

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121238.7 - phase: 0
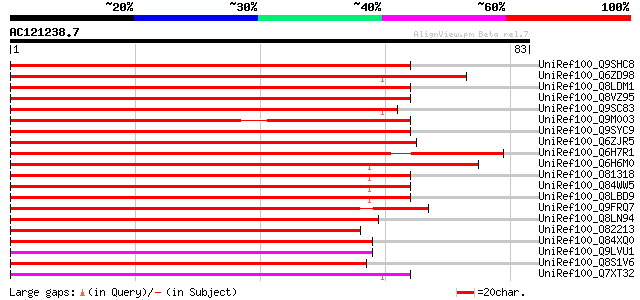
(83 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SHC8 Putative VAMP-associated protein (Putative VAMP... 88 5e-17
UniRef100_Q6ZD98 Putative vesicle-associated membrane protein-as... 86 2e-16
UniRef100_Q8LDM1 Putative VAMP-associated protein [Arabidopsis t... 86 3e-16
UniRef100_Q8VZ95 Hypothetical protein At3g60600; T4C21_10 [Arabi... 85 4e-16
UniRef100_Q9SC83 VAP27 [Nicotiana plumbaginifolia] 83 1e-15
UniRef100_Q9M003 Hypothetical protein T4C21_10 [Arabidopsis thal... 75 4e-13
UniRef100_Q9SYC9 F11M15.13 protein [Arabidopsis thaliana] 74 7e-13
UniRef100_Q6ZJR5 Putative 27k vesicle-associated membrane protei... 72 3e-12
UniRef100_Q6H7R1 Putative vesicle-associated membrane protein-as... 72 4e-12
UniRef100_Q6H6M0 Vesicle-associated membraneprotein-like [Oryza ... 72 4e-12
UniRef100_O81318 F6N15.21 protein [Arabidopsis thaliana] 70 1e-11
UniRef100_Q84WW5 Putative proline-rich protein [Arabidopsis thal... 70 1e-11
UniRef100_Q8LBD9 Putative proline-rich protein [Arabidopsis thal... 70 1e-11
UniRef100_Q9FRQ7 F22O13.31 [Arabidopsis thaliana] 67 1e-10
UniRef100_Q8LN94 Putative vesicle-associated membrane protein [O... 66 2e-10
UniRef100_O82213 Hypothetical protein At2g23830 [Arabidopsis tha... 63 2e-09
UniRef100_Q84XQ0 Putative VAMP-associated protein [Brassica rapa... 60 1e-08
UniRef100_Q9LVU1 VAMP (Vesicle-associated membrane protein)-asso... 59 4e-08
UniRef100_Q8S1V6 P0504E02.2 protein [Oryza sativa] 57 1e-07
UniRef100_Q7XT32 OSJNBa0010H02.20 protein [Oryza sativa] 56 3e-07
>UniRef100_Q9SHC8 Putative VAMP-associated protein (Putative VAMP (Vesicle-associated
membrane protein)-associated protein) [Arabidopsis
thaliana]
Length = 239
Score = 88.2 bits (217), Expect = 5e-17
Identities = 40/64 (62%), Positives = 51/64 (79%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +EAP D+QCKDKFLLQ +VA PGAT KD+T +MF K++G+ V+E+K R V VAPP
Sbjct: 70 MQAQKEAPADLQCKDKFLLQCVVASPGATPKDVTHEMFSKEAGHRVEETKLRVVYVAPPR 129
Query: 61 SPNP 64
P+P
Sbjct: 130 PPSP 133
>UniRef100_Q6ZD98 Putative vesicle-associated membrane protein-associated protein
[Oryza sativa]
Length = 243
Score = 86.3 bits (212), Expect = 2e-16
Identities = 44/75 (58%), Positives = 52/75 (68%), Gaps = 2/75 (2%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP-- 58
MQA EAPPDMQCKDKFL+QS++A G T KDIT +MF K+SG +V+E K R +AP
Sbjct: 77 MQAQREAPPDMQCKDKFLVQSVIAPSGVTVKDITGEMFTKESGNKVEEVKLRVTYIAPPQ 136
Query: 59 PPSPNPPLSVHASES 73
PPSP P S S S
Sbjct: 137 PPSPVPEESEEGSPS 151
>UniRef100_Q8LDM1 Putative VAMP-associated protein [Arabidopsis thaliana]
Length = 240
Score = 85.5 bits (210), Expect = 3e-16
Identities = 37/64 (57%), Positives = 49/64 (75%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +EAP DMQCKDKFLLQ ++A PG T K++T +MF K++G+ V+E+K R VAPP
Sbjct: 71 MQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEMFSKEAGHRVEETKLRVTYVAPPQ 130
Query: 61 SPNP 64
P+P
Sbjct: 131 PPSP 134
>UniRef100_Q8VZ95 Hypothetical protein At3g60600; T4C21_10 [Arabidopsis thaliana]
Length = 256
Score = 85.1 bits (209), Expect = 4e-16
Identities = 37/64 (57%), Positives = 49/64 (75%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +EAP DMQCKDKFLLQ ++A PG T K++T +MF K++G+ V+E+K R VAPP
Sbjct: 87 MQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEMFSKEAGHRVEETKLRVTYVAPPR 146
Query: 61 SPNP 64
P+P
Sbjct: 147 PPSP 150
>UniRef100_Q9SC83 VAP27 [Nicotiana plumbaginifolia]
Length = 240
Score = 83.2 bits (204), Expect = 1e-15
Identities = 41/64 (64%), Positives = 48/64 (74%), Gaps = 2/64 (3%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP-- 58
MQA +EAP DMQCKDKFLLQS+VA PGAT KDIT +MF K+ G V++ K R + V P
Sbjct: 73 MQAQKEAPADMQCKDKFLLQSVVASPGATAKDITPEMFNKEEGNHVEDCKLRVIYVPPQQ 132
Query: 59 PPSP 62
PPSP
Sbjct: 133 PPSP 136
>UniRef100_Q9M003 Hypothetical protein T4C21_10 [Arabidopsis thaliana]
Length = 250
Score = 75.1 bits (183), Expect = 4e-13
Identities = 35/64 (54%), Positives = 46/64 (71%), Gaps = 4/64 (6%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +EAP DMQCKDKFLLQ ++A PG T K++T +M +G+ V+E+K R VAPP
Sbjct: 87 MQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEM----AGHRVEETKLRVTYVAPPR 142
Query: 61 SPNP 64
P+P
Sbjct: 143 PPSP 146
>UniRef100_Q9SYC9 F11M15.13 protein [Arabidopsis thaliana]
Length = 571
Score = 74.3 bits (181), Expect = 7e-13
Identities = 32/64 (50%), Positives = 45/64 (70%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQAL+EAP DMQC+DK L Q V P T KD+TS+MF K++G+ +E++ + + V PP
Sbjct: 241 MQALKEAPADMQCRDKLLFQCKVVEPETTAKDVTSEMFSKEAGHPAEETRLKVMYVTPPQ 300
Query: 61 SPNP 64
P+P
Sbjct: 301 PPSP 304
Score = 73.2 bits (178), Expect = 2e-12
Identities = 33/64 (51%), Positives = 45/64 (69%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQAL+EAP D QCKDK L Q V PG K++TS+MF K++G+ V+E+ F+ + VAPP
Sbjct: 71 MQALKEAPADRQCKDKLLFQCKVVEPGTMDKEVTSEMFSKEAGHRVEETIFKIIYVAPPQ 130
Query: 61 SPNP 64
+P
Sbjct: 131 PQSP 134
>UniRef100_Q6ZJR5 Putative 27k vesicle-associated membrane protein-associated protein
[Oryza sativa]
Length = 237
Score = 72.4 bits (176), Expect = 3e-12
Identities = 37/65 (56%), Positives = 45/65 (68%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA EAPPDMQCKDKFL+QS+ A GATT+DI+++MF K +G V+E K R V V
Sbjct: 70 MQAQREAPPDMQCKDKFLVQSVAAENGATTQDISAEMFNKVAGKVVEEFKLRVVYVPTTT 129
Query: 61 SPNPP 65
S P
Sbjct: 130 SSAMP 134
>UniRef100_Q6H7R1 Putative vesicle-associated membrane protein-associated protein
[Oryza sativa]
Length = 259
Score = 71.6 bits (174), Expect = 4e-12
Identities = 39/79 (49%), Positives = 48/79 (60%), Gaps = 3/79 (3%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA EAPPDMQCKDKFL+QS + T KDIT DMF K+SG V E K + V P P
Sbjct: 107 MQAQREAPPDMQCKDKFLVQSAIVTQELTPKDITGDMFTKESGNVVDEVKLKVVYTQPHP 166
Query: 61 SPNPPLSVHASESLDQVSF 79
+ L+ + E L +S+
Sbjct: 167 T---SLNGGSEEGLGSLSY 182
>UniRef100_Q6H6M0 Vesicle-associated membraneprotein-like [Oryza sativa]
Length = 136
Score = 71.6 bits (174), Expect = 4e-12
Identities = 39/76 (51%), Positives = 47/76 (61%), Gaps = 1/76 (1%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV-APP 59
MQ +E P D CKDKFL+QS+V G T KDI SDMF K++G V+E K R V + A P
Sbjct: 1 MQPPKEIPTDYNCKDKFLIQSVVVEDGTTQKDIHSDMFSKEAGKVVEEFKLRVVYIPANP 60
Query: 60 PSPNPPLSVHASESLD 75
PSP P +SLD
Sbjct: 61 PSPVPEEEEDEIDSLD 76
>UniRef100_O81318 F6N15.21 protein [Arabidopsis thaliana]
Length = 399
Score = 70.1 bits (170), Expect = 1e-11
Identities = 33/65 (50%), Positives = 48/65 (73%), Gaps = 1/65 (1%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV-APP 59
MQA +EAP DMQCKDKFL+Q++V G T+K++ ++MF K++G +++ K R V + A P
Sbjct: 84 MQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNKEAGRVIEDFKLRVVYIPANP 143
Query: 60 PSPNP 64
PSP P
Sbjct: 144 PSPVP 148
>UniRef100_Q84WW5 Putative proline-rich protein [Arabidopsis thaliana]
Length = 239
Score = 70.1 bits (170), Expect = 1e-11
Identities = 33/65 (50%), Positives = 48/65 (73%), Gaps = 1/65 (1%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV-APP 59
MQA +EAP DMQCKDKFL+Q++V G T+K++ ++MF K++G +++ K R V + A P
Sbjct: 72 MQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNKEAGRVIEDFKLRVVYIPANP 131
Query: 60 PSPNP 64
PSP P
Sbjct: 132 PSPVP 136
>UniRef100_Q8LBD9 Putative proline-rich protein [Arabidopsis thaliana]
Length = 239
Score = 70.1 bits (170), Expect = 1e-11
Identities = 33/65 (50%), Positives = 48/65 (73%), Gaps = 1/65 (1%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV-APP 59
MQA +EAP DMQCKDKFL+Q++V G T+K++ ++MF K++G +++ K R V + A P
Sbjct: 72 MQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNKEAGRVIEDFKLRVVYIPANP 131
Query: 60 PSPNP 64
PSP P
Sbjct: 132 PSPVP 136
>UniRef100_Q9FRQ7 F22O13.31 [Arabidopsis thaliana]
Length = 432
Score = 67.0 bits (162), Expect = 1e-10
Identities = 34/67 (50%), Positives = 42/67 (61%), Gaps = 2/67 (2%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +E PPDM CKDKFL+QS T +DIT+ MF K G ++E+K R V PP
Sbjct: 70 MQAFKEPPPDMVCKDKFLIQSTAVSAETTDEDITASMFSKAEGKHIEENKLRVTLV--PP 127
Query: 61 SPNPPLS 67
S +P LS
Sbjct: 128 SDSPELS 134
>UniRef100_Q8LN94 Putative vesicle-associated membrane protein [Oryza sativa]
Length = 275
Score = 65.9 bits (159), Expect = 2e-10
Identities = 32/59 (54%), Positives = 39/59 (65%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPP 59
MQA APPD+QCKDKFL+QS+V G + KDITS MF +D V+E K + V PP
Sbjct: 72 MQAQIVAPPDLQCKDKFLVQSVVVDDGLSAKDITSQMFLRDENNMVEEVKLKVSYVMPP 130
>UniRef100_O82213 Hypothetical protein At2g23830 [Arabidopsis thaliana]
Length = 149
Score = 63.2 bits (152), Expect = 2e-09
Identities = 28/56 (50%), Positives = 41/56 (73%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV 56
MQA +E P DMQ +KF++QS++A PG T K++T +MF K+SG+ V+E+K R V
Sbjct: 71 MQAQKEVPSDMQSFEKFMIQSVLASPGVTAKEVTREMFSKESGHVVEETKLRVTYV 126
>UniRef100_Q84XQ0 Putative VAMP-associated protein [Brassica rapa subsp.
pekinensis]
Length = 165
Score = 60.5 bits (145), Expect = 1e-08
Identities = 28/58 (48%), Positives = 35/58 (60%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP 58
+QA E PPDMQCKDKFLLQS + P D+ D F KDSG + E K + ++P
Sbjct: 21 LQAQREYPPDMQCKDKFLLQSTIVPPHTDVDDLPQDTFTKDSGKTLTECKLKVSYISP 78
>UniRef100_Q9LVU1 VAMP (Vesicle-associated membrane protein)-associated protein-like
[Arabidopsis thaliana]
Length = 220
Score = 58.5 bits (140), Expect = 4e-08
Identities = 27/58 (46%), Positives = 34/58 (58%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP 58
+QA E PPDMQCKDKFLLQS + P ++ D F KDSG + E K + + P
Sbjct: 74 LQAQREYPPDMQCKDKFLLQSTIVPPHTDVDELPQDTFTKDSGKTLTECKLKVSYITP 131
>UniRef100_Q8S1V6 P0504E02.2 protein [Oryza sativa]
Length = 394
Score = 57.0 bits (136), Expect = 1e-07
Identities = 28/57 (49%), Positives = 37/57 (64%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVA 57
MQA APPDMQ KDKFL+Q+ V G + +D+ F K+SG ++ESK R V V+
Sbjct: 70 MQAQRTAPPDMQLKDKFLVQTTVVPYGTSDEDLVPSYFSKESGRYIEESKLRVVLVS 126
>UniRef100_Q7XT32 OSJNBa0010H02.20 protein [Oryza sativa]
Length = 244
Score = 55.8 bits (133), Expect = 3e-07
Identities = 31/65 (47%), Positives = 38/65 (57%), Gaps = 1/65 (1%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP-P 59
MQA E D CKDKFL+QS+ GAT +D ++F K G ++E K R V VA P
Sbjct: 70 MQAPVEMLSDYHCKDKFLVQSVAVGYGATMRDFVPELFTKAPGRVIEEFKLRVVYVAANP 129
Query: 60 PSPNP 64
PSP P
Sbjct: 130 PSPVP 134
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 146,599,322
Number of Sequences: 2790947
Number of extensions: 4945561
Number of successful extensions: 22573
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 22536
Number of HSP's gapped (non-prelim): 47
length of query: 83
length of database: 848,049,833
effective HSP length: 59
effective length of query: 24
effective length of database: 683,383,960
effective search space: 16401215040
effective search space used: 16401215040
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC121238.7


