

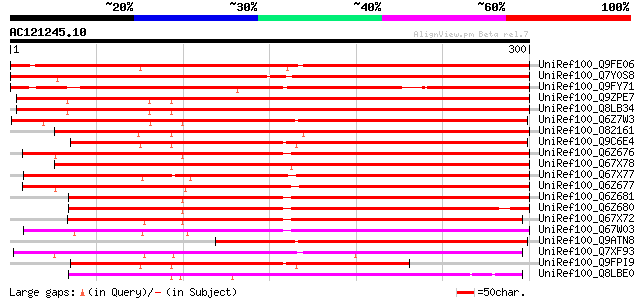
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121245.10 - phase: 0
(300 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FE06 Putative phi-1 protein [Arabidopsis thaliana] 437 e-121
UniRef100_Q7Y0S8 Erg-1 [Solanum tuberosum] 419 e-116
UniRef100_Q9FY71 Hypothetical protein T5E8_240 [Arabidopsis thal... 358 1e-97
UniRef100_Q9ZPE7 Hypothetical protein At4g08950; T3H13.3 [Arabid... 354 1e-96
UniRef100_Q8LB34 Putative phi-1-like phosphate-induced protein [... 354 2e-96
UniRef100_Q6Z7W3 Putative phi-1 [Oryza sativa] 353 4e-96
UniRef100_O82161 Phi-1 protein [Nicotiana tabacum] 350 3e-95
UniRef100_Q9C6E4 Phosphate-induced (Phi-1) protein, putative [Ar... 338 1e-91
UniRef100_Q6Z676 Putative phi-1 [Oryza sativa] 321 2e-86
UniRef100_Q67X78 Putative phi-1 [Oryza sativa] 320 2e-86
UniRef100_Q67X77 Putative phi-1 [Oryza sativa] 317 3e-85
UniRef100_Q6Z677 Putative phi-1 [Oryza sativa] 307 3e-82
UniRef100_Q6Z681 Putative phi-1 [Oryza sativa] 290 3e-77
UniRef100_Q6Z680 Putative phi-1 [Oryza sativa] 275 1e-72
UniRef100_Q67X72 Putative phi-1 [Oryza sativa] 273 3e-72
UniRef100_Q67W03 Putative phi-1 [Oryza sativa] 267 3e-70
UniRef100_Q9ATN8 Phosphate-induced protein 1-like protein [Penni... 256 5e-67
UniRef100_Q7XF93 Putative phosphate-induced protein [Oryza sativa] 250 4e-65
UniRef100_Q9FPI9 At1g35140 [Arabidopsis thaliana] 237 2e-61
UniRef100_Q8LBE0 Hypothetical protein [Arabidopsis thaliana] 191 3e-47
>UniRef100_Q9FE06 Putative phi-1 protein [Arabidopsis thaliana]
Length = 305
Score = 437 bits (1123), Expect = e-121
Identities = 221/309 (71%), Positives = 246/309 (79%), Gaps = 13/309 (4%)
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 60
MASNY FAI L+ L S LV+EQPLV+KYHNG LLKG +TVNL+WYG FTPI
Sbjct: 1 MASNYRFAIFLT--LFFATAGFSAAALVEEQPLVMKYHNGVLLKGNITVNLVWYGKFTPI 58
Query: 61 QRSIIVDFINSLST----TGAALPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNL 116
QRS+IVDFI+SL++ + AA+PS ++WWKTTEKYK GSS L VGKQ L Y LGK+L
Sbjct: 59 QRSVIVDFIHSLNSKDVASSAAVPSVASWWKTTEKYKGGSSTLVVGKQLLLENYPLGKSL 118
Query: 117 KGKDLLALATKFNE-LSSITVVLTAKDVNVEGFCMSRCGTHGSV----RRGSGGARTPYI 171
K L AL+TK N L SITVVLTAKDV VE FCMSRCGTHGS RR + GA Y+
Sbjct: 119 KNPYLRALSTKLNGGLRSITVVLTAKDVTVERFCMSRCGTHGSSGSNPRRAANGAA--YV 176
Query: 172 WVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNG 231
WVGN+ET CPG CAWPFHQPIYGPQTPPLVAPNGDVGVDGM+INLATLLA TVTNPFNNG
Sbjct: 177 WVGNSETQCPGYCAWPFHQPIYGPQTPPLVAPNGDVGVDGMIINLATLLANTVTNPFNNG 236
Query: 232 YFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDP 291
Y+QGP APLEAV+AC G+FGSG+YPGYAGRVLVD +GSSYNA G GRKYLLPAMWDP
Sbjct: 237 YYQGPPTAPLEAVSACPGIFGSGSYPGYAGRVLVDKTTGSSYNARGLAGRKYLLPAMWDP 296
Query: 292 QTSACKTLV 300
Q+S CKTLV
Sbjct: 297 QSSTCKTLV 305
>UniRef100_Q7Y0S8 Erg-1 [Solanum tuberosum]
Length = 301
Score = 419 bits (1077), Expect = e-116
Identities = 207/305 (67%), Positives = 242/305 (78%), Gaps = 9/305 (2%)
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGE----LVQEQPLVLKYHNGQLLKGKLTVNLIWYGT 56
M SNY A V S +L L+ + FG LVQEQPLVLKYHNG LLKG +TVNLIWYG
Sbjct: 1 MVSNYISATVSSILLFLLCSSSLFGASMAALVQEQPLVLKYHNGALLKGTVTVNLIWYGK 60
Query: 57 FTPIQRSIIVDFINSLSTTGAALPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNL 116
FTPIQRSIIVDF+ SL++ A PSA++WWKTTEKYK G+S +T+GKQ L +LGK+L
Sbjct: 61 FTPIQRSIIVDFLQSLNSPKAPNPSAASWWKTTEKYKTGASTVTLGKQILDENCSLGKSL 120
Query: 117 KGKDLLALATKFNELS-SITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGN 175
K ++ LA+K + S+ +VLTAKDV VEGFC SRCG+HGS R G R Y WVGN
Sbjct: 121 KNSHIVYLASKGGYMGRSVNLVLTAKDVFVEGFC-SRCGSHGSTR---GKVRFTYAWVGN 176
Query: 176 AETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQG 235
+ET C GQCAWPFHQPIYGPQTPPL+APNGDVGVDGM+IN+AT+LAGTVTNPFNNGYFQG
Sbjct: 177 SETQCAGQCAWPFHQPIYGPQTPPLLAPNGDVGVDGMIINVATVLAGTVTNPFNNGYFQG 236
Query: 236 PAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSA 295
PA APLEAV+ACTG+FGSG+YPGY G+ LVD ++G+SYNAHG NGR++LLPAMWDP SA
Sbjct: 237 PATAPLEAVSACTGMFGSGSYPGYPGQTLVDKSTGASYNAHGVNGRRFLLPAMWDPTKSA 296
Query: 296 CKTLV 300
C LV
Sbjct: 297 CSALV 301
>UniRef100_Q9FY71 Hypothetical protein T5E8_240 [Arabidopsis thaliana]
Length = 278
Score = 358 bits (918), Expect = 1e-97
Identities = 190/303 (62%), Positives = 219/303 (71%), Gaps = 28/303 (9%)
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 60
MA NY FAI+L +L+ T+ F + P+ + L G +T+NLIWYG FTPI
Sbjct: 1 MAYNYRFAILL----VLLSATVGFTAAALKPPV-------RTLNGNITLNLIWYGKFTPI 49
Query: 61 QRSIIVDFINSLSTTGAAL-PSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNLKGK 119
QRSIIVDFI S+S+ AA PS ++WWKTTEKYK G S L VGKQ L Y LGK+LK
Sbjct: 50 QRSIIVDFIRSISSVTAAKGPSVASWWKTTEKYKTGVSTLVVGKQLLLENYPLGKSLKSP 109
Query: 120 DLLALATKFNE--LSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAE 177
L AL++K N SITVVLTAKDV VEG CM+RCGTHGS + S Y+WVGN+E
Sbjct: 110 YLRALSSKLNAGGARSITVVLTAKDVTVEGLCMNRCGTHGS--KSSSVNSGAYVWVGNSE 167
Query: 178 TLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPA 237
T CPG CAWPFHQPIYGPQ+PPLVAPNGDVGVDGM+IN+ATLL TVTNP
Sbjct: 168 TQCPGYCAWPFHQPIYGPQSPPLVAPNGDVGVDGMIINIATLLVNTVTNP---------- 217
Query: 238 AAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACK 297
+P EAV+ACTG+FGSGAYPGYAGRVLVD SG+SYNA G GRKYLLPA+WDPQTS CK
Sbjct: 218 -SP-EAVSACTGIFGSGAYPGYAGRVLVDKTSGASYNALGLAGRKYLLPALWDPQTSTCK 275
Query: 298 TLV 300
T+V
Sbjct: 276 TMV 278
>UniRef100_Q9ZPE7 Hypothetical protein At4g08950; T3H13.3 [Arabidopsis thaliana]
Length = 314
Score = 354 bits (909), Expect = 1e-96
Identities = 178/313 (56%), Positives = 220/313 (69%), Gaps = 17/313 (5%)
Query: 5 YYFAIVLSAVLLLVLPTLSFGELVQEQP---LVLKYHNGQLLKGKLTVNLIWYGTFTPIQ 61
Y L L L+ ++S L ++P +LKYH G LL GK++VNLIWYG F P Q
Sbjct: 2 YLLVFKLFLFLSLLQISVSARNLASQEPNQFQLLKYHKGALLSGKISVNLIWYGKFKPSQ 61
Query: 62 RSIIVDFINSLSTTGAAL-----PSASAWWKTTEKY--------KVGSSALTVGKQFLHP 108
R+II DFI SL+ T PS + WWKTTEKY +LT+GKQ +
Sbjct: 62 RAIISDFITSLTHTSPTSKTLHQPSVATWWKTTEKYYKLATPSKNSSPLSLTLGKQIIDE 121
Query: 109 AYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRR-GSGGAR 167
+ +LGK+L K + LA+K ++ ++I VVLT+ DV V GF MSRCGTHG R G G++
Sbjct: 122 SCSLGKSLTDKKIQTLASKGDQRNAINVVLTSADVTVTGFGMSRCGTHGHARGLGKRGSK 181
Query: 168 TPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNP 227
YIWVGN+ET CPGQCAWPFH P+YGPQ+PPLVAPN DVG+DGMVINLA+LLAGT TNP
Sbjct: 182 FAYIWVGNSETQCPGQCAWPFHAPVYGPQSPPLVAPNNDVGLDGMVINLASLLAGTATNP 241
Query: 228 FNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPA 287
F NGY+QGP APLEA +AC GV+G GAYPGYAG +LVD +G S+NA+GANGRK+LLPA
Sbjct: 242 FGNGYYQGPQNAPLEAASACPGVYGKGAYPGYAGDLLVDTTTGGSFNAYGANGRKFLLPA 301
Query: 288 MWDPQTSACKTLV 300
++DP TSAC T+V
Sbjct: 302 LYDPTTSACSTMV 314
>UniRef100_Q8LB34 Putative phi-1-like phosphate-induced protein [Arabidopsis
thaliana]
Length = 314
Score = 354 bits (908), Expect = 2e-96
Identities = 178/313 (56%), Positives = 220/313 (69%), Gaps = 17/313 (5%)
Query: 5 YYFAIVLSAVLLLVLPTLSFGELVQEQP---LVLKYHNGQLLKGKLTVNLIWYGTFTPIQ 61
Y L L L+ ++S L ++P +LKYH G LL GK++VNLIWYG F P Q
Sbjct: 2 YSLVFKLFLFLSLLQISVSARNLASQEPNQFQLLKYHKGALLSGKISVNLIWYGKFKPSQ 61
Query: 62 RSIIVDFINSLSTTGAAL-----PSASAWWKTTEKY--------KVGSSALTVGKQFLHP 108
R+II DFI SL+ T PS + WWKTTEKY +LT+GKQ +
Sbjct: 62 RAIISDFITSLTHTSPTSKTLHQPSVATWWKTTEKYYKLATPSKNSSPLSLTLGKQIIDE 121
Query: 109 AYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRR-GSGGAR 167
+ +LGK+L K + LA+K ++ ++I VVLT+ DV V GF MSRCGTHG R G G++
Sbjct: 122 SCSLGKSLTDKKIQTLASKGDQRNAINVVLTSADVTVTGFGMSRCGTHGHARGLGKRGSK 181
Query: 168 TPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNP 227
YIWVGN+ET CPGQCAWPFH P+YGPQ+PPLVAPN DVG+DGMVINLA+LLAGT TNP
Sbjct: 182 FAYIWVGNSETQCPGQCAWPFHAPVYGPQSPPLVAPNNDVGLDGMVINLASLLAGTATNP 241
Query: 228 FNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPA 287
F NGY+QGP APLEA +AC GV+G GAYPGYAG +LVD +G S+NA+GANGRK+LLPA
Sbjct: 242 FGNGYYQGPQNAPLEAASACPGVYGKGAYPGYAGDLLVDTTTGGSFNAYGANGRKFLLPA 301
Query: 288 MWDPQTSACKTLV 300
++DP TSAC T+V
Sbjct: 302 LYDPTTSACSTMV 314
>UniRef100_Q6Z7W3 Putative phi-1 [Oryza sativa]
Length = 311
Score = 353 bits (905), Expect = 4e-96
Identities = 176/309 (56%), Positives = 221/309 (70%), Gaps = 12/309 (3%)
Query: 2 ASNYYFAIVLSAVLLLV---LPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFT 58
AS+ FA L A LL + + LVQ+QP+ + YH G LL G++ VNLIWYG F+
Sbjct: 3 ASSAAFAACLLACALLFQMCVASRKLTALVQDQPITMTYHKGALLSGRIAVNLIWYGNFS 62
Query: 59 PIQRSIIVDFINSLSTTGAALP----SASAWWKTTEKYKVGSSA----LTVGKQFLHPAY 110
QR++I DF++SLST + P S ++W+KT +KY S A L++G+ L +Y
Sbjct: 63 APQRAVITDFVSSLSTPPSPQPQPEPSVASWFKTAQKYYANSKARFPALSLGQHVLDQSY 122
Query: 111 TLGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPY 170
+LGK L KDL+ LA + + +I VVLTA DV V+GFCMSRCGTHG+ R G R Y
Sbjct: 123 SLGKRLGEKDLVRLAARGSPSRAINVVLTADDVAVDGFCMSRCGTHGASPRSRAG-RFAY 181
Query: 171 IWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNN 230
+WVGN T CPGQCAWP+HQP+YGPQ PL PNGDVGVDGMVI+LA+++ GTVTNPF N
Sbjct: 182 VWVGNPATQCPGQCAWPYHQPVYGPQAAPLTPPNGDVGVDGMVISLASMIVGTVTNPFGN 241
Query: 231 GYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWD 290
G+FQG A APLEA TAC GV+G GAYPGYAG +LVD ASG+SYNA+GA+GRKYL+PA+ D
Sbjct: 242 GFFQGDADAPLEAATACAGVYGKGAYPGYAGSLLVDPASGASYNANGAHGRKYLVPALVD 301
Query: 291 PQTSACKTL 299
P TSAC T+
Sbjct: 302 PDTSACSTV 310
>UniRef100_O82161 Phi-1 protein [Nicotiana tabacum]
Length = 313
Score = 350 bits (898), Expect = 3e-95
Identities = 176/285 (61%), Positives = 210/285 (72%), Gaps = 11/285 (3%)
Query: 27 LVQE-QPLVLKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLS--TTGAALPSAS 83
LVQE + +L+YH G LL GK++VNLIWYG F P QR+I+ DFI SLS T PS +
Sbjct: 29 LVQEPENQLLQYHKGALLFGKISVNLIWYGKFKPSQRAIVSDFITSLSSSTPSKTDPSVA 88
Query: 84 AWWKTTEKY-----KVGSSALTVGKQFLHPAYTLGKNLKGKDLLALATKFNELSSITVVL 138
WWKTTEKY S +L +GKQ L Y+LGK+L K ++ LA+K + +I +VL
Sbjct: 89 KWWKTTEKYYHLANSKKSLSLYLGKQVLVENYSLGKSLTQKQIVQLASKGEQKDAINIVL 148
Query: 139 TAKDVNVEGFCMSRCGTHGSVRRGSGGART---PYIWVGNAETLCPGQCAWPFHQPIYGP 195
TA DV V+GFC++RCGTHGS + +T YIWVGN+ET C G CAWPFHQPIYGP
Sbjct: 149 TASDVAVDGFCVNRCGTHGSSKGAIIRGKTYKFAYIWVGNSETQCAGYCAWPFHQPIYGP 208
Query: 196 QTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGA 255
Q+PPLVAPN DVGVDGMVINLA+LLAGT TNPF NGY+QG A APLEA +AC GV+ GA
Sbjct: 209 QSPPLVAPNNDVGVDGMVINLASLLAGTATNPFGNGYYQGEADAPLEAASACPGVYAKGA 268
Query: 256 YPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTLV 300
YPGYAG +LVD +G+SYNAHG NGRKYLLPA++DP TS C TLV
Sbjct: 269 YPGYAGDLLVDKTTGASYNAHGTNGRKYLLPALYDPSTSTCSTLV 313
>UniRef100_Q9C6E4 Phosphate-induced (Phi-1) protein, putative [Arabidopsis thaliana]
Length = 309
Score = 338 bits (867), Expect = 1e-91
Identities = 161/277 (58%), Positives = 204/277 (73%), Gaps = 14/277 (5%)
Query: 36 KYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLST----TGAALPSASAWWKTTEK 91
+YH G LL G +++NLIWYG F P QR+I+ DF+ SLS+ T A PS + WWKT EK
Sbjct: 33 QYHKGALLTGDVSINLIWYGKFKPSQRAIVTDFVASLSSSRRSTMAQNPSVATWWKTVEK 92
Query: 92 Y-------KVGSSALTVGKQFLHPAYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVN 144
Y +L++G+Q L Y++GK+L K+L LA K + ++ VVLT+ DV
Sbjct: 93 YYQFRKMTTTRGLSLSLGEQILDQGYSMGKSLTEKNLKDLAAKGGQSYAVNVVLTSADVT 152
Query: 145 VEGFCMSRCGTHGSVRRGSG--GARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVA 202
V+GFCM+RCG+HG+ GSG G+R YIWVGN+ET CPGQCAWPFH P+YGPQ+PPLVA
Sbjct: 153 VQGFCMNRCGSHGT-GSGSGKKGSRFAYIWVGNSETQCPGQCAWPFHAPVYGPQSPPLVA 211
Query: 203 PNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGR 262
PN DVG+DGMVINLA+L+A T TNPF +GY+QGP APLEA +ACTGV+G G+YPGYAG
Sbjct: 212 PNNDVGLDGMVINLASLMAATATNPFGDGYYQGPKTAPLEAGSACTGVYGKGSYPGYAGE 271
Query: 263 VLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTL 299
+LVD +G SYN G NGRKYLLPA++DP+T +C TL
Sbjct: 272 LLVDATTGGSYNVKGLNGRKYLLPALFDPKTDSCSTL 308
>UniRef100_Q6Z676 Putative phi-1 [Oryza sativa]
Length = 316
Score = 321 bits (822), Expect = 2e-86
Identities = 166/316 (52%), Positives = 214/316 (67%), Gaps = 27/316 (8%)
Query: 8 AIVLSAVLLLVLPTLSFG-----ELVQEQPLV-LKYHNGQLLKGKLTVNLIWYGTFTPIQ 61
AIVL +VLLL S G EL + P L YHNG +L+G + V+++WYG FTP Q
Sbjct: 5 AIVLVSVLLLCSAHPSAGARRLMELYKPPPSEQLTYHNGTVLRGDIPVSVVWYGRFTPAQ 64
Query: 62 RSIIVDFINSLSTTGAA-LPSASAWWKTTEKYKVGSSA---------------LTVGKQF 105
++++ DF+ L+ A PS S WW T + + +A + + Q
Sbjct: 65 KAVVSDFLLLLTVASPAPTPSVSQWWNTINQLYLSKAAAQGKNGGGGGKITTQVRLAGQL 124
Query: 106 LHPAYTLGKNLKGKDLLALATKFN-ELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSG 164
+LGK+LK L ALA + + I +VLTA+DV+VEGFCMSRCGTH S +
Sbjct: 125 TDDQCSLGKSLKLSQLPALAARAKPKKGGIALVLTAQDVSVEGFCMSRCGTHAS----NA 180
Query: 165 GARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTV 224
ART Y+WVGN+ T CPGQCAWPFHQP+YGPQTP LV P+GDVG+DGMV+N+A+++AG V
Sbjct: 181 KARTAYVWVGNSATQCPGQCAWPFHQPVYGPQTPALVPPSGDVGMDGMVMNIASMVAGVV 240
Query: 225 TNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYL 284
TNPF +G++QGP APLEA TAC GV+GSGAYPGYAG + VD A+G+SYNA+GA+GRKYL
Sbjct: 241 TNPFGDGFYQGPKEAPLEAATACPGVYGSGAYPGYAGNLAVDPATGASYNANGAHGRKYL 300
Query: 285 LPAMWDPQTSACKTLV 300
LPA++DP TS C TLV
Sbjct: 301 LPALFDPATSTCSTLV 316
>UniRef100_Q67X78 Putative phi-1 [Oryza sativa]
Length = 314
Score = 320 bits (821), Expect = 2e-86
Identities = 156/278 (56%), Positives = 194/278 (69%), Gaps = 4/278 (1%)
Query: 27 LVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLSTTG-AALPSASAW 85
LV+ P+VLK H+G +L G +TVN+++YG FTP QR+++ F+ S S +PS +AW
Sbjct: 37 LVKPDPIVLKDHHGVVLSGNVTVNVLYYGRFTPAQRAVVAGFVRSASAAQHPRVPSVAAW 96
Query: 86 WKTTEKYKVGSSALTVGKQFLHPAYTLGKNLKGKDLLALATKF-NELSSITVVLTAKDVN 144
W TT Y+ G + L +G Q + +LG++L ++ AL + ++T VLTA DV
Sbjct: 97 WSTTSLYRGGGARLRLGMQVMDERMSLGRSLSLDNVTALTRAAGHHRGAVTAVLTAPDVL 156
Query: 145 VEGFCMSRCGTHGSVRR--GSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVA 202
V FCMSRCG HG G AR Y+W GN CPGQCAWPFHQP+YGPQ PPLV
Sbjct: 157 VAPFCMSRCGVHGHGGGVGAHGRARYAYLWAGNPAQQCPGQCAWPFHQPVYGPQAPPLVP 216
Query: 203 PNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGR 262
PNGDVGVDGMVI+LA LLAGTVTNPF +GY+QG A A +EA TAC GVFGSGA+PGY G+
Sbjct: 217 PNGDVGVDGMVISLAALLAGTVTNPFGDGYYQGDAGAGMEAATACAGVFGSGAFPGYPGK 276
Query: 263 VLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTLV 300
+L D +G+SYNA G GRKYLLPA+WDP TS CKTLV
Sbjct: 277 LLKDPVTGASYNAVGLAGRKYLLPALWDPTTSQCKTLV 314
>UniRef100_Q67X77 Putative phi-1 [Oryza sativa]
Length = 324
Score = 317 bits (811), Expect = 3e-85
Identities = 161/317 (50%), Positives = 207/317 (64%), Gaps = 30/317 (9%)
Query: 9 IVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDF 68
+V++ + + + LV+ QP L YHNG +L G + V+++WYG FTP Q++++ DF
Sbjct: 13 LVVAGLAVSAMADRKLMSLVKPQPNQLTYHNGAVLSGDIPVSILWYGRFTPAQKAVVTDF 72
Query: 69 INSLSTT--GAALPSASAWWKTTEKYKVGSSALTVGK----------------------Q 104
+ SL+ A PS S WW + + + S A+ VGK Q
Sbjct: 73 VLSLAAPLQAAPAPSVSQWWGSIHRLYL-SKAVAVGKNGGAHGGGGGGRAKNARVVLSGQ 131
Query: 105 FLHPAYTLGKNLKGKDLLALATKFNE-LSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGS 163
+LGK+LK L LA + + +VLTA+DV VEGFCMSRCGTHG V R
Sbjct: 132 VSDEGCSLGKSLKLSQLPTLAARARPGKGGVALVLTAQDVAVEGFCMSRCGTHGPVSR-- 189
Query: 164 GGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGT 223
A Y WVGN+ T CPGQCAWPFHQP+YGPQ PLV P+GDVG+DGMVIN+A+++AG
Sbjct: 190 --AGAAYAWVGNSATQCPGQCAWPFHQPVYGPQAAPLVPPSGDVGMDGMVINVASMVAGA 247
Query: 224 VTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKY 283
VTNPF +G++QG A LEA TACTGV+G GAYPGYAG +LVD A+G+SYNAHGA+GRKY
Sbjct: 248 VTNPFGDGFYQGERGAALEAATACTGVYGKGAYPGYAGALLVDKATGASYNAHGAHGRKY 307
Query: 284 LLPAMWDPQTSACKTLV 300
LLPA++DP TSAC TLV
Sbjct: 308 LLPALFDPDTSACSTLV 324
>UniRef100_Q6Z677 Putative phi-1 [Oryza sativa]
Length = 313
Score = 307 bits (786), Expect = 3e-82
Identities = 159/313 (50%), Positives = 208/313 (65%), Gaps = 24/313 (7%)
Query: 8 AIVLSAVLLLVLPTLSFG-----ELVQEQPL-VLKYHNGQLLKGKLTVNLIWYGTFTPIQ 61
A++L+ ++L+ +S G EL + P +L YH+G +L+G + V++ WYG FTP Q
Sbjct: 5 AVMLALLVLVSTAQVSMGARRRMELYKPDPADMLSYHSGAVLQGNIPVSIYWYGKFTPAQ 64
Query: 62 RSIIVDFINSLSTTG-AALPSASAWWKTTEKYKVGSSALT------------VGKQFLHP 108
+SI+ DF+ SLS AA PS + WW + ++ + + T V Q
Sbjct: 65 KSILFDFLLSLSVAPYAAAPSVAQWWSSIDELYLSKAVQTNSNGQSKKTQVLVASQVSDI 124
Query: 109 AYTLGKNLKGKDLLALATKFN-ELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGAR 167
++GK+L + ALA + + I +V TA+DV VEGF MSRCG HGS +
Sbjct: 125 NCSMGKSLTLAQVAALAAQAKPKKGGIALVFTAQDVTVEGFGMSRCGLHGSDAKSG---- 180
Query: 168 TPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNP 227
T YIWVGN T CPG+CAWPFHQP+YGPQ PLVAPNGD+G DGMV+NLA++LAGTVTNP
Sbjct: 181 TAYIWVGNPATQCPGECAWPFHQPMYGPQGAPLVAPNGDIGADGMVMNLASMLAGTVTNP 240
Query: 228 FNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPA 287
F +GY+QG APLEA TAC GVFGSGAYPG+AG + VD A+G+SYNA+GANGRKYLLPA
Sbjct: 241 FGDGYYQGSRDAPLEAATACPGVFGSGAYPGFAGELKVDQATGASYNANGANGRKYLLPA 300
Query: 288 MWDPQTSACKTLV 300
+++P T C TLV
Sbjct: 301 LYNPSTGTCNTLV 313
>UniRef100_Q6Z681 Putative phi-1 [Oryza sativa]
Length = 308
Score = 290 bits (742), Expect = 3e-77
Identities = 144/276 (52%), Positives = 189/276 (68%), Gaps = 14/276 (5%)
Query: 35 LKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLS-TTGAALPSASAWWKTTEKYK 93
L YH+G +L G + V+++WYG FTP Q SII DF+ SL+ AA PS WW T E+
Sbjct: 37 LTYHHGSVLSGDIPVSILWYGKFTPTQMSIIADFVVSLTGAPNAATPSVGQWWGTIEQLY 96
Query: 94 VGSSA--------LTVGKQFLHPAYTLGKNLKGKDLLALATKFN-ELSSITVVLTAKDVN 144
+ ++A + + +Q +LGK+L + LA + + + +V T +DV
Sbjct: 97 LSNAATNSQTSTRVLLDEQVSDEQCSLGKSLTLAQIDQLAARVGTKRGGVALVFTDEDVT 156
Query: 145 VEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPN 204
VEGFC SRCG HGS A T +IWVGN+ CPGQCAWPF QP+YGPQ PLVAPN
Sbjct: 157 VEGFCSSRCGKHGS----DASAGTTHIWVGNSAKQCPGQCAWPFAQPVYGPQGTPLVAPN 212
Query: 205 GDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVL 264
DVG DGMV+ LA+++AGTVTNP+ +G++QGP APLEA +AC GV+GSGAYPG AG++L
Sbjct: 213 NDVGADGMVMILASMVAGTVTNPYGDGFYQGPQDAPLEACSACPGVYGSGAYPGNAGKLL 272
Query: 265 VDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTLV 300
VD +G+SYNA+GAN RKYLLPA+++P TS+C TLV
Sbjct: 273 VDATTGASYNANGANRRKYLLPALYNPATSSCDTLV 308
>UniRef100_Q6Z680 Putative phi-1 [Oryza sativa]
Length = 302
Score = 275 bits (702), Expect = 1e-72
Identities = 140/276 (50%), Positives = 183/276 (65%), Gaps = 20/276 (7%)
Query: 35 LKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLS-TTGAALPSASAWWKTTEKYK 93
L YH+G +L G + V+++WYG FTP Q SII DF+ SL+ AA PS WW T E+
Sbjct: 37 LTYHHGSVLSGDIPVSILWYGKFTPTQMSIIADFVVSLTGAPNAATPSVGQWWGTIEQLY 96
Query: 94 VGSSA--------LTVGKQFLHPAYTLGKNLKGKDLLALATKFN-ELSSITVVLTAKDVN 144
+ ++A + + +Q +LGK+L + LA + + + +V T +DV
Sbjct: 97 LSNAATNSQTSTRVLLDEQVSDEQCSLGKSLTLAQIDQLAARVGTKRGGVALVFTDEDVT 156
Query: 145 VEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPN 204
VEGFC SRCG HGS A T +IWVGN+ CPGQCAWPF QP+YGPQ PLVAPN
Sbjct: 157 VEGFCSSRCGKHGS----DASAGTTHIWVGNSAKQCPGQCAWPFAQPVYGPQGTPLVAPN 212
Query: 205 GDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVL 264
DVG DGMV+ LA+++AGTVTNP+ +G++QGP APLEA +AC GV+GSGAYPG AG++L
Sbjct: 213 NDVGADGMVMILASMVAGTVTNPYGDGFYQGPQDAPLEACSACPGVYGSGAYPGNAGKLL 272
Query: 265 VDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTLV 300
VD +G+SYNA+GAN RKY +P TS+C TLV
Sbjct: 273 VDATTGASYNANGANRRKY------NPATSSCDTLV 302
>UniRef100_Q67X72 Putative phi-1 [Oryza sativa]
Length = 321
Score = 273 bits (699), Expect = 3e-72
Identities = 139/281 (49%), Positives = 176/281 (62%), Gaps = 22/281 (7%)
Query: 34 VLKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLSTTG-----AALPSASAWWKT 88
VL YH G +L G + V+++WYG F P Q+ I+VDF+ SL++T AA PSA+ WW T
Sbjct: 40 VLSYHGGAVLSGDIPVSIVWYGKFAPSQKDIVVDFVQSLTSTSSSSQRAATPSAAQWWST 99
Query: 89 TEKYKVGSSA------------LTVGKQFLHPAYTLGKNLKGKDLLALAT-KFNELSSIT 135
+ ++ + + Q Y+LGK L + LA + ++
Sbjct: 100 LATVYLSNATTGGGGKPAAATRVVLSGQVSDEEYSLGKTLTLVQVFQLAAGAAPKRGAVV 159
Query: 136 VVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGP 195
+VLT DV VEGFC RCG HGS GA Y WVGNAE CPGQCAWPF P YGP
Sbjct: 160 LVLTDPDVVVEGFCSVRCGVHGS----DAGAGYAYAWVGNAERQCPGQCAWPFAAPPYGP 215
Query: 196 QTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGA 255
Q PL APNGDVG DGMV+ LA+ LAG VTNPF + Y+QG A LEA TAC GV+GSG+
Sbjct: 216 QGSPLGAPNGDVGTDGMVVTLASTLAGAVTNPFGDAYYQGDKDAALEACTACAGVYGSGS 275
Query: 256 YPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSAC 296
YPGYAG+VLVD A+G SYNA G G+++LLPA+++P T+ C
Sbjct: 276 YPGYAGKVLVDEANGGSYNAIGGGGKRFLLPAIYNPATTGC 316
>UniRef100_Q67W03 Putative phi-1 [Oryza sativa]
Length = 316
Score = 267 bits (682), Expect = 3e-70
Identities = 146/312 (46%), Positives = 185/312 (58%), Gaps = 24/312 (7%)
Query: 9 IVLSAVLLLVLPTLSFGELVQEQPLVLK--------YHNGQLLKGKLTVNLIWYGTFTPI 60
+V L+V+ + P +LK YH G +L G + V L+WYG F P
Sbjct: 9 VVAMVAALVVMSLAGVSMAARRVPALLKSHVGDGISYHGGAVLGGDIPVTLVWYGKFKPA 68
Query: 61 QRSIIVDFINSLSTT--GAALPSASAWWKTTEKYKVGSSALTV--------GKQFLHPAY 110
Q++I+VDF+ SL+ T A PSA+ WW + S+A V Q Y
Sbjct: 69 QKAIVVDFLLSLTATPPNATTPSAAQWWGAIAAGYLSSNATNVTTAARVVLANQTSDEEY 128
Query: 111 TLGKNLKGKDLLALATKF-NELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTP 169
+LGK+L ++ LA + + VVLT +DV VEGFC +RCG HGS GA
Sbjct: 129 SLGKSLTLVEVFQLAAGVVPDRGDLVVVLTDRDVAVEGFCSARCGVHGS----DAGAGYA 184
Query: 170 YIWVGNAETLCPGQCAWPFHQPIYGPQ-TPPLVAPNGDVGVDGMVINLATLLAGTVTNPF 228
Y W G+AE CPGQCAWPF +P YGP+ LV PNGDVG DG+V LA +LAG VTNPF
Sbjct: 185 YAWAGDAERQCPGQCAWPFAKPPYGPKGEAALVPPNGDVGADGVVATLAGVLAGAVTNPF 244
Query: 229 NNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAM 288
+GY+ G A LEA +AC G +GS +YPGYAG+VLVD +G SYNA GA+GRKYLLPA+
Sbjct: 245 GDGYYLGDKDAALEACSACAGAYGSDSYPGYAGKVLVDETTGGSYNAVGAHGRKYLLPAV 304
Query: 289 WDPQTSACKTLV 300
+DP TS C TLV
Sbjct: 305 YDPATSRCTTLV 316
>UniRef100_Q9ATN8 Phosphate-induced protein 1-like protein [Pennisetum ciliare]
Length = 315
Score = 256 bits (654), Expect = 5e-67
Identities = 121/181 (66%), Positives = 143/181 (78%), Gaps = 2/181 (1%)
Query: 120 DLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETL 179
+LL LA + +I VVLTA DV V+GFCMSRCGTHG+ RR G R Y+WVGN T
Sbjct: 135 NLLKLAASGSPSHAINVVLTADDVAVDGFCMSRCGTHGASRRSRSG-RFAYVWVGNPATQ 193
Query: 180 CPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQ-GPAA 238
CPGQCAWPFHQP+YGPQ PL PNGDVG DGMVI+LA+++ GTVTNPF NG++Q G A
Sbjct: 194 CPGQCAWPFHQPVYGPQAAPLTPPNGDVGADGMVISLASMIVGTVTNPFGNGFYQEGSAD 253
Query: 239 APLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKT 298
APLEA TAC GV+G GAYPGYAG +LVD ASG+S+NA+GA+GRKYL+PA+ DP TS+C T
Sbjct: 254 APLEAATACAGVYGKGAYPGYAGSLLVDQASGASFNANGAHGRKYLVPALVDPDTSSCAT 313
Query: 299 L 299
L
Sbjct: 314 L 314
>UniRef100_Q7XF93 Putative phosphate-induced protein [Oryza sativa]
Length = 334
Score = 250 bits (638), Expect = 4e-65
Identities = 136/330 (41%), Positives = 184/330 (55%), Gaps = 39/330 (11%)
Query: 3 SNYYFAIVLSAVLLLVLPTLSF---GELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTP 59
S++ A ++AV+++V+ + L Q P + YH+G +L+G + V++++YG F P
Sbjct: 4 SHHLLAAAVAAVVVVVMGWSARPCEASLYQPPPPAMAYHDGAVLEGAVPVSVLYYGAFPP 63
Query: 60 IQRSIIVDFINSLSTTG-------------AALPSASAWWKTTEKYK--------VGSSA 98
R+++ DF+ SLS G A P+ + WW T E+Y G +
Sbjct: 64 HHRAVVADFLMSLSPRGRDHQPHTFGAPGPAPPPTVARWWGTVERYVRKAGRGGGAGVAR 123
Query: 99 LTVGKQFLHPAYTLGKNLKGKDLLALATKFNEL-SSITVVLTAKDVNVEGFCMSRCGTHG 157
+ + Q +LG+ L + LA + + VVLTA DV VEGFC S CG HG
Sbjct: 124 VVLASQVDDEGCSLGRRLSRAQVERLAARLGVAPGGVAVVLTAADVAVEGFCSSACGAHG 183
Query: 158 SVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTP-----------PLVAPNGD 206
S G G ++WVG+A CPG+CAWPFH YG L APNGD
Sbjct: 184 SSAPGGGAV---HVWVGDASAQCPGRCAWPFHAADYGDADAGRHRRAHGHDVALRAPNGD 240
Query: 207 VGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVD 266
GVDG+VINLA L+AG VTNP+ GYFQG AAAP+E AC GV+G GAYPGY G V VD
Sbjct: 241 AGVDGVVINLAALMAGAVTNPYGRGYFQGDAAAPVEVAGACPGVYGRGAYPGYPGAVRVD 300
Query: 267 GASGSSYNAHGANGRKYLLPAMWDPQTSAC 296
A+G+ YN G NGR+YL+PA+ DP +C
Sbjct: 301 AATGAGYNVVGRNGRRYLVPALVDPDNYSC 330
>UniRef100_Q9FPI9 At1g35140 [Arabidopsis thaliana]
Length = 244
Score = 237 bits (605), Expect = 2e-61
Identities = 116/209 (55%), Positives = 149/209 (70%), Gaps = 14/209 (6%)
Query: 36 KYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLST----TGAALPSASAWWKTTEK 91
+YH G LL G +++NLIWYG F P QR+I+ DF+ SLS+ T A PS + WWKT EK
Sbjct: 33 QYHKGALLTGDVSINLIWYGKFKPSQRAIVTDFVASLSSSRRSTMAQNPSVATWWKTVEK 92
Query: 92 Y-------KVGSSALTVGKQFLHPAYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVN 144
Y +L++G+Q L Y++GK+L K+L LA K + ++ VVLT+ DV
Sbjct: 93 YYQFRKMTTTRGLSLSLGEQILDQGYSMGKSLTEKNLKDLAAKGGQSYAVNVVLTSADVT 152
Query: 145 VEGFCMSRCGTHGSVRRGSG--GARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVA 202
V+GFCM+RCG+HG+ GSG G+R YIWVGN+ET CPGQCAWPFH P+YGPQ+PPLVA
Sbjct: 153 VQGFCMNRCGSHGT-GSGSGKKGSRFAYIWVGNSETQCPGQCAWPFHAPVYGPQSPPLVA 211
Query: 203 PNGDVGVDGMVINLATLLAGTVTNPFNNG 231
PN DVG+DGMVINLA+L+A T TNPF +G
Sbjct: 212 PNNDVGLDGMVINLASLMAATATNPFGDG 240
>UniRef100_Q8LBE0 Hypothetical protein [Arabidopsis thaliana]
Length = 337
Score = 191 bits (484), Expect = 3e-47
Identities = 107/278 (38%), Positives = 154/278 (54%), Gaps = 18/278 (6%)
Query: 35 LKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLSTTGAALPSASAWWKTTEKY-- 92
L+YH G +L +TV+ IWYGT+ Q+ II +FINS+S G+ PS S WWKT + Y
Sbjct: 55 LRYHMGPVLTNNITVHPIWYGTWQKSQKKIIREFINSISAVGSKHPSVSGWWKTVQLYTD 114
Query: 93 KVGSS---ALTVGKQFLHPAYTLGKNLKGKDLLALATK----------FNELSSITVVLT 139
+ GS+ + +G++ Y+ GK+L + ++ N S + ++LT
Sbjct: 115 QTGSNITGTVRLGEEKNDRFYSHGKSLTRLSIQSVIKSAVTSRSRPLPVNPKSGLYLLLT 174
Query: 140 AKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPP 199
A DV V+ FC CG H G PY WVGN+ LCPG CA+PF P + P P
Sbjct: 175 ADDVYVQDFCGQVCGFHYFTFPSIVGFTLPYAWVGNSAKLCPGVCAYPFXVPAFIPGLKP 234
Query: 200 LVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGP-AAAPLEAVTACTGVFGSGAYPG 258
+ +PNGDVGVDGM+ +A +A TNP N ++ GP AP+E C G++G+G
Sbjct: 235 VKSPNGDVGVDGMISVIAHEIAELATNPLVNAWYAGPDPVAPVEIADLCEGIYGTGGGGS 294
Query: 259 YAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSAC 296
Y G++L D SG++YN +G R+YL+ +W S C
Sbjct: 295 YTGQMLND-HSGATYNVNGIR-RRYLIQWLWSHVVSYC 330
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 531,664,624
Number of Sequences: 2790947
Number of extensions: 22916609
Number of successful extensions: 59220
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 59080
Number of HSP's gapped (non-prelim): 61
length of query: 300
length of database: 848,049,833
effective HSP length: 126
effective length of query: 174
effective length of database: 496,390,511
effective search space: 86371948914
effective search space used: 86371948914
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC121245.10


