

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121241.8 - phase: 0
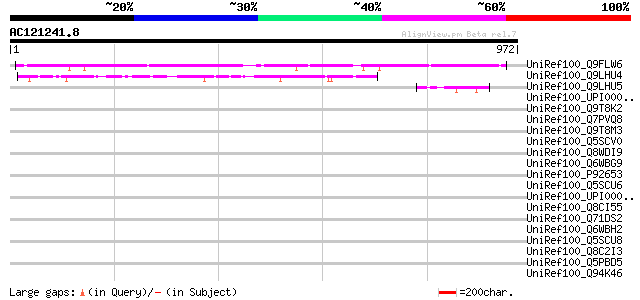
(972 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FLW6 Dbj|BAA94533.1 [Arabidopsis thaliana] 503 e-141
UniRef100_Q9LHU4 Hypothetical protein [Oryza sativa] 188 6e-46
UniRef100_Q9LHU5 Hypothetical protein [Oryza sativa] 51 1e-04
UniRef100_UPI0000364A29 UPI0000364A29 UniRef100 entry 41 0.20
UniRef100_Q9T8K2 NADH dehydrogenase subunit 2 [Liolaemus salinic... 40 0.26
UniRef100_Q7PVQ8 ENSANGP00000005315 [Anopheles gambiae str. PEST] 40 0.45
UniRef100_Q9T8M3 NADH dehydrogenase subunit 2 [Liolaemus cuyanus] 39 1.00
UniRef100_Q5SCV0 NADH dehydrogenase subunit 2 [Liolaemus cf. mel... 38 1.3
UniRef100_Q8WDI9 NADH dehydrogenase subunit 2 [Anolis transversa... 38 1.7
UniRef100_Q6WBG9 NADH dehydrogenase subunit 2 [Liolaemus canqueli] 38 1.7
UniRef100_P92653 NADH dehydrogenase subunit II [Euprepis auratus] 38 1.7
UniRef100_Q5SCU6 NADH dehydrogenase subunit 2 [Liolaemus xanthov... 38 1.7
UniRef100_UPI0000434691 UPI0000434691 UniRef100 entry 37 2.2
UniRef100_Q8CI55 Ikbke protein [Mus musculus] 37 2.2
UniRef100_Q71DS2 NADH dehydrogenase subunit 2 [Microlophus ataca... 37 2.2
UniRef100_Q6WBH2 NADH dehydrogenase subunit 2 [Liolaemus xanthov... 37 2.2
UniRef100_Q5SCU8 NADH dehydrogenase subunit 2 [Liolaemus canqueli] 37 2.2
UniRef100_Q8C2I3 Mus musculus 2 days neonate thymus thymic cells... 37 2.9
UniRef100_Q5PBD5 Hypothetical protein [Anaplasma marginale str. ... 37 2.9
UniRef100_Q94K46 Putative retroelement pol polyprotein [Arabidop... 37 2.9
>UniRef100_Q9FLW6 Dbj|BAA94533.1 [Arabidopsis thaliana]
Length = 945
Score = 503 bits (1296), Expect = e-141
Identities = 358/994 (36%), Positives = 518/994 (52%), Gaps = 110/994 (11%)
Query: 11 YSHTNRIVLLIDLDPLLLLLHNTTTTTNYIKNILSTSKTLFSFPPLSTSLFAFKFFFSSL 70
Y+ T R VLLIDL+PLL+ + Y+ ++S ++ L FPPLS SLF+FKFFFSSL
Sbjct: 7 YAKTQRFVLLIDLNPLLI----KPNSEQYLAVVISAAEKLLLFPPLSASLFSFKFFFSSL 62
Query: 71 PPHLSFSKLHPF-LPKHSFSFDHPFSTFNLLSKTLSTFPNFPL--------TYNPKAANL 121
LS SKL + SFD P T L + + + L +P+ N+
Sbjct: 63 SSLLSSSKLSSLSISSSPLSFDLPNPTLVSLKRAIDAVKGYELRSSSTVATAASPRGVNV 122
Query: 122 IDSLTQLLHDYPWEPN-SDAD-------TDTTTPLVSPNLILLFTPFFNSFNSLAGFFD- 172
+L Q+++DY WEP D + TD +V NL+++F+P + ++ F D
Sbjct: 123 AANLRQIVYDYAWEPVVRDPEIGMIPGFTDGGLDVVRSNLVVMFSPISRDLDWVSEFLDV 182
Query: 173 -SDEDSLRIENSFCDRFLGFFGNVSRRFRSKGVHCSWIGVNSDDKEDEVGMIRGLFEIGT 231
S ++ + F + F V+ F + + SWI V S D+ E+G+ G F+ G
Sbjct: 183 KSGDECFSDLDLFKSKLREIFYCVNELFDDRDIQLSWIDVKSGDERCELGLKSGFFDSGI 242
Query: 232 GKLGWGFCSLDSILLGSALVPFGLIYPKIGVSWISVRCCSREVKVQLTLEILDVNGSPIE 291
+LGWG CS DSI+ GS++VPFGLIYP IG+S S++ +Q +LEI D+NG P+E
Sbjct: 243 RELGWGHCSTDSIVFGSSVVPFGLIYPAIGIS--PKLSTSQKFTLQASLEIADINGKPME 300
Query: 292 YNC-CDLEVLDFRVFGR---GEDVNLQGGG---RKERLWNVCSDGMAKLKVTVVRKCDAF 344
C +LE +F E +NL G + L DG KL + +R CD
Sbjct: 301 CKCGGELEFSSSEIFSGKRCDEFINLASGTEPVNHDSLVEQFCDGSTKLSIKALRMCDDL 360
Query: 345 VKFRSCLSDSVLVREVLGECMKG--DSGGFFADRVLELLATEFGCQGRRKSVPVWEMLLS 402
++ D+ +V +V + + + F+AD V ++L E G + ++S P+W++LLS
Sbjct: 361 IELERYTCDTFVVHQVSQDSDQDQEEESAFWADLVFQILGKETGERVAKRSSPIWQILLS 420
Query: 403 YLYKEDCWALVSVDSGKGGGSCVGILRPFTVSSALLSVLEDPQSASDFGAANMNSFIRTG 462
YLY+E ALVS S G GIL+PFT SSAL+ V ++ S
Sbjct: 421 YLYREGFSALVSF-SNSNGSLRTGILKPFTFSSALICVFDNGVSP--------------- 464
Query: 463 ILKSDRIFHKNRDLVDSQVKDVVGIKGEQKKKMTDLSALRNLTWSSFYDLVYDQFEMDLH 522
+ D DS+ K + + K+ ++L +++W F V D ++DL
Sbjct: 465 ---------QTVDHEDSRKK----VSCSEYKRKPRKNSLNDISWEEFCRSVKDYGQIDLE 511
Query: 523 EVYYAMECNKSKKLKFLKCWMKQVKK------SSCHDLNLSEYPKPNQIFAEGTDSKLNE 576
+VY+ + +KSKKLKFLKCWMKQ+ K S + N E N I + S E
Sbjct: 512 DVYF-FKYSKSKKLKFLKCWMKQISKPRGCSLSVASNCNALEDVDANPIEEKNNSS---E 567
Query: 577 LPQNGEQPIPQVVMSAGINA-EVDTKKDDAVLDCGLETSEAFFRNLSNRIQQGIESDVID 635
P+P ++ + K++ E+SE FF +L ++I+QGIES+ ID
Sbjct: 568 ETDKASSPLPLAEEDIALSGNRISGKQESNTYVHASESSENFFVSLPSKIKQGIESEEID 627
Query: 636 LVALAERLVNSSIYWLCQKVDRETIPLIQVHSPLKDNNACGS----MVVSELVKQLLKDP 691
L ALAERLV S +++ Q+ +++ +C S +V+ EL K LLKDP
Sbjct: 628 LSALAERLVKSCVFYSSQRAEKD--------------YSCESGTLLLVIDELTKMLLKDP 673
Query: 692 KDIAAKHKSRDSSSQA----FDAAGPTIRYELQILFRLEILQSEVGRGIEDSSKQKFVKQ 747
KD+ AK K +DSSS A D AGPTIRYELQILFR+EIL+ + G G E+S QKF KQ
Sbjct: 674 KDLVAKFKKKDSSSVASERNCDEAGPTIRYELQILFRMEILRCKKGLGNEESVTQKFAKQ 733
Query: 748 ICLLLENIQCHMEGGFFGDWNLENYVAKIIKSRYSHTLEDIVHKIYNKMDLLLFVNEDEA 807
IC+ LE IQC ++GGFFG+W+L+ YV K IK+RY H L V+ IY +MDLL+F +ED
Sbjct: 734 ICMFLEGIQCKLDGGFFGEWSLDKYVDKTIKARYQHILGGAVNIIYTEMDLLMFTDED-L 792
Query: 808 PDCSFNSEDSSKSLDRKFYGD-EMGENDVGNGPFSAENKPFHLQKNVRGKLQRNIEGGHN 866
D N+EDSS+S + + + + N +K + +++ K + +G
Sbjct: 793 EDSFMNNEDSSQSGRENIHSNFKNHHHSQSNEDVPGTSKQKNTEESREAKKEVEAQGMRE 852
Query: 867 KT-LIEALKIRMPAL-RVWAPKQ-KGMKSKKDHLSKIPKR-KDRTSACYDTVCETPMT-- 920
+ + MP L RVWAPKQ K K K D + KR K++ S YD VCETPMT
Sbjct: 853 RARRFSSFTSWMPDLCRVWAPKQTKNSKGKADQQQRTAKRKKEQRSVEYDRVCETPMTTI 912
Query: 921 RNTRSMTRNT---VCETPMTRSTRSSPQSIGSDD 951
R T N CET + RS S P+++ DD
Sbjct: 913 ETKRIRTGNKDDYECET-LPRS--SVPKALFQDD 943
>UniRef100_Q9LHU4 Hypothetical protein [Oryza sativa]
Length = 684
Score = 188 bits (478), Expect = 6e-46
Identities = 196/732 (26%), Positives = 336/732 (45%), Gaps = 120/732 (16%)
Query: 16 RIVLLIDLDPLLLLLHNTTTT-----TNYIKNILSTSKTLFSFPPLSTSLFAFKFFFSSL 70
R+VLL+D+DPLL + T ++Y+ +L + +L S P + SL A + FFSSL
Sbjct: 9 RVVLLVDVDPLLPSPKASAPTAQPLASHYLAAVLPAATSLLSASP-AASLSAARLFFSSL 67
Query: 71 PPHLSFSKLHPFLPKHSFSFDHPFSTFNLLSKTLSTF--------PNFPLTYNPKAANLI 122
P LSFS L LP + P S F+L +TL++ P P ++++
Sbjct: 68 SPILSFSLLPGPLP----AAPAPLS-FHLHGETLASLAPLRRLALPACAHRRVPPSSSIA 122
Query: 123 DSLTQLLHDYPWEPNSDADTDTTTPLVSPNLILLFTPFFNSFNSLAGFFDSDEDSLRIEN 182
S+ QL HDYPW+ + ++ +PNL++LFT F G D+D
Sbjct: 123 KSILQLEHDYPWDDDPESIRRRRVFQQTPNLVVLFTAAAE-FEEFGG--DAD-------- 171
Query: 183 SFCDRFLGFFGNVSRRFRSKGVHCSWIGVNSDDKEDEVGMIRGLFEIGTGKLGWGFCSLD 242
F RF G F V R ++GV W+ V + G+ R + E LGW F + D
Sbjct: 172 -FGGRFRGVFRPVRDRLAARGVQVCWVAVGGCGE----GVRRAVTE-----LGWWFTAAD 221
Query: 243 SILLGSALVPFGLIYPKIGVSWISVRCCSREVKVQLTLEILDVNGSPIEYNCCDLEVLDF 302
++ LGSA+ GL++ +G+ + ++ LEI DV G P+ C++EV+
Sbjct: 222 AVALGSAIATPGLVWGCLGLGGEEGGS-----RGEVVLEIADVEGKPLVCKGCEVEVIG- 275
Query: 303 RVFGRGEDVNLQGGGRKERLWNVCSDGMAKLKVTVVRKCDAFVKFRSCLSDSVLVREVLG 362
W + D + K+ V V + + + + D+V+VR
Sbjct: 276 -----------------STPWRLRGDSVFKMHVKAVCEVGNWEQLITGDGDAVMVRGCFQ 318
Query: 363 ECMKGDSGG-----FFADRVLEL-LATEFGCQGRRKSVPVWEMLLSYLYKEDCWALVSVD 416
E K D FFA +++EL L + G K P+W+++L +L++++ A+VS+
Sbjct: 319 EAGKIDGEEAAEKEFFAHKIVELMLGDDKDKLGGGK--PIWQLILVFLHRKNYCAMVSIS 376
Query: 417 SGKGGGSCVGILRPFTVSSALLSVLEDPQSASDFGAANMNSFIRTGILKSDRIFHKNRDL 476
G G G++ P +++ ALL V ++ + FG ++ K L
Sbjct: 377 DGDGN-PLDGVIVPLSMNYALLHVAKN---GAGFG----------------QVVAKGPAL 416
Query: 477 VDSQVKDVVGIKGEQKKKMTDLSAL-RNLTWSSFYDLVYDQFE-----MDLHEVYYAMEC 530
+DS + D + +KK+ +S L TW SF D++ + +DL ++Y++
Sbjct: 417 LDSCMSDTSKEQSARKKRSKLVSKLFEATTWISFCDVLLKSADGSMPVVDLEDLYFSRYA 476
Query: 531 NKSKKLKFLKCWMKQVKK---SSCHDLNLSEYPKPNQIFAEGTDSKLNELPQNGEQPIPQ 587
SKK++FLKCWMKQVK+ S+ + + + + ++K L ++ P+
Sbjct: 477 ATSKKMRFLKCWMKQVKQQCLSTSSSIVAVAEEEKHLSSKDEAETKSPVLEEDASAPLVN 536
Query: 588 VVMSAGINAEVDTKKDDAVLDC-------GLETS--------EAFFRNLSNRIQQGIESD 632
+ + + D D+ ++C G ETS EAF ++ +I+Q + S+
Sbjct: 537 FSVDELVCDKEDKPMDE--INCNKVDKPVGDETSDFSSMEDLEAFLDSVPQKIEQSLCSE 594
Query: 633 VIDLVALAERLVNSSIYWLCQKVDRETIPLIQVHSPLKDNNACGSMVVSELVKQLLKDPK 692
DL LA RLV S++ L K + + ++ + + + + E LL PK
Sbjct: 595 DADLGNLAGRLVGLSVHALMIKHGKIS---VRYSNRGEVEDVSDGKIACEASGILLVKPK 651
Query: 693 DIAAKHKSRDSS 704
++ AK+K R+++
Sbjct: 652 ELVAKYKDRNTA 663
>UniRef100_Q9LHU5 Hypothetical protein [Oryza sativa]
Length = 182
Score = 51.2 bits (121), Expect = 1e-04
Identities = 45/154 (29%), Positives = 65/154 (41%), Gaps = 27/154 (17%)
Query: 780 RYSHTLEDIVHKIYNKMDLLLFVNEDEAPDCSFNSEDSSKSLDRKFYGDEMGENDVGNGP 839
RY ++ED++ KIY +M+ LF D+ +CS + SS + D N
Sbjct: 6 RYIESMEDVIKKIYTEMEFDLF---DDEVECSESLPSSSNH-----------DVDGSNSR 51
Query: 840 FSAENKPFHLQKNVRG-------KLQRNIEGGHNKTLIEALKIRMPALR-VWAPKQKGMK 891
N HL + G +L R E + + + +P LR VWA K G +
Sbjct: 52 RHRSNSAPHLLRRDHGGGSSHEERLARAEERRNRDRRLSSFTSWVPDLRRVWALKHPGKE 111
Query: 892 SK-----KDHLSKIPKRKDRTSACYDTVCETPMT 920
+ +RK R +AC D VCETPMT
Sbjct: 112 PAAAAPPQSRQGASKRRKRRRAACTDMVCETPMT 145
>UniRef100_UPI0000364A29 UPI0000364A29 UniRef100 entry
Length = 1157
Score = 40.8 bits (94), Expect = 0.20
Identities = 32/133 (24%), Positives = 54/133 (40%), Gaps = 8/133 (6%)
Query: 803 NEDEAPDCSFNSEDSSKSLDRKFYGDEMGENDVGNGPFSAENKPFHLQKNVRGKLQRNIE 862
++D+ D S SK+ D + + DE N + + +K + Q V + R+
Sbjct: 790 DDDDDDDDDAESSRLSKTSDSRSFPDEQRRNYLDKRAYGCVSKVYLKQGFVEFQALRHDA 849
Query: 863 GGHNKTLIEALKIRMPALRVWAPKQKGMKSKKDHLSKIPKRKDRTSA-CYDTVCETPMTR 921
G N T + P R W + ++ K H +IP+ + RTSA ++P
Sbjct: 850 GPQNWTAL-------PETRTWTSAEANIQEKSKHRVQIPQHESRTSAPAIQQRTQSPALV 902
Query: 922 NTRSMTRNTVCET 934
TVC+T
Sbjct: 903 TGELPALVTVCDT 915
>UniRef100_Q9T8K2 NADH dehydrogenase subunit 2 [Liolaemus salinicola]
Length = 344
Score = 40.4 bits (93), Expect = 0.26
Identities = 41/149 (27%), Positives = 62/149 (41%), Gaps = 15/149 (10%)
Query: 18 VLLIDLDPLLLLLHNTTTTTNYIKNILSTSKTL------FSFPPLSTSLFAFKFF-FSSL 70
+L I+L LL+ TTT ++ I++ SKT+ +S P+ T+ A L
Sbjct: 200 ILAINLAIYLLM-----TTTMFLMLIMTKSKTIQDLSLSWSLSPMITTFMALTLLSLGGL 254
Query: 71 PPHLSFSKLHPFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLH 130
PP F + + + P +T +S LS + LTY + L S +
Sbjct: 255 PPLTGFMPKWLIIEELTLQNLTPMATMLAMSALLSLYFYLRLTYT---STLTMSPNSTMT 311
Query: 131 DYPWEPNSDADTDTTTPLVSPNLILLFTP 159
Y W N T T PL + L+L TP
Sbjct: 312 KYKWRYNMKNHTIFTIPLPAALLLLPITP 340
>UniRef100_Q7PVQ8 ENSANGP00000005315 [Anopheles gambiae str. PEST]
Length = 236
Score = 39.7 bits (91), Expect = 0.45
Identities = 42/173 (24%), Positives = 73/173 (41%), Gaps = 19/173 (10%)
Query: 564 QIFAEGTDSKLNELPQNGEQPIPQVVMSAGINAEVDTKKDDAVL-DCGLETSEAFFRNLS 622
Q+ A GT K NE Q G P + A +NAE+D K + + D L +E R+L
Sbjct: 44 QLAAGGTAGKPNEPTQPGVPPEEINAVRAELNAEIDKLKSEILAKDQKLHLTEELKRSLE 103
Query: 623 NRIQ---------QGIESDVIDLVALAERLVNSSIYWLCQKVDRETIPLIQVHSPLKDNN 673
N IQ + + + + ++E + I +L +VD ++ L + L+
Sbjct: 104 NEIQNLHEKVADAERAAASKLSALTISEECLKEQINYLEHRVDEQSDQLTVKDAELEKQY 163
Query: 674 ACGSMVVSELVKQL------LKDPKDIAAKHKSRDSSSQAFDAAGPTIRYELQ 720
SE ++L L+ KD+ A+ RD Q + +R +++
Sbjct: 164 LALKNAESEQEERLAALQDELRSKKDLLAE---RDQQVQLLEQTVEELRADVR 213
>UniRef100_Q9T8M3 NADH dehydrogenase subunit 2 [Liolaemus cuyanus]
Length = 345
Score = 38.5 bits (88), Expect = 1.00
Identities = 38/140 (27%), Positives = 60/140 (42%), Gaps = 11/140 (7%)
Query: 28 LLLHNTTTTTNYIKNILSTSKTL------FSFPPLSTSLFAFKFF-FSSLPPHLSFSKLH 80
L+++ TTT ++ I++ SKT+ +S P+ TSL LPP F
Sbjct: 205 LIIYLLMTTTMFLMLIMTKSKTIQDLSLSWSMSPMITSLTTLVLLSLGGLPPLTGFIPKW 264
Query: 81 PFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLHDYPWEPNSDA 140
+ + + P +T +S LS + LTY + L S + + W N+
Sbjct: 265 LIIEELTLQSLTPLATTLAMSALLSLYFYLRLTY---TSTLTMSPNSAMTKHKWRFNTKT 321
Query: 141 DTDT-TTPLVSPNLILLFTP 159
T TTPL + L+L TP
Sbjct: 322 PTTIFTTPLPAALLLLPITP 341
>UniRef100_Q5SCV0 NADH dehydrogenase subunit 2 [Liolaemus cf. melanops]
Length = 345
Score = 38.1 bits (87), Expect = 1.3
Identities = 38/140 (27%), Positives = 60/140 (42%), Gaps = 11/140 (7%)
Query: 28 LLLHNTTTTTNYIKNILSTSKTL------FSFPPLSTSLFAFKFF-FSSLPPHLSFSKLH 80
L+++ TTT ++ I++ SKT+ +S P+ TSL LPP F
Sbjct: 205 LIIYLLMTTTMFLMMIMTKSKTIQDLSLSWSMSPMITSLTTLVLLSLGGLPPLTGFIPKW 264
Query: 81 PFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLHDYPWEPNSDA 140
+ + + P +T +S LS + LTY + L S + + W N+
Sbjct: 265 LIIEELTLQSLTPLATTLAMSALLSLYFYLRLTY---TSALTMSPNSAMTKHKWRFNTKT 321
Query: 141 DTDT-TTPLVSPNLILLFTP 159
T TTPL + L+L TP
Sbjct: 322 PTTIFTTPLPAALLLLPITP 341
>UniRef100_Q8WDI9 NADH dehydrogenase subunit 2 [Anolis transversalis]
Length = 345
Score = 37.7 bits (86), Expect = 1.7
Identities = 44/148 (29%), Positives = 61/148 (40%), Gaps = 16/148 (10%)
Query: 28 LLLHNTTTTTNYIKNILSTSKTL-------FSFPPLSTSLFAFKFFFSSLPPHLSFSKLH 80
LL++ T TTT + ILS SKT+ + P ++T + LPP F
Sbjct: 205 LLMYLTMTTTMFCLLILSKSKTIQDTANTWTTSPTMTTIMMLTLLSLGGLPPLTGFLPKW 264
Query: 81 PFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLHDYPWEPNSDA 140
L + + P +T LS LS F LTY +A L S Y W S
Sbjct: 265 LILEELTMQNLLPMATIMALSSLLSLFFYLRLTY---SATLTLSPNNAQTKYKWRFKS-- 319
Query: 141 DTDTTTPLV--SPNLILLFTPFFNSFNS 166
T T PL+ +P +LL P + N+
Sbjct: 320 -TMNTLPLILMTPASLLLM-PILPTMNN 345
>UniRef100_Q6WBG9 NADH dehydrogenase subunit 2 [Liolaemus canqueli]
Length = 345
Score = 37.7 bits (86), Expect = 1.7
Identities = 38/140 (27%), Positives = 60/140 (42%), Gaps = 11/140 (7%)
Query: 28 LLLHNTTTTTNYIKNILSTSKTL------FSFPPLSTSLFAFKFF-FSSLPPHLSFSKLH 80
L+++ TTT ++ I++ SKT+ +S P+ TSL LPP F
Sbjct: 205 LIIYLLMTTTMFLMLIMTKSKTIQDLSLSWSMSPMITSLTTLVLLSLGGLPPLTGFIPKW 264
Query: 81 PFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLHDYPWEPNSDA 140
+ + + P +T +S LS + LTY + L S + + W N+
Sbjct: 265 LIIEELTLQSLTPLATTLAMSALLSLYFYLRLTY---TSALTMSPNSAMTKHKWRFNTKT 321
Query: 141 DTDT-TTPLVSPNLILLFTP 159
T TTPL + L+L TP
Sbjct: 322 PTTIFTTPLPAALLLLPITP 341
>UniRef100_P92653 NADH dehydrogenase subunit II [Euprepis auratus]
Length = 345
Score = 37.7 bits (86), Expect = 1.7
Identities = 41/165 (24%), Positives = 71/165 (42%), Gaps = 21/165 (12%)
Query: 12 SHTNRIVLLIDLDP----LLLLLHNTTTTTNYIKNILSTSKTLFSF-------PPLSTSL 60
+H + ++ L+P L LLL+ TT ++ L+TSKT+ PP++ ++
Sbjct: 185 AHLGWMASILTLNPNILILNLLLYIIMTTPMFLMLELTTSKTIKDLTTSWTISPPMTATM 244
Query: 61 FAFKFFFSSLPPHLSFSKLHPFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAAN 120
A LPP F L + + +T +LS LS F L+Y
Sbjct: 245 MALLMSLGGLPPLTGFMPKWLVLQELTTHNLTMTATIMVLSALLSLFFYLRLSYT-STLT 303
Query: 121 LIDSLTQLLHDYPWEPNSDADTDTTTPLVSPN----LILLFTPFF 161
+ + T+ + + ++P TT P+++P L+L TP F
Sbjct: 304 VYPTTTKSSYKWRFQPKL-----TTMPIMAPTVLSLLLLPMTPMF 343
>UniRef100_Q5SCU6 NADH dehydrogenase subunit 2 [Liolaemus xanthoviridis]
Length = 345
Score = 37.7 bits (86), Expect = 1.7
Identities = 38/140 (27%), Positives = 60/140 (42%), Gaps = 11/140 (7%)
Query: 28 LLLHNTTTTTNYIKNILSTSKTL------FSFPPLSTSLFAFKFF-FSSLPPHLSFSKLH 80
L+++ TTT ++ I++ SKT+ +S P+ TSL LPP F
Sbjct: 205 LIIYLLMTTTMFLMLIMTKSKTIQDLSLSWSISPMITSLTTLVLLSLGGLPPLTGFIPKW 264
Query: 81 PFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLHDYPWEPNSDA 140
+ + + P +T +S LS + LTY + L S + + W N+
Sbjct: 265 LIIEELTLQSLTPLATTLAMSALLSLYFYLRLTY---TSALTMSPNSAMTKHKWRFNTKT 321
Query: 141 DTDT-TTPLVSPNLILLFTP 159
T TTPL + L+L TP
Sbjct: 322 PTTIFTTPLPAALLLLPITP 341
>UniRef100_UPI0000434691 UPI0000434691 UniRef100 entry
Length = 693
Score = 37.4 bits (85), Expect = 2.2
Identities = 31/125 (24%), Positives = 54/125 (42%), Gaps = 7/125 (5%)
Query: 419 KGGGSCVGILRPFTVSSALLSVLEDPQSASDFGAANMNSFIRTGI-----LKSDRIFHKN 473
+GG ++ + S +LLSVLEDP++ +R + L+ + I H
Sbjct: 52 QGGSRQKVLIMEYCSSGSLLSVLEDPENTFGLSEEEFLVVLRCVVAGMNHLRENGIVH-- 109
Query: 474 RDLVDSQVKDVVGIKGEQKKKMTDLSALRNLTWSSFYDLVYDQFEMDLHEVYYAMECNKS 533
RD+ + +VG +G+ K++D A R L + VY E ++Y K
Sbjct: 110 RDIKPGNIMRLVGEEGQSIYKLSDFGAARKLDDDEKFVSVYGTEEYLHPDMYERAVLRKP 169
Query: 534 KKLKF 538
++ F
Sbjct: 170 QQKAF 174
>UniRef100_Q8CI55 Ikbke protein [Mus musculus]
Length = 693
Score = 37.4 bits (85), Expect = 2.2
Identities = 31/125 (24%), Positives = 54/125 (42%), Gaps = 7/125 (5%)
Query: 419 KGGGSCVGILRPFTVSSALLSVLEDPQSASDFGAANMNSFIRTGI-----LKSDRIFHKN 473
+GG ++ + S +LLSVLEDP++ +R + L+ + I H
Sbjct: 52 QGGSRQKVLIMEYCSSGSLLSVLEDPENTFGLSEEEFLVVLRCVVAGMNHLRENGIVH-- 109
Query: 474 RDLVDSQVKDVVGIKGEQKKKMTDLSALRNLTWSSFYDLVYDQFEMDLHEVYYAMECNKS 533
RD+ + +VG +G+ K++D A R L + VY E ++Y K
Sbjct: 110 RDIKPGNIMRLVGEEGQSIYKLSDFGAARKLDDDEKFVSVYGTEEYLHPDMYERAVLRKP 169
Query: 534 KKLKF 538
++ F
Sbjct: 170 QQKAF 174
>UniRef100_Q71DS2 NADH dehydrogenase subunit 2 [Microlophus atacamensis]
Length = 344
Score = 37.4 bits (85), Expect = 2.2
Identities = 47/161 (29%), Positives = 69/161 (42%), Gaps = 25/161 (15%)
Query: 14 TNRIVLLIDLDPLLLLLHNTTTTTNYIKNILSTSKTL-------FSFPPLSTSLFAFKFF 66
T +L +L L L+++ T TTT + ILS SKT+ + P L+T +
Sbjct: 191 TTASFMLTNLMVLNLIIYITMTTTMFSTLILSNSKTIKDTTTLPTTSPALTTIMALTLLS 250
Query: 67 FSSLPPHLSFSKLHPFLPKHSFSFD------HPFSTFNLLSKTLSTFPNFPLTYNPKAAN 120
LPP L F+PK + P ++ +S LS F LTY
Sbjct: 251 LGGLPP------LSGFMPKWLIMEELINQSLIPAASLMAMSALLSLFFYLRLTYT-TTMT 303
Query: 121 LIDSLTQLLHDYPWEPNSDADT--DTTTPLVSPNLILLFTP 159
L + T +H + + PN + + TTTPL L+L TP
Sbjct: 304 LSPNQTTAMHKWRF-PNQNMNVLISTTTPLTL--LLLPMTP 341
>UniRef100_Q6WBH2 NADH dehydrogenase subunit 2 [Liolaemus xanthoviridis]
Length = 345
Score = 37.4 bits (85), Expect = 2.2
Identities = 38/140 (27%), Positives = 59/140 (42%), Gaps = 11/140 (7%)
Query: 28 LLLHNTTTTTNYIKNILSTSKTL------FSFPPLSTSLFAFKFF-FSSLPPHLSFSKLH 80
L+++ TTT ++ I++ SKT+ +S P+ TSL LPP F
Sbjct: 205 LIIYLLMTTTMFLMLIMTKSKTIQDLSLSWSMSPMITSLTTLVLLSLGGLPPLTGFIPKW 264
Query: 81 PFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLHDYPWEPNSDA 140
+ + + P +T +S LS + LTY +S T + W N+
Sbjct: 265 LIIEELTLQSLTPLATALAMSALLSLYFYLRLTYTSALTMSPNSATT---KHKWRFNTKT 321
Query: 141 DTDT-TTPLVSPNLILLFTP 159
T TTPL + L+L TP
Sbjct: 322 PTTIFTTPLPAALLLLPITP 341
>UniRef100_Q5SCU8 NADH dehydrogenase subunit 2 [Liolaemus canqueli]
Length = 345
Score = 37.4 bits (85), Expect = 2.2
Identities = 38/140 (27%), Positives = 59/140 (42%), Gaps = 11/140 (7%)
Query: 28 LLLHNTTTTTNYIKNILSTSKTL------FSFPPLSTSLFAFKFF-FSSLPPHLSFSKLH 80
L+++ TTT ++ I++ SKT+ +S P+ TSL LPP F
Sbjct: 205 LIIYLLMTTTMFLMLIMTKSKTIQDLSLSWSMSPMITSLTTLVLLSLGGLPPLTGFIPKW 264
Query: 81 PFLPKHSFSFDHPFSTFNLLSKTLSTFPNFPLTYNPKAANLIDSLTQLLHDYPWEPNSDA 140
+ + + P +T +S LS + LTY +S T + W N+
Sbjct: 265 LIIEELTLQSLTPLATTLAMSALLSLYFYLRLTYTSALTMSPNSATT---KHKWRFNTKT 321
Query: 141 DTDT-TTPLVSPNLILLFTP 159
T TTPL + L+L TP
Sbjct: 322 PTTIFTTPLPAALLLLPITP 341
>UniRef100_Q8C2I3 Mus musculus 2 days neonate thymus thymic cells cDNA, RIKEN full-
length enriched library, clone:E430020M09
product:inhibitor of kappaB kinase epsilon, full insert
sequence [Mus musculus]
Length = 717
Score = 37.0 bits (84), Expect = 2.9
Identities = 31/124 (25%), Positives = 53/124 (42%), Gaps = 7/124 (5%)
Query: 420 GGGSCVGILRPFTVSSALLSVLEDPQSASDFGAANMNSFIRTGI-----LKSDRIFHKNR 474
GG ++ + S +LLSVLEDP++ +R + L+ + I H R
Sbjct: 77 GGSRQKVLIMEYCSSGSLLSVLEDPENTFGLSEEEFLVVLRCVVAGMNHLRENGIVH--R 134
Query: 475 DLVDSQVKDVVGIKGEQKKKMTDLSALRNLTWSSFYDLVYDQFEMDLHEVYYAMECNKSK 534
D+ + +VG +G+ K++D A R L + VY E ++Y K +
Sbjct: 135 DIKPGNIMRLVGEEGQSIYKLSDFGAARKLDDDEKFVSVYGTEEYLHPDMYERAVLRKPQ 194
Query: 535 KLKF 538
+ F
Sbjct: 195 QKAF 198
>UniRef100_Q5PBD5 Hypothetical protein [Anaplasma marginale str. St. Maries]
Length = 382
Score = 37.0 bits (84), Expect = 2.9
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 10/103 (9%)
Query: 120 NLIDSLTQLLHD--YPWEPNSDADTDTTTPLVSPNLILLFTPFFNSFNSLAGFFDSDEDS 177
NL+ SL L D Y W+ + SPNL +L F SFN LA FF+ + D
Sbjct: 36 NLLTSLLLSLGDGFYLWDEKKRVER------FSPNLQMLLNSVFCSFNELANFFE-ESDQ 88
Query: 178 LRIENSFCDRFLGFFGNVSRRFRSKGVHCSWIGVNSDDKEDEV 220
LR +N R + + + + ++CS G + D +D++
Sbjct: 89 LR-KNFTEARKINKSFTMDLKGKDAEIYCSCYGQSIVDDQDKI 130
>UniRef100_Q94K46 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 405
Score = 37.0 bits (84), Expect = 2.9
Identities = 30/105 (28%), Positives = 47/105 (44%), Gaps = 5/105 (4%)
Query: 784 TLEDIVHKIYNKMDLLLFVNEDEA--PDCSFN---SEDSSKSLDRKFYGDEMGENDVGNG 838
+LED++ K+ D L E+ A P +FN S+DS+ S Y G+++ G
Sbjct: 206 SLEDVIPKLTGYDDRLQGYLEETAVSPHVAFNITTSDDSNASGYFNAYNRGKGKSNRGRN 265
Query: 839 PFSAENKPFHLQKNVRGKLQRNIEGGHNKTLIEALKIRMPALRVW 883
FS + FH Q + + GG + K+ PAL+ W
Sbjct: 266 SFSTRGRGFHQQISSTNSSSGSQSGGTSVVCQICGKMGHPALKCW 310
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,645,819,122
Number of Sequences: 2790947
Number of extensions: 72237437
Number of successful extensions: 160347
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 160302
Number of HSP's gapped (non-prelim): 44
length of query: 972
length of database: 848,049,833
effective HSP length: 137
effective length of query: 835
effective length of database: 465,690,094
effective search space: 388851228490
effective search space used: 388851228490
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC121241.8


