPE 181
Query: 188 VIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARR 247
VI +KGYDG KADIWSCGV+L+VLLAG+LPF D NL+ MY+KI + DFK P WF+ E RR
Sbjct: 182 VINRKGYDGTKADIWSCGVVLFVLLAGYLPFHDSNLMEMYRKIGKADFKAPSWFAPEVRR 241
Query: 248 LITKLLDPNPNTRISISKIMDSSWFKKPV-PKSIAMRKKEKE----------EEEDKVFE 296
L+ K+LDPNP TRI+I++I +SSWF+K + K M K+ KE E+
Sbjct: 242 LLCKMLDPNPETRITIARIRESSWFRKGLHMKQKKMEKRVKEINSVEAGTAGTNENGAGP 301
Query: 297 FMECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAK 356
T +NAF +I+LS GFDL+ LF + + + E RF + S +ISKLE+VA+
Sbjct: 302 SENGADEPTNLNAFDLIALSAGFDLAGLFGD---DNKRESRFTSQKPASVIISKLEEVAQ 358
Query: 357 AVKFDVKSSDTKV-RLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCS 415
+K ++ + + +L+ + GRKG L++ A+I+ VTP+F +VEVKK GDTLEY + +
Sbjct: 359 RLKLSIRKREAGLFKLERLKEGRKGILSMDAEIFQVTPNFHLVEVKKSNGDTLEYQKLVA 418
Query: 416 KELRPALKDIFW 427
++LRPAL DI W
Sbjct: 419 EDLRPALSDIVW 430
>UniRef100_Q6ZLP5 Putative CBL-interacting protein kinase 23 [Oryza sativa]
Length = 450
Score = 473 bits (1218), Expect = e-132
Identities = 232/423 (54%), Positives = 316/423 (73%), Gaps = 12/423 (2%)
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
G+YELGR LG GTFAKV A+N ++G+NVA+K++ K+KV+K MI QIKREIS MK+++H
Sbjct: 11 GRYELGRTLGEGTFAKVKFARNADSGENVAIKILDKDKVLKHKMIAQIKREISTMKLIRH 70
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLISAVDFCHSR 134
PN++++HEVMASK+KIYI MELV GGELF+KI +GRLKED AR YFQQLI+AVD+CHSR
Sbjct: 71 PNVIRMHEVMASKTKIYIVMELVTGGELFDKIASRGRLKEDDARKYFQQLINAVDYCHSR 130
Query: 135 GVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGY 194
GVYHRDLKPENLLLD G LKVSDFGL S+ +R+DGLLHTTCGTP YV+PEVI KGY
Sbjct: 131 GVYHRDLKPENLLLDASGTLKVSDFGLSALSQQVREDGLLHTTCGTPNYVAPEVINNKGY 190
Query: 195 DGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLD 254
DGAKAD+WSCGVIL+VL+AG+LPF+D NL+++YKKI++ DF CP WFS A++LI K+LD
Sbjct: 191 DGAKADLWSCGVILFVLMAGYLPFEDSNLMSLYKKIFKADFSCPSWFSTSAKKLIKKILD 250
Query: 255 PNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFE---------FMECEKSST 305
PNP+TRI+I++++++ WFKK ++ + +F E+ +
Sbjct: 251 PNPSTRITIAELINNEWFKKGYQPPRFETADVNLDDINSIFNESGDQTQLVVERREERPS 310
Query: 306 TMNAFHIISLSEGFDLSPLFEEKKR-EEREEMRFATGGTPSRVISKLEQVAKAVKFDVKS 364
MNAF +IS S+G +L LFE++ + + E RFA+ + ++SK+E A + F+V+
Sbjct: 311 VMNAFELISTSQGLNLGTLFEKQSQGSVKRETRFASRLPANEILSKIEAAAGPMGFNVQK 370
Query: 365 SDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKELRPALKD 424
+ K++LQG+ GRKG+LAIA +++ VTPS +VE++K GDTLE+++F + LKD
Sbjct: 371 RNYKLKLQGENPGRKGQLAIATEVFEVTPSLYMVELRKSNGDTLEFHKF-YHNISNGLKD 429
Query: 425 IFW 427
+ W
Sbjct: 430 VMW 432
>UniRef100_Q94A54 AT5g25110/T11H3_120 [Arabidopsis thaliana]
Length = 487
Score = 473 bits (1217), Expect = e-132
Identities = 241/434 (55%), Positives = 317/434 (72%), Gaps = 13/434 (2%)
Query: 5 ENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIK 64
E +L KYE+GR+LG GTF KVY+ K + TG++VA+K++ K++V + GM+EQIK
Sbjct: 30 EEEQQQLRVLFAKYEMGRLLGKGTFGKVYYGKEITTGESVAIKIINKDQVKREGMMEQIK 89
Query: 65 REISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQL 124
REIS+M++V+HPNIV+L EVMA+K+KI+ ME V+GGELF+KIVKG+LKED AR YFQQL
Sbjct: 90 REISIMRLVRHPNIVELKEVMATKTKIFFIMEYVKGGELFSKIVKGKLKEDSARKYFQQL 149
Query: 125 ISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYV 184
ISAVDFCHSRGV HRDLKPENLL+DE+G+LKVSDFGL E + QDGLLHT CGTPAYV
Sbjct: 150 ISAVDFCHSRGVSHRDLKPENLLVDENGDLKVSDFGLSALPEQILQDGLLHTQCGTPAYV 209
Query: 185 SPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLE 244
+PEV+ KKGYDGAK DIWSCG+ILYVLLAGFLPFQD+NL+ MY+KI++ +F+ PPWFS E
Sbjct: 210 APEVLRKKGYDGAKGDIWSCGIILYVLLAGFLPFQDENLMKMYRKIFKSEFEYPPWFSPE 269
Query: 245 ARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEE--EDKVFEFMECEK 302
++RLI+KLL +PN RISI IM + WF+K + I + E E + ED+
Sbjct: 270 SKRLISKLLVVDPNKRISIPAIMRTPWFRKNINSPIEFKIDELEIQNVEDETPTTTATTA 329
Query: 303 SSTT------MNAFHII-SLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVA 355
++TT NAF I S+S GFDLS LFE K+ + F + + S ++ KLE +
Sbjct: 330 TTTTPVSPKFFNAFEFISSMSSGFDLSSLFESKR---KLRSMFTSRWSASEIMGKLEGIG 386
Query: 356 KAVKFDVK-SSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFC 414
K + VK + D KV+L G+ GRKG++A+ A+++ V P VVE+ K GDTLEYN+
Sbjct: 387 KEMNMKVKRTKDFKVKLFGKTEGRKGQIAVTAEVFEVAPEVAVVELCKSAGDTLEYNRLY 446
Query: 415 SKELRPALKDIFWT 428
+ +RPAL++I W+
Sbjct: 447 EEHVRPALEEIVWS 460
>UniRef100_Q8W1D5 CBL-interacting protein kinase CIPK25 [Arabidopsis thaliana]
Length = 488
Score = 473 bits (1216), Expect = e-132
Identities = 241/435 (55%), Positives = 317/435 (72%), Gaps = 14/435 (3%)
Query: 5 ENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIK 64
E +L KYE+GR+LG GTF KVY+ K + TG++VA+K++ K++V + GM+EQIK
Sbjct: 30 EEEQQQLRVLFAKYEMGRLLGKGTFGKVYYGKEITTGESVAIKIINKDQVKREGMMEQIK 89
Query: 65 REISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQL 124
REIS+M++V+HPNIV+L EVMA+K+KI+ ME V+GGELF+KIVKG+LKED AR YFQQL
Sbjct: 90 REISIMRLVRHPNIVELKEVMATKTKIFFIMEYVKGGELFSKIVKGKLKEDSARKYFQQL 149
Query: 125 ISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYV 184
ISAVDFCHSRGV HRDLKPENLL+DE+G+LKVSDFGL E + QDGLLHT CGTPAYV
Sbjct: 150 ISAVDFCHSRGVSHRDLKPENLLVDENGDLKVSDFGLSALPEQILQDGLLHTQCGTPAYV 209
Query: 185 SPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLE 244
+PEV+ KKGYDGAK DIWSCG+ILYVLLAGFLPFQD+NL+ MY+KI++ +F+ PPWFS E
Sbjct: 210 APEVLRKKGYDGAKGDIWSCGIILYVLLAGFLPFQDENLMKMYRKIFKSEFEYPPWFSPE 269
Query: 245 ARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEE--EDKVFEFMECEK 302
++RLI+KLL +PN RISI IM + WF+K + I + E E + ED+
Sbjct: 270 SKRLISKLLVVDPNKRISIPAIMRTPWFRKNINSPIEFKIDELEIQNVEDETPTTTATTA 329
Query: 303 SSTT-------MNAFHII-SLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQV 354
++TT NAF I S+S GFDLS LFE K+ + F + + S ++ KLE +
Sbjct: 330 TTTTTPVSPKFFNAFEFISSMSSGFDLSSLFESKR---KLRSMFTSRWSASEIMGKLEGI 386
Query: 355 AKAVKFDVK-SSDTKVRLQGQERGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQF 413
K + VK + D KV+L G+ GRKG++A+ A+++ V P VVE+ K GDTLEYN+
Sbjct: 387 GKEMNMKVKRTKDFKVKLFGKTEGRKGQIAVTAEVFEVAPEVAVVELCKSAGDTLEYNRL 446
Query: 414 CSKELRPALKDIFWT 428
+ +RPAL++I W+
Sbjct: 447 YEEHVRPALEEIVWS 461
>UniRef100_Q9C562 SOS2-like protein kinase PKS2 [Arabidopsis thaliana]
Length = 479
Score = 473 bits (1216), Expect = e-132
Identities = 242/448 (54%), Positives = 326/448 (72%), Gaps = 33/448 (7%)
Query: 8 NSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREI 67
N S L D KY++GR+LG GTFAKVY+ +++ T ++VA+K++ KEKV+KVG+IEQIKREI
Sbjct: 3 NKPSVLTD-KYDVGRLLGQGTFAKVYYGRSILTNQSVAIKMIDKEKVMKVGLIEQIKREI 61
Query: 68 SVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVKGRLKEDVARVYFQQLISA 127
SVM++ +HPN+V+L+EVMA+K++IY ME +GGELFNK+ KG+L++DVA YF QLI+A
Sbjct: 62 SVMRIARHPNVVELYEVMATKTRIYFVMEYCKGGELFNKVAKGKLRDDVAWKYFYQLINA 121
Query: 128 VDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPE 187
VDFCHSR VYHRD+KPENLLLD++ NLKVSDFGL ++ RQDGLLHTTCGTPAYV+PE
Sbjct: 122 VDFCHSREVYHRDIKPENLLLDDNENLKVSDFGLSALADCKRQDGLLHTTCGTPAYVAPE 181
Query: 188 VIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARR 247
VI +KGYDG KADIWSCGV+L+VLLAG+LPF D NL+ MY+KI + DFK P WF+ E RR
Sbjct: 182 VINRKGYDGTKADIWSCGVVLFVLLAGYLPFHDSNLMEMYRKIGKADFKAPSWFAPEVRR 241
Query: 248 LITKLLDPNPNTRISISKIMDSSWFKKPV-PKSIAMRKKEKE------------------ 288
L+ K+LDPNP TRI+I++I +SSWF+K + K M K+ KE
Sbjct: 242 LLCKMLDPNPETRITIARIRESSWFRKGLHMKQKKMEKRVKEINSVEAGTAGTNENGAGP 301
Query: 289 --------EEEDKVFEFMECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFAT 340
E D+V E ++ T +NAF +I+LS GFDL+ LF + + + E RF +
Sbjct: 302 SENGAGPSENGDRVTEENHTDE-PTNLNAFDLIALSAGFDLAGLFGD---DNKRESRFTS 357
Query: 341 GGTPSRVISKLEQVAKAVKFDVKSSDTKV-RLQGQERGRKGKLAIAADIYAVTPSFLVVE 399
S +ISKLE+VA+ +K ++ + + +L+ + GRKG L++ A+I+ VTP+F +VE
Sbjct: 358 QKPASVIISKLEEVAQRLKLSIRKREAGLFKLERLKEGRKGILSMDAEIFQVTPNFHLVE 417
Query: 400 VKKDKGDTLEYNQFCSKELRPALKDIFW 427
VKK GDTLEY + +++LRPAL DI W
Sbjct: 418 VKKSNGDTLEYQKLVAEDLRPALSDIVW 445
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 727,861,286
Number of Sequences: 2790947
Number of extensions: 30838026
Number of successful extensions: 144098
Number of sequences better than 10.0: 19454
Number of HSP's better than 10.0 without gapping: 13644
Number of HSP's successfully gapped in prelim test: 5810
Number of HSP's that attempted gapping in prelim test: 91030
Number of HSP's gapped (non-prelim): 22911
length of query: 435
length of database: 848,049,833
effective HSP length: 130
effective length of query: 305
effective length of database: 485,226,723
effective search space: 147994150515
effective search space used: 147994150515
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC122160.9


