

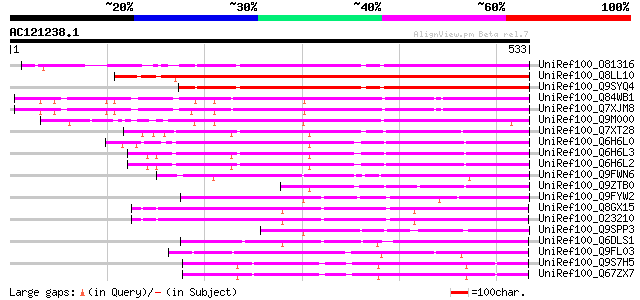
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121238.1 + phase: 0
(533 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O81316 F6N15.20 protein [Arabidopsis thaliana] 389 e-106
UniRef100_Q8LL10 Hairy meristem [Petunia hybrida] 371 e-101
UniRef100_Q9SYQ4 Scarecrow-like 6 [Arabidopsis thaliana] 364 4e-99
UniRef100_Q84WB1 Putative SCARECROW gene regulator [Arabidopsis ... 356 8e-97
UniRef100_Q7XJM8 Putative SCARECROW gene regulator [Arabidopsis ... 356 8e-97
UniRef100_Q9M000 Putative scarecrow protein [Arabidopsis thaliana] 331 4e-89
UniRef100_Q7XT28 OSJNBa0010H02.25 protein [Oryza sativa] 300 7e-80
UniRef100_Q6H6L0 Putative Scl1 protein [Oryza sativa] 295 3e-78
UniRef100_Q6H6L3 Scl1 protein [Oryza sativa] 294 4e-78
UniRef100_Q6H6L2 Scl1 protein [Oryza sativa] 294 4e-78
UniRef100_Q9FWN6 Putative Scl1 protein [Oryza sativa] 280 1e-73
UniRef100_Q9ZTB0 Scl1 protein [Oryza sativa] 204 7e-51
UniRef100_Q9FYW2 BAC19.14 [Lycopersicon esculentum] 197 5e-49
UniRef100_Q8GX15 Ap2 SCARECROW-like protein [Arabidopsis thaliana] 183 9e-45
UniRef100_O23210 SCARECROW-like protein [Arabidopsis thaliana] 182 2e-44
UniRef100_Q9SPP3 Scl1 protein [Oryza sativa] 181 4e-44
UniRef100_Q6DLS1 SCARECROW-like protein [Brassica napus] 179 1e-43
UniRef100_Q9FL03 SCARECROW gene regulator [Arabidopsis thaliana] 145 4e-33
UniRef100_Q9S7H5 Putative SCARECROW gene regulator [Arabidopsis ... 135 2e-30
UniRef100_Q67ZX7 Putative SCARECROW gene regulator [Arabidopsis ... 135 4e-30
>UniRef100_O81316 F6N15.20 protein [Arabidopsis thaliana]
Length = 558
Score = 389 bits (998), Expect = e-106
Identities = 239/530 (45%), Positives = 313/530 (58%), Gaps = 66/530 (12%)
Query: 13 DDNSEEKCGGGGMRMEDWEGQ-----DQSLLRLIMGDVEDPSAGLNKILQNSGYGSQNVD 67
D ++E+CG G+ DWE Q +QS+L LIMGD DPS LN ILQ S +
Sbjct: 83 DATTDEQCGAIGLG--DWEEQVPHDHEQSILGLIMGDSTDPSLELNSILQTSPTFHDSDY 140
Query: 68 FHGGFGVLDHQQQQGLTMMDASVQQGNYNVFPFIPENYNVLPLLDSGQEVF-ARRHQQQE 126
GFGV+D F ++++V P SG + ++ H Q
Sbjct: 141 SSPGFGVVDTG---------------------FGLDHHSVPPSHVSGLLINQSQTHYTQN 179
Query: 127 TQLPLFPHHYLQHQQQQQQSSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIE 186
+ HH+ P AK +++ G I +QL K AE+IE
Sbjct: 180 PAAIFYGHHH----------HTPPPAK--RLNPGPVG----------ITEQLVKAAEVIE 217
Query: 187 AGNPVQAQGILARLNHQLS-PIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKI 245
+ + AQGILARLN QLS P+G P +RA+FY KEAL +LH+ L +P S IFKI
Sbjct: 218 SDTCL-AQGILARLNQQLSSPVGKPLERAAFYFKEALNNLLHNVSQTL---NPYSLIFKI 273
Query: 246 GAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNG 305
AYKSFSEISPVLQFANFT NQ+L+E+ F R+H+IDFDIG+G QW+S MQE+VLR N
Sbjct: 274 AAYKSFSEISPVLQFANFTSNQALLESFHGFHRLHIIDFDIGYGGQWASLMQELVLRDNA 333
Query: 306 KP-SLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHF 364
P SLKIT SP++ +++EL FTQ+NL +A ++NI + VL+++ L S P
Sbjct: 334 APLSLKITVFASPANHDQLELGFTQDNLKHFASEINISLDIQVLSLDLLGSISWPNS--- 390
Query: 365 FDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVL 424
EA+ VN +SF S P+ L F+K L P I+V D+ C+R D+P + H L
Sbjct: 391 -SEKEAVAVNISAASF----SHLPLVLRFVKHLSPTIIVCSDRGCERTDLPFSQQLAHSL 445
Query: 425 QCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQRDK-LPPWRNMFLQSGFS 483
++AL ESLDAVN NLD +QKIER IQP I K+VL ++ + W+ MFLQ GFS
Sbjct: 446 HSHTALFESLDAVNANLDAMQKIERFLIQPEIEKLVLDRSRPIERPMMTWQAMFLQMGFS 505
Query: 484 PFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
P + SNFTE+QAECLVQR PVRGF VE+K +SL+LCWQR EL+ VS WRC
Sbjct: 506 PVTHSNFTESQAECLVQRTPVRGFHVEKKHNSLLLCWQRTELVGVSAWRC 555
>UniRef100_Q8LL10 Hairy meristem [Petunia hybrida]
Length = 721
Score = 371 bits (952), Expect = e-101
Identities = 201/437 (45%), Positives = 280/437 (63%), Gaps = 19/437 (4%)
Query: 108 LPLLDSGQEVFARRHQQQETQLPLFPHHYLQHQQQQQQSSVVPFAKQQKVSSSTTGDDAS 167
+P D ++ RR E Q + L QQ Q + +VP K+ ++ G + S
Sbjct: 293 VPFFDPSGDLSMRRQLLGEMQQHF---NLLPPQQFQPKPLIVP-----KLEAACGGGNGS 344
Query: 168 I-------QLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKE 220
+ Q QQ I+DQ+F+ +EL+ AG+ AQ ILARLN QLSPIG PF+RA+FY KE
Sbjct: 345 LMVPRHQQQEQQFIYDQIFQASELLLAGHFSNAQMILARLNQQLSPIGKPFKRAAFYFKE 404
Query: 221 ALQL--MLHSNGNNLTAFSPISF--IFKIGAYKSFSEISPVLQFANFTCNQSLIEALERF 276
ALQL +L L SP F + K+ AYKSFSE+SP++QF NFT NQ+++EAL
Sbjct: 405 ALQLPFLLPCTSTFLPPRSPTPFDCVLKMDAYKSFSEVSPLIQFMNFTSNQAILEALGDV 464
Query: 277 DRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYA 336
+RIH+IDFDIGFG QWSSFMQE+ + SLKITA SPS+ + +E+ E+L+Q+A
Sbjct: 465 ERIHIIDFDIGFGAQWSSFMQELPSSNRKATSLKITAFASPSTHHSVEIAIMHESLTQFA 524
Query: 337 KDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQ 396
D I FE V+N+++ + S PL EAI +NFP+ S S FP LH++KQ
Sbjct: 525 NDAGIRFELEVINLDTFDPKSYPLSSLRSSDCEAIAINFPIWSISSCLFAFPSLLHYMKQ 584
Query: 397 LRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTI 456
L PKI+V+L++ C+R ++PL +++H LQ Y LL S+DA NV ++ +KIE+ + P+I
Sbjct: 585 LSPKIIVSLERGCERTELPLKHHLLHALQYYEILLASIDAANVTPEIGKKIEKSLLLPSI 644
Query: 457 NKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSL 516
+VL D++P WRN+F +GFSP +FSN TE QAEC+V+R V GF VE++ SSL
Sbjct: 645 ENMVLGRLRSPDRMPQWRNLFASAGFSPVAFSNLTEIQAECVVKRTQVGGFHVEKRQSSL 704
Query: 517 VLCWQRKELISVSTWRC 533
VLCW+++EL+S TWRC
Sbjct: 705 VLCWKQQELLSALTWRC 721
Score = 37.4 bits (85), Expect = 1.1
Identities = 20/42 (47%), Positives = 24/42 (56%), Gaps = 4/42 (9%)
Query: 32 GQDQSLLRLIMGDVEDPSAGLNKILQNSGYGSQNVDFHGGFG 73
G DQ+LLR I GD+EDPS L ++L G FHG G
Sbjct: 129 GADQTLLRWISGDMEDPSVSLKQLL----LGGNANGFHGSSG 166
>UniRef100_Q9SYQ4 Scarecrow-like 6 [Arabidopsis thaliana]
Length = 378
Score = 364 bits (934), Expect = 4e-99
Identities = 195/363 (53%), Positives = 249/363 (67%), Gaps = 15/363 (4%)
Query: 174 IFDQLFKTAELIEAGNPVQAQGILARLNHQLS-PIGNPFQRASFYMKEALQLMLHSNGNN 232
I +QL K AE+IE+ + AQGILARLN QLS P+G P +RA+FY KEAL +LH+
Sbjct: 25 ITEQLVKAAEVIESDTCL-AQGILARLNQQLSSPVGKPLERAAFYFKEALNNLLHNVSQT 83
Query: 233 LTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQW 292
L +P S IFKI AYKSFSEISPVLQFANFT NQ+L+E+ F R+H+IDFDIG+G QW
Sbjct: 84 L---NPYSLIFKIAAYKSFSEISPVLQFANFTSNQALLESFHGFHRLHIIDFDIGYGGQW 140
Query: 293 SSFMQEIVLRSNGKP-SLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIE 351
+S MQE+VLR N P SLKIT SP++ +++EL FTQ+NL +A ++NI + VL+++
Sbjct: 141 ASLMQELVLRDNAAPLSLKITVFASPANHDQLELGFTQDNLKHFASEINISLDIQVLSLD 200
Query: 352 SLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDR 411
L S P EA+ VN +SF S P+ L F+K L P I+V D+ C+R
Sbjct: 201 LLGSISWPNS----SEKEAVAVNISAASF----SHLPLVLRFVKHLSPTIIVCSDRGCER 252
Query: 412 MDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQRDK-L 470
D+P + H L ++AL ESLDAVN NLD +QKIER IQP I K+VL ++ +
Sbjct: 253 TDLPFSQQLAHSLHSHTALFESLDAVNANLDAMQKIERFLIQPEIEKLVLDRSRPIERPM 312
Query: 471 PPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVST 530
W+ MFLQ GFSP + SNFTE+QAECLVQR PVRGF VE+K +SL+LCWQR EL+ VS
Sbjct: 313 MTWQAMFLQMGFSPVTHSNFTESQAECLVQRTPVRGFHVEKKHNSLLLCWQRTELVGVSA 372
Query: 531 WRC 533
WRC
Sbjct: 373 WRC 375
>UniRef100_Q84WB1 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 640
Score = 356 bits (914), Expect = 8e-97
Identities = 244/573 (42%), Positives = 316/573 (54%), Gaps = 66/573 (11%)
Query: 6 NLAHLSNDDNSEEKCGGGGMRMEDW------EGQDQSLLRLIMGD-----VEDPSAGLNK 54
N + DDN+ KC G+ D GQ+QS+LRLIM V DP G
Sbjct: 89 NTTVTAGDDNNNNKCSQMGLDDLDGVLSASSPGQEQSILRLIMDPGSAFGVFDPGFGFG- 147
Query: 55 ILQNSGYGSQNVDFHGGFGVLDHQQQQGLTM-MDASVQQGNYNVF---PFIP--ENYN-- 106
SG G + +L + Q +T +A + N+ +F P P + +N
Sbjct: 148 ----SGSGPVSAPVSDNSNLLCNFPFQEITNPAEALINPSNHCLFYNPPLSPPAKRFNSG 203
Query: 107 -----VLPLLDS--GQEVFARRHQQQETQLPLFP-HHYLQHQQQQQQSSVVPFAKQQKVS 158
V PL D G + R+HQ Q FP +H Q QQ SS A S
Sbjct: 204 SLHQPVFPLSDPDPGHDPVRRQHQFQ------FPFYHNNQQQQFPSSSSSTAVAMVPVPS 257
Query: 159 SSTTGDDASIQLQQSIFDQLFKTAELI-EAGN-----PVQAQGILARLNHQLSPIGN--- 209
GDD S+ I +QLF AELI GN V AQGILARLNH L+ N
Sbjct: 258 PGMAGDDQSV-----IIEQLFNAAELIGTTGNNNGDQTVLAQGILARLNHHLNTSSNHKS 312
Query: 210 PFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSL 269
PFQRA+ ++ EAL ++H N ++ +P + I +I AY+SFSE SP LQF NFT NQS+
Sbjct: 313 PFQRAASHIAEALLSLIH-NESSPPLITPENLILRIAAYRSFSETSPFLQFVNFTANQSI 371
Query: 270 IEALER--FDRIHVIDFDIGFGVQWSSFMQEIVL-----RSNGKPSLKITAVVSPSSC-- 320
+E+ FDRIH+IDFD+G+G QWSS MQE+ R N SLK+T P S
Sbjct: 372 LESCNESGFDRIHIIDFDVGYGGQWSSLMQELASGVGGRRRNRASSLKLTVFAPPPSTVS 431
Query: 321 NEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSF 380
+E EL FT+ENL +A ++ I FE +L++E L P+ EAI VN PV+S
Sbjct: 432 DEFELRFTEENLKTFAGEVKIPFEIELLSVELLLNPAYWPLSLRSSEKEAIAVNLPVNSV 491
Query: 381 ISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVN 440
S P+ L FLKQL P IVV D+ CDR D P P V+H LQ +++LLESLDA N N
Sbjct: 492 ASG--YLPLILRFLKQLSPNIVVCSDRGCDRNDAPFPNAVIHSLQYHTSLLESLDA-NQN 548
Query: 441 LDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQ 500
D IER ++QP+I K+++ H ++ PPWR +F Q GFSP S S EAQAECL+Q
Sbjct: 549 QDD-SSIERFWVQPSIEKLLMKRHRWIERSPPWRILFTQCGFSPASLSQMAEAQAECLLQ 607
Query: 501 RAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
R PVRGF VE++ SSLV+CWQRKEL++VS W+C
Sbjct: 608 RNPVRGFHVEKRQSSLVMCWQRKELVTVSAWKC 640
>UniRef100_Q7XJM8 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 640
Score = 356 bits (914), Expect = 8e-97
Identities = 242/573 (42%), Positives = 315/573 (54%), Gaps = 66/573 (11%)
Query: 6 NLAHLSNDDNSEEKCGGGGMRMEDW------EGQDQSLLRLIMGD-----VEDPSAGLNK 54
N + DDN+ KC G+ D GQ+QS+LRLIM V DP G
Sbjct: 89 NTTVTAGDDNNNNKCSQMGLDDLDGVLSASSPGQEQSILRLIMDPGSAFGVFDPGFGFG- 147
Query: 55 ILQNSGYGSQNVDFHGGFGVLDHQQQQGLTM-MDASVQQGNYNVF---PFIP--ENYN-- 106
SG G + +L + Q +T +A + N+ +F P P + +N
Sbjct: 148 ----SGSGPVSAPVSDNSNLLCNFPFQEITNPAEALINPSNHCLFYNPPLSPPAKRFNSG 203
Query: 107 -----VLPLLDS--GQEVFARRHQQQETQLPLFP-HHYLQHQQQQQQSSVVPFAKQQKVS 158
V PL D G + R+HQ Q FP +H Q QQ SS A S
Sbjct: 204 SLHQPVFPLSDPDPGHDPVRRQHQFQ------FPFYHNNQQQQFPSSSSSTAVAMVPVPS 257
Query: 159 SSTTGDDASIQLQQSIFDQLFKTAELI------EAGNPVQAQGILARLNHQLSPIGN--- 209
GDD S+ I +QLF AELI + V AQGILARLNH L+ N
Sbjct: 258 PGMAGDDQSV-----IIEQLFNAAELIGTTGNNNGDHTVLAQGILARLNHHLNTSSNHKS 312
Query: 210 PFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSL 269
PFQRA+ ++ EAL ++H N ++ +P + I +I AY+SFSE SP LQF NFT NQS+
Sbjct: 313 PFQRAASHIAEALLSLIH-NESSPPLITPENLILRIAAYRSFSETSPFLQFVNFTANQSI 371
Query: 270 IEALER--FDRIHVIDFDIGFGVQWSSFMQEIVL-----RSNGKPSLKITAVVSPSSC-- 320
+E+ FDRIH+IDFD+G+G QWSS MQE+ R N SLK+T P S
Sbjct: 372 LESCNESGFDRIHIIDFDVGYGGQWSSLMQELASGVGGRRRNRASSLKLTVFAPPPSTVS 431
Query: 321 NEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSF 380
+E EL FT+ENL +A ++ I FE +L++E L P+ EAI VN PV+S
Sbjct: 432 DEFELRFTEENLKTFAGEVKIPFEIELLSVELLLNPAYWPLSLRSSEKEAIAVNLPVNSV 491
Query: 381 ISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVN 440
S P+ L FLKQL P IVV D+ CDR D P P V+H LQ +++LLESLDA N N
Sbjct: 492 ASG--YLPLILRFLKQLSPNIVVCSDRGCDRNDAPFPNAVIHSLQYHTSLLESLDA-NQN 548
Query: 441 LDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQ 500
D IER ++QP+I K+++ H ++ PPWR +F Q GFSP S S EAQAECL+Q
Sbjct: 549 QDD-SSIERFWVQPSIEKLLMKRHRWIERSPPWRILFTQCGFSPASLSQMAEAQAECLLQ 607
Query: 501 RAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
R PVRGF VE++ SSLV+CWQRKEL++VS W+C
Sbjct: 608 RNPVRGFHVEKRQSSLVMCWQRKELVTVSAWKC 640
>UniRef100_Q9M000 Putative scarecrow protein [Arabidopsis thaliana]
Length = 623
Score = 331 bits (848), Expect = 4e-89
Identities = 220/531 (41%), Positives = 307/531 (57%), Gaps = 53/531 (9%)
Query: 32 GQDQSLLRLIM-GDVEDPSAGLNKILQNSG----YGSQNVDFHGGFGVLDHQQQQGLTMM 86
GQ+QS+ RLIM GDV DP + SG + N F GF + +++ +
Sbjct: 117 GQEQSIFRLIMAGDVVDPGSEFVGFDIGSGSDPVIDNPNPLFGYGFPFQNAPEEEKF-QI 175
Query: 87 DASVQQGNYNVFPFIPENYNVLPLLDSGQEVFARRHQQQETQLPLFPHHYLQHQQQQQQS 146
+ G ++ P P L+SGQ +H Q +FP H+
Sbjct: 176 SINPNPGFFSDPPSSPPAKR----LNSGQP--GSQHLQW-----VFPFSDPGHESHD--- 221
Query: 147 SVVPFAKQQKVSSSTTGDDASIQLQQS-IFDQLFKTA-ELIEAG---NPVQAQGILARLN 201
PF K++ G+D + Q Q + I DQLF A EL G NPV AQGILARLN
Sbjct: 222 ---PFLTPPKIA----GEDQNDQDQSAVIIDQLFSAAAELTTNGGDNNPVLAQGILARLN 274
Query: 202 HQLSPIGN--------PFQRASFYMKEALQLMLHSNGNNLTAFSPI-SFIFKIGAYKSFS 252
H L+ + PF RA+ Y+ EAL +L + + + SP + IF+I AY++FS
Sbjct: 275 HNLNNNNDDTNNNPKPPFHRAASYITEALHSLLQDSSLSPPSLSPPQNLIFRIAAYRAFS 334
Query: 253 EISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIV---LRSNGKPSL 309
E SP LQF NFT NQ+++E+ E FDRIH++DFDIG+G QW+S +QE+ RS+ PSL
Sbjct: 335 ETSPFLQFVNFTANQTILESFEGFDRIHIVDFDIGYGGQWASLIQELAGKRNRSSSAPSL 394
Query: 310 KITAVVSPSS-CNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPS-CPLPGHFFDS 367
KITA SPS+ +E EL FT+ENL +A + + FE +LN+E L P+ PL
Sbjct: 395 KITAFASPSTVSDEFELRFTEENLRSFAGETGVSFEIELLNMEILLNPTYWPLSLFRSSE 454
Query: 368 NEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDR-MDVPLPTNVVHVLQC 426
EAI VN P+SS +S P+ L FLKQ+ P +VV D++CDR D P P V++ LQ
Sbjct: 455 KEAIAVNLPISSMVS--GYLPLILRFLKQISPNVVVCSDRSCDRNNDAPFPNGVINALQY 512
Query: 427 YSALLESLDAVNV-NLDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPF 485
Y++LLESLD+ N+ N + IER +QP+I K++ + + ++ PPWR++F Q GF+P
Sbjct: 513 YTSLLESLDSGNLNNAEAATSIERFCVQPSIQKLLTNRYRWMERSPPWRSLFGQCGFTPV 572
Query: 486 SFSNFTEAQAECLVQRAPVRGFQVERKPS---SLVLCWQRKELISVSTWRC 533
+ S E QAE L+QR P+RGF +E++ S SLVLCWQRKEL++VS W+C
Sbjct: 573 TLSQTAETQAEYLLQRNPMRGFHLEKRQSSSPSLVLCWQRKELVTVSAWKC 623
>UniRef100_Q7XT28 OSJNBa0010H02.25 protein [Oryza sativa]
Length = 711
Score = 300 bits (768), Expect = 7e-80
Identities = 187/449 (41%), Positives = 250/449 (55%), Gaps = 43/449 (9%)
Query: 118 FARRHQQQETQLPLFPHH------YLQHQQQQQQS-------------SVVPFAKQQKV- 157
F+ QQ Q P HH +L H Q Q S VPF Q +
Sbjct: 273 FSDHQQQPLLQPPPKRHHSVPDNLFLLHNQPQPPPPAPAQCLPFPTLHSAVPFQLQPSMQ 332
Query: 158 ----SSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQR 213
+ +T A+ Q QQ + D+L A+ E GN + A+ ILARLN QL PIG PF R
Sbjct: 333 HPRNAMKSTAAAAAAQ-QQHLLDELAAAAKATEVGNSIGAREILARLNQQLPPIGKPFLR 391
Query: 214 ASFYMKEALQLML---HSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLI 270
++ Y+K+AL L L H LT SP+ K+ AYKSFS++SPVLQFANFT Q+L+
Sbjct: 392 SASYLKDALLLALADGHHAATRLT--SPLDVALKLTAYKSFSDLSPVLQFANFTVTQALL 449
Query: 271 EALERFDR--IHVIDFDIGFGVQWSSFMQEIVLRSNGK----PSLKITAVVSPSSCNEIE 324
+ + I VIDFD+G G QW+SF+QE+ R P LK+TA VS +S + +E
Sbjct: 450 DEIASTTASCIRVIDFDLGVGGQWASFLQELAHRCGSGGVSLPMLKLTAFVSAASHHPLE 509
Query: 325 LNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNP 384
L+ TQ+NLSQ+A DL I FEFN +N+++ + P ++E + V+ PV P
Sbjct: 510 LHLTQDNLSQFAADLGIPFEFNAINLDAFDPMELIAP----TADEVVAVSLPVGCSARTP 565
Query: 385 SCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVL 444
P L +KQL PKIVV +D DR D+P + ++ LQ LLESLDA + D +
Sbjct: 566 --LPAMLQLVKQLAPKIVVAIDYGSDRSDLPFSQHFLNCLQSCLCLLESLDAAGTDADAV 623
Query: 445 QKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPV 504
KIER IQP + VL + DK WR + +GF+P SN EAQA+CL++R V
Sbjct: 624 SKIERFLIQPRVEDAVLGRR-RADKAIAWRTVLTSAGFAPQPLSNLAEAQADCLLKRVQV 682
Query: 505 RGFQVERKPSSLVLCWQRKELISVSTWRC 533
RGF VE++ + L L WQR EL+SVS WRC
Sbjct: 683 RGFHVEKRGAGLALYWQRGELVSVSAWRC 711
>UniRef100_Q6H6L0 Putative Scl1 protein [Oryza sativa]
Length = 715
Score = 295 bits (754), Expect = 3e-78
Identities = 185/457 (40%), Positives = 259/457 (56%), Gaps = 39/457 (8%)
Query: 99 PFIPENYNVLPLLDSG--------QEVFARRHQQQETQ-----LPLFPHHYLQHQQQQQQ 145
P PE+ + PLL +++ R+Q LP P H Q Q
Sbjct: 276 PSFPEHNHQSPLLQPPPKRHHSMPDDIYLARNQLPPAAAAAQGLPFSPLH--ASVPFQLQ 333
Query: 146 SSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLS 205
S P K TT +A+ QQ + D+L A+ EAGN V A+ ILARLN QL
Sbjct: 334 PSPPPIRGAMK----TTAAEAA---QQQLLDELAAAAKATEAGNSVGAREILARLNQQLP 386
Query: 206 PIGNPFQRASFYMKEALQLMLHSNGNNLTAFS-PISFIFKIGAYKSFSEISPVLQFANFT 264
P+G PF R++ Y++EAL L L + + +++ + P+ K+ AYKSFS++SPVLQFANFT
Sbjct: 387 PLGKPFLRSASYLREALLLALADSHHGVSSVTTPLDVALKLAAYKSFSDLSPVLQFANFT 446
Query: 265 CNQSLIEAL--ERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGK----PSLKITAVVSPS 318
Q+L++ + IHVIDFD+G G QW+SF+QE+ R P LK+TA VS +
Sbjct: 447 ATQALLDEIGGTATSCIHVIDFDLGVGGQWASFLQELAHRRAAGGVTLPLLKLTAFVSTA 506
Query: 319 SCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDS--NEAIGVNFP 376
S + +EL+ TQ+NLSQ+A DL I FEFN +++++ N PG S +E + V+ P
Sbjct: 507 SHHPLELHLTQDNLSQFAADLGIPFEFNAVSLDAFN------PGELISSTGDEVVAVSLP 560
Query: 377 VSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDA 436
V P P L +KQL PKIVV +D DR D+ + ++ Q LL+SLDA
Sbjct: 561 VGCSARAPP-LPAILRLVKQLSPKIVVAIDHGADRADLSFSQHFLNCFQSCVFLLDSLDA 619
Query: 437 VNVNLDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAE 496
++ D KIER IQP ++ +VL H K WR++F +GF P SN EAQA+
Sbjct: 620 AGIDADSACKIERFLIQPRVHDMVLGRHKVH-KAIAWRSVFAAAGFKPVPPSNLAEAQAD 678
Query: 497 CLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
CL++R VRGF VE+ ++L L WQR EL+S+S+WRC
Sbjct: 679 CLLKRVQVRGFHVEKCGAALTLYWQRGELVSISSWRC 715
>UniRef100_Q6H6L3 Scl1 protein [Oryza sativa]
Length = 709
Score = 294 bits (753), Expect = 4e-78
Identities = 179/446 (40%), Positives = 251/446 (56%), Gaps = 47/446 (10%)
Query: 122 HQQQETQLPLFPHHYLQHQ----QQQQQSSVV-------------------PFAKQQKVS 158
HQ Q PL HH + + QQQSS V P + +
Sbjct: 277 HQSPHLQPPLKRHHAIPDDLYLARNQQQSSAVAPGLAYSPPLHGPAPFQLHPSPPPIRGA 336
Query: 159 SSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYM 218
+T +A+ QQ + D+L A+ EAGN V A+ ILARLN QL +G PF R++ Y+
Sbjct: 337 MKSTAAEAA---QQQLLDELAAAAKATEAGNSVGAREILARLNQQLPQLGKPFLRSASYL 393
Query: 219 KEALQLML---HSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALER 275
KEAL L L H + +T SP+ K+ AYKSFS++SPVLQF NFT Q+L++ +
Sbjct: 394 KEALLLALADSHHGSSGVT--SPLDVALKLAAYKSFSDLSPVLQFTNFTATQALLDEIGG 451
Query: 276 FDR--IHVIDFDIGFGVQWSSFMQEIVLRSNGK----PSLKITAVVSPSSCNEIELNFTQ 329
IHVIDFD+G G QW+SF+QE+ R P LK+TA +S +S + +EL+ TQ
Sbjct: 452 MATSCIHVIDFDLGVGGQWASFLQELAHRRGAGGMALPLLKLTAFMSTASHHPLELHLTQ 511
Query: 330 ENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSN--EAIGVNFPVSSFISNPSCF 387
+NLSQ+A +L I FEFN +++++ N P S+ E + V+ PV P
Sbjct: 512 DNLSQFAAELRIPFEFNAVSLDAFN------PAELISSSGDEVVAVSLPVGCSARAPP-L 564
Query: 388 PVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKI 447
P L +KQL PK+VV +D DR D+P + ++ Q LL+SLDA ++ D KI
Sbjct: 565 PAILRLVKQLCPKVVVAIDHGGDRADLPFSQHFLNCFQSCVFLLDSLDAAGIDADSACKI 624
Query: 448 ERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGF 507
ER IQP + V+ H + K WR++F +GF P SN EAQA+CL++R VRGF
Sbjct: 625 ERFLIQPRVEDAVIGRHKAQ-KAIAWRSVFAATGFKPVQLSNLAEAQADCLLKRVQVRGF 683
Query: 508 QVERKPSSLVLCWQRKELISVSTWRC 533
VE++ ++L L WQR EL+S+S+WRC
Sbjct: 684 HVEKRGAALTLYWQRGELVSISSWRC 709
>UniRef100_Q6H6L2 Scl1 protein [Oryza sativa]
Length = 531
Score = 294 bits (753), Expect = 4e-78
Identities = 179/446 (40%), Positives = 251/446 (56%), Gaps = 47/446 (10%)
Query: 122 HQQQETQLPLFPHHYLQHQ----QQQQQSSVV-------------------PFAKQQKVS 158
HQ Q PL HH + + QQQSS V P + +
Sbjct: 99 HQSPHLQPPLKRHHAIPDDLYLARNQQQSSAVAPGLAYSPPLHGPAPFQLHPSPPPIRGA 158
Query: 159 SSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYM 218
+T +A+ QQ + D+L A+ EAGN V A+ ILARLN QL +G PF R++ Y+
Sbjct: 159 MKSTAAEAA---QQQLLDELAAAAKATEAGNSVGAREILARLNQQLPQLGKPFLRSASYL 215
Query: 219 KEALQLML---HSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALER 275
KEAL L L H + +T SP+ K+ AYKSFS++SPVLQF NFT Q+L++ +
Sbjct: 216 KEALLLALADSHHGSSGVT--SPLDVALKLAAYKSFSDLSPVLQFTNFTATQALLDEIGG 273
Query: 276 FDR--IHVIDFDIGFGVQWSSFMQEIVLRSNGK----PSLKITAVVSPSSCNEIELNFTQ 329
IHVIDFD+G G QW+SF+QE+ R P LK+TA +S +S + +EL+ TQ
Sbjct: 274 MATSCIHVIDFDLGVGGQWASFLQELAHRRGAGGMALPLLKLTAFMSTASHHPLELHLTQ 333
Query: 330 ENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSN--EAIGVNFPVSSFISNPSCF 387
+NLSQ+A +L I FEFN +++++ N P S+ E + V+ PV P
Sbjct: 334 DNLSQFAAELRIPFEFNAVSLDAFN------PAELISSSGDEVVAVSLPVGCSARAPP-L 386
Query: 388 PVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKI 447
P L +KQL PK+VV +D DR D+P + ++ Q LL+SLDA ++ D KI
Sbjct: 387 PAILRLVKQLCPKVVVAIDHGGDRADLPFSQHFLNCFQSCVFLLDSLDAAGIDADSACKI 446
Query: 448 ERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGF 507
ER IQP + V+ H + K WR++F +GF P SN EAQA+CL++R VRGF
Sbjct: 447 ERFLIQPRVEDAVIGRHKAQ-KAIAWRSVFAATGFKPVQLSNLAEAQADCLLKRVQVRGF 505
Query: 508 QVERKPSSLVLCWQRKELISVSTWRC 533
VE++ ++L L WQR EL+S+S+WRC
Sbjct: 506 HVEKRGAALTLYWQRGELVSISSWRC 531
>UniRef100_Q9FWN6 Putative Scl1 protein [Oryza sativa]
Length = 582
Score = 280 bits (715), Expect = 1e-73
Identities = 160/393 (40%), Positives = 236/393 (59%), Gaps = 21/393 (5%)
Query: 151 FAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPI--- 207
F K + D+A+ ++Q L + A+L EAG+ A+ ILARLN++L
Sbjct: 201 FVPALKPKAEAANDEAAAAVEQ-----LAEAAKLAEAGDAFGAREILARLNYRLPAAPTA 255
Query: 208 GNPFQRASFYMKEALQLMLHSNGNNL--TAFSPISFIFKIGAYKSFSEISPVLQFANFTC 265
G P R++FY KEAL+L L G+ +A +P + K+GAYK+FSE+SPVLQFA+ TC
Sbjct: 256 GTPLLRSAFYFKEALRLTLSPTGDAPAPSASTPYDVVVKLGAYKAFSEVSPVLQFAHLTC 315
Query: 266 NQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIEL 325
Q++++ L IHV+DFDIG G QW+S MQE+ + +LK+TA+VSP+S + +EL
Sbjct: 316 VQAVLDELGGAGCIHVLDFDIGMGEQWASLMQELA-QLRPAAALKVTALVSPASHHPLEL 374
Query: 326 NFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPS 385
ENLS +A +L + F F V NI++L+ P+ L + +A+ V+ PV ++ +
Sbjct: 375 QLIHENLSGFAAELGVFFHFTVFNIDTLD-PAELLAN--ATAGDAVAVHLPVGP--AHAA 429
Query: 386 CFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQ 445
P L +K+L K+VV++D+ CDR D+P ++ H LLES+DAV + D
Sbjct: 430 ATPAVLRLVKRLGAKVVVSVDRGCDRSDLPFAAHLFHSFHSAVYLLESIDAVGTDPDTAS 489
Query: 446 KIERHYIQPTINKIVLSHHNQRDKL-----PPWRNMFLQSGFSPFSFSNFTEAQAECLVQ 500
KIER+ I P I + V++ H + PPWR F +GF+P + F E+QAE L+
Sbjct: 490 KIERYLIHPAIEQCVVASHRAASAMDKAPPPPWRAAFAAAGFAPVQATTFAESQAESLLS 549
Query: 501 RAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
+ VRGF+VE++ SL L WQR EL+SVS WRC
Sbjct: 550 KVHVRGFRVEKRAGSLCLYWQRGELVSVSAWRC 582
>UniRef100_Q9ZTB0 Scl1 protein [Oryza sativa]
Length = 261
Score = 204 bits (518), Expect = 7e-51
Identities = 111/261 (42%), Positives = 157/261 (59%), Gaps = 15/261 (5%)
Query: 279 IHVIDFDIGFGVQWSSFMQEIVLRSNGK----PSLKITAVVSPSSCNEIELNFTQENLSQ 334
IHVIDFD+G G QW+SF+QE+ R P LK+TA +S +S + +EL+ TQ+NLSQ
Sbjct: 10 IHVIDFDLGVGGQWASFLQELAHRRGAGGMALPLLKLTAFMSTASHHPLELHLTQDNLSQ 69
Query: 335 YAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFL 394
+A +L I FEFN +++++ N P +E + V+ PV P P L +
Sbjct: 70 FAAELRIPFEFNAVSLDAFN----PAESISSSGDEVVAVSLPVGCSARAPP-LPAILRLV 124
Query: 395 KQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSA--LLESLDAVNVNLDVLQKIERHYI 452
KQL PK+VV +D DR D+P H L C+ + L+SLDA ++ D KIER I
Sbjct: 125 KQLCPKVVVAIDHGGDRADLPFSQ---HFLNCFQSCVFLDSLDAAGIDADSACKIERFLI 181
Query: 453 QPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERK 512
QP + V+ H + K WR++F +GF P SN EAQA+CL++R VRGF VE++
Sbjct: 182 QPRVEDAVIGRHKAQ-KAIAWRSVFAATGFKPVQLSNLAEAQADCLLKRVQVRGFHVEKR 240
Query: 513 PSSLVLCWQRKELISVSTWRC 533
++L L WQR EL+S+S+WRC
Sbjct: 241 GAALTLYWQRGELVSISSWRC 261
>UniRef100_Q9FYW2 BAC19.14 [Lycopersicon esculentum]
Length = 536
Score = 197 bits (502), Expect = 5e-49
Identities = 132/369 (35%), Positives = 195/369 (52%), Gaps = 19/369 (5%)
Query: 176 DQLFKTAELIEAGNPVQAQGILARLNHQL-SPIGNPFQRASFYMKEALQLMLHSNGNNLT 234
D+L + AE E A ILARLN +L S G P QRA+FY KEALQ L +
Sbjct: 176 DELIRFAECFETNAFQLAHVILARLNQRLRSAAGKPLQRAAFYFKEALQAQLAGSARQTR 235
Query: 235 AFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSS 294
+ S I I +YK S ISP+ F++FT NQ+++EA++ +HVIDFDIG G W+S
Sbjct: 236 SSSSSDVIQTIKSYKILSNISPIPMFSSFTANQAVLEAVDGSMLVHVIDFDIGLGGHWAS 295
Query: 295 FMQEIV----LRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNI 350
FM+E+ R P L+ITA+V +E +ENL+Q+A++LNI FE + + I
Sbjct: 296 FMKELADKAECRKANAPILRITALVPEEYA--VESRLIRENLTQFARELNIGFEIDFVLI 353
Query: 351 ESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKN-- 408
+ L S F E V + F S F ++ L+++ P +VV +D
Sbjct: 354 RTFELLS--FKAIKFMEGEKTAVLLSPAIFRRVGSGF---VNELRRISPNVVVHVDSEGL 408
Query: 409 CDRMDVPLPTNVVHVLQCYSALLESLDAVNVN----LDVLQKIERHYIQPTINKIVLSHH 464
+ V+ L+ YS LLESL+A N+ D ++KIE + P I ++
Sbjct: 409 MGYGAMSFRQTVIDGLEFYSTLLESLEAANIGGGNCGDWMRKIENFVLFPKIVDMI-GAV 467
Query: 465 NQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKE 524
+R WR+ + +GF P S F + QA+CL+ R VRGF V ++ + ++LCW +
Sbjct: 468 GRRGGGGSWRDAMVDAGFRPVGLSQFADFQADCLLGRVQVRGFHVAKRQAEMLLCWHDRA 527
Query: 525 LISVSTWRC 533
L++ S WRC
Sbjct: 528 LVATSAWRC 536
>UniRef100_Q8GX15 Ap2 SCARECROW-like protein [Arabidopsis thaliana]
Length = 486
Score = 183 bits (465), Expect = 9e-45
Identities = 129/417 (30%), Positives = 214/417 (50%), Gaps = 17/417 (4%)
Query: 126 ETQLPLFPHHYLQHQQQQQQSSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELI 185
++ LP FP +Q + S V P + Q D+ + L + + +
Sbjct: 76 DSNLPGFPDQ-IQPSDFESSSDVYP-GQNQTTGYGFNSLDSVDNGGFDFIEDLIRVVDCV 133
Query: 186 EAGNPVQAQGILARLNHQL-SPIGNPFQRASFYMKEAL-QLMLHSNGNNLTAFSPISFIF 243
E+ AQ +L+RLN +L SP G P QRA+FY KEAL + SN N + S +
Sbjct: 134 ESDELQLAQVVLSRLNQRLRSPAGRPLQRAAFYFKEALGSFLTGSNRNPIRLSSWSEIVQ 193
Query: 244 KIGAYKSFSEISPVLQFANFTCNQSLIEALERFDR---IHVIDFDIGFGVQWSSFMQEIV 300
+I A K +S ISP+ F++FT NQ+++++L +HV+DF+IGFG Q++S M+EI
Sbjct: 194 RIRAIKEYSGISPIPLFSHFTANQAILDSLSSQSSSPFVHVVDFEIGFGGQYASLMREIT 253
Query: 301 LRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPL 360
+S L++TAVV+ +E +ENL+Q+A ++ I F+ + +++ + S
Sbjct: 254 EKSVSGGFLRVTAVVAEECA--VETRLVKENLTQFAAEMKIRFQIEFVLMKTFEMLSFKA 311
Query: 361 PGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDV---PLP 417
+ ++ + +S + F ++ L+++ PK+VV +D
Sbjct: 312 IRFVEGERTVVLISPAIFRRLSGITDF---VNNLRRVSPKVVVFVDSEGWTEIAGSGSFR 368
Query: 418 TNVVHVLQCYSALLESLDAVNVNLDVLQKI-ERHYIQPTINKIVLSHHNQRDKLP-PWRN 475
V L+ Y+ +LESLDA D+++KI E ++P I+ V + ++R WR
Sbjct: 369 REFVSALEFYTMVLESLDAAAPPGDLVKKIVEAFVLRPKISAAVETAADRRHTGEMTWRE 428
Query: 476 MFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
F +G P S F + QAECL+++A VRGF V ++ LVLCW + L++ S WR
Sbjct: 429 AFCAAGMRPIQLSQFADFQAECLLEKAQVRGFHVAKRQGELVLCWHGRALVATSAWR 485
>UniRef100_O23210 SCARECROW-like protein [Arabidopsis thaliana]
Length = 486
Score = 182 bits (462), Expect = 2e-44
Identities = 129/417 (30%), Positives = 214/417 (50%), Gaps = 17/417 (4%)
Query: 126 ETQLPLFPHHYLQHQQQQQQSSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELI 185
++ LP FP +Q + S V P + Q D+ + L + + +
Sbjct: 76 DSNLPGFPDQ-IQPSDFESSSDVYP-GQNQTTGYGFNSLDSVDNGGFDFIEDLIRVVDCV 133
Query: 186 EAGNPVQAQGILARLNHQL-SPIGNPFQRASFYMKEAL-QLMLHSNGNNLTAFSPISFIF 243
E+ AQ +L+RLN +L SP G P QRA+FY KEAL + SN N + S +
Sbjct: 134 ESDELQLAQVVLSRLNQRLRSPAGRPLQRAAFYFKEALGSFLTGSNRNPIRLSSWSEIVQ 193
Query: 244 KIGAYKSFSEISPVLQFANFTCNQSLIEALERFDR---IHVIDFDIGFGVQWSSFMQEIV 300
+I A K +S ISP+ F++FT NQ+++++L +HV+DF+IGFG Q++S M+EI
Sbjct: 194 RIRAIKEYSGISPIPLFSHFTANQAILDSLSSQSSSPFVHVVDFEIGFGGQYASLMREIT 253
Query: 301 LRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPL 360
+S L++TAVV+ +E +ENL+Q+A ++ I F+ + +++ + S
Sbjct: 254 EKSVSGGFLRVTAVVAEECA--VETRLVKENLTQFAAEMKIRFQIEFVLMKTFEMLSFKA 311
Query: 361 PGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDV---PLP 417
+ ++ + +S + F ++ L+++ PK+VV +D
Sbjct: 312 IRFVEGERTVVLISPAIFRRLSGITDF---VNNLRRVSPKVVVFVDSEGWTEIAGSGSFR 368
Query: 418 TNVVHVLQCYSALLESLDAVNVNLDVLQKI-ERHYIQPTINKIVLSHHNQRDKLP-PWRN 475
V L+ Y+ +LESLDA D+++KI E ++P I+ V + ++R WR
Sbjct: 369 REFVSALEFYTMVLESLDAAAPPGDLVKKIVEAFVLRPKISAAVETAADRRHTGEMTWRE 428
Query: 476 MFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
F +G P S F + QAECL+++A VRGF V ++ LVLCW + L++ S WR
Sbjct: 429 AFCAAGMRPIQQSQFADFQAECLLEKAQVRGFHVAKRQGELVLCWHGRALVATSAWR 485
>UniRef100_Q9SPP3 Scl1 protein [Oryza sativa]
Length = 360
Score = 181 bits (460), Expect = 4e-44
Identities = 103/280 (36%), Positives = 161/280 (56%), Gaps = 18/280 (6%)
Query: 258 LQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIV---LRSNGKP-SLKITA 313
+Q FT Q+ ++A R+H++ D+GFG W MQE+ R+ G P +LK+TA
Sbjct: 85 VQXTTFTSTQAFLDAXGSARRLHLLXXDVGFGAHWPPLMQELAHHWRRAXGPPPNLKVTA 144
Query: 314 VVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGV 373
+VSP S + EL+ T E+L+++A +L I FEF L + L+ S PL G +EA+ V
Sbjct: 145 LVSPGSSHPXELHLTNESLTRFAAELGIPFEFTALVFDPLSSASPPL-GLSAAPDEAVAV 203
Query: 374 NFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLES 433
+ S +P+ P L +K+LRP +VV +D C+R +++L+ +ALLES
Sbjct: 204 HLTAGSGAFSPA--PAHLRVVKELRPAVVVCVDHGCER-------GALNLLRSCAALLES 254
Query: 434 LDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEA 493
LDA + DV+ K+E+ ++P + ++ + PP ++M +GF+ +N A
Sbjct: 255 LDAAGASPDVVSKVEQFVLRPRVERLAVGVGGG----PPLQSMLASAGFAALQVNNAAXA 310
Query: 494 QAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
QAECL++R GF VE +P++L L W R EL+SVS WRC
Sbjct: 311 QAECLLRRTAXHGFHVEXRPAALALWWXRSELVSVSXWRC 350
>UniRef100_Q6DLS1 SCARECROW-like protein [Brassica napus]
Length = 461
Score = 179 bits (455), Expect = 1e-43
Identities = 123/375 (32%), Positives = 200/375 (52%), Gaps = 33/375 (8%)
Query: 176 DQLFKTAELIEAGNPVQAQGILARLNHQL-SPIGNPFQRASFYMKEAL-QLMLHSNGNNL 233
+ L + + IE+ A +L++LN +L + G P QRA+FY KEAL L+ +N N L
Sbjct: 101 EDLIRVVDCIESDELHLAHVVLSQLNQRLQTSAGRPLQRAAFYFKEALGSLLTGTNRNQL 160
Query: 234 TAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDR---IHVIDFDIGFGV 290
++S I + KI A K FS ISP+ F++FT NQ+++++L +HV+DF+IGFG
Sbjct: 161 FSWSDI--VQKIRAIKEFSGISPIPLFSHFTANQAILDSLSSQSSSPFVHVVDFEIGFGG 218
Query: 291 QWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNI 350
Q++S M+EI +S L++TAVV+ +E +ENL+Q+A ++ I F+ + +
Sbjct: 219 QYASLMREIAEKSANGGFLRVTAVVAEDCA--VETRLVKENLTQFAAEMKIRFQIEFVLM 276
Query: 351 ESLNLPSCPLPGHFFDSNEAIGVNFP--------VSSFISNPSCFPVALHFLKQLRPKIV 402
++ + S F D + + P ++ F++N L ++ P +V
Sbjct: 277 KTFEILSFKAI-RFVDGERTVVLISPAIFRRVIGIAEFVNN----------LGRVSPNVV 325
Query: 403 VTLD-KNCDRM--DVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKI-ERHYIQPTINK 458
V +D + C V + Y+ +LESLDA D+++KI E ++P I+
Sbjct: 326 VFVDSEGCTETAGSGSFRREFVSAFEFYTMVLESLDAAAPPGDLVKKIVETFLLRPKISA 385
Query: 459 IVLSHHNQRDK-LPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLV 517
V + N+R WR M +G P S F + QAECL+++A VRGF V ++ LV
Sbjct: 386 AVETAANRRSAGQMTWREMLCAAGMRPVQLSQFADFQAECLLEKAQVRGFHVAKRQGELV 445
Query: 518 LCWQRKELISVSTWR 532
LCW + L++ S WR
Sbjct: 446 LCWHGRALVATSAWR 460
>UniRef100_Q9FL03 SCARECROW gene regulator [Arabidopsis thaliana]
Length = 584
Score = 145 bits (365), Expect = 4e-33
Identities = 107/386 (27%), Positives = 190/386 (48%), Gaps = 25/386 (6%)
Query: 164 DDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQ 223
+D L+ + ++ A + ++ +P +A L ++ +S +G+P +R +FY EAL
Sbjct: 207 EDDDFDLEPPLLKAIYDCARISDS-DPNEASKTLLQIRESVSELGDPTERVAFYFTEALS 265
Query: 224 LMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVID 283
L N + T+ S S I +YK+ ++ P +FA+ T NQ+++EA E+ ++IH++D
Sbjct: 266 NRLSPN-SPATSSSSSSTEDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHIVD 324
Query: 284 FDIGFGVQWSSFMQEIVLRSNGKPS-LKITAVVSPS--SCNEIELNFTQENLSQYAKDLN 340
F I G+QW + +Q + R++GKP+ ++++ + +PS E L T L +AK L+
Sbjct: 325 FGIVQGIQWPALLQALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKVLD 384
Query: 341 ILFEFNVLNIESLNLPSCPLPGHFF--DSNEAIGVNFPVSSF---ISNPSCFPVALHFLK 395
+ F+F + P L G F D +E + VNF + + P+ AL K
Sbjct: 385 LNFDF-----IPILTPIHLLNGSSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTALRLAK 439
Query: 396 QLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQ--KIERHYIQ 453
L P++V + V V + LQ YSA+ ESL+ N+ D + ++ER
Sbjct: 440 SLNPRVVTLGEYEVSLNRVGFANRVKNALQFYSAVFESLEP-NLGRDSEERVRVERELFG 498
Query: 454 PTINKIVLS-----HHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQ 508
I+ ++ H + ++ WR + +GF SN+ +QA+ L+
Sbjct: 499 RRISGLIGPEKTGIHRERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLY 558
Query: 509 --VERKPSSLVLCWQRKELISVSTWR 532
VE KP + L W L+++S+WR
Sbjct: 559 SIVESKPGFISLAWNDLPLLTLSSWR 584
>UniRef100_Q9S7H5 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 413
Score = 135 bits (341), Expect = 2e-30
Identities = 102/372 (27%), Positives = 170/372 (45%), Gaps = 29/372 (7%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNN----L 233
L A+ + N + A+ + L +S G P QR YM E L L ++G++ L
Sbjct: 54 LVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVARLAASGSSIYKSL 113
Query: 234 TAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWS 293
+ P S+ F Y E+ P +F + N ++ EA++ +RIH+IDF IG G QW
Sbjct: 114 QSREPESYEFLSYVYV-LHEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGSQWI 172
Query: 294 SFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESL 353
+ +Q R G P+++IT V S L ++ L + AK ++ F FN ++
Sbjct: 173 ALIQAFAARPGGAPNIRITGVGDGS-----VLVTVKKRLEKLAKKFDVPFRFN-----AV 222
Query: 354 NLPSCPLPGHFFD--SNEAIGVNFPV------SSFISNPSCFPVALHFLKQLRPKIVVTL 405
+ PSC + D EA+GVNF +S + L +K L PK+V +
Sbjct: 223 SRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMVKSLSPKVVTLV 282
Query: 406 DKNCDRMDVPLPTNVVHVLQCYSALLESLDA-VNVNLDVLQKIERHYIQPTINKIVLSHH 464
++ C+ P + L Y+A+ ES+D + N IE+H + + I+
Sbjct: 283 EQECNTNTSPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMARDVVNIIACEG 342
Query: 465 NQR----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCW 520
+R + L W++ F +GF P+ S+ A L+ R G+ +E + +L L W
Sbjct: 343 AERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALL-RDYSNGYAIEERDGALYLGW 401
Query: 521 QRKELISVSTWR 532
+ L+S W+
Sbjct: 402 MDRILVSSCAWK 413
>UniRef100_Q67ZX7 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 413
Score = 135 bits (339), Expect = 4e-30
Identities = 102/372 (27%), Positives = 170/372 (45%), Gaps = 29/372 (7%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNN----L 233
L A+ + N + A+ + L +S G P QR YM E L L ++G++ L
Sbjct: 54 LVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVARLAASGSSIYKSL 113
Query: 234 TAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWS 293
+ P S+ F Y E+ P +F + N ++ EA++ +RIH+IDF IG G QW
Sbjct: 114 QSREPESYEFLSYVYV-LHEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGSQWI 172
Query: 294 SFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESL 353
+ +Q R G P+++IT V S L ++ L + AK ++ F FN ++
Sbjct: 173 ALIQAFAARPGGAPNIRITGVGDGS-----VLVTVKKRLEKLAKKFDVPFRFN-----AV 222
Query: 354 NLPSCPLPGHFFD--SNEAIGVNFPV------SSFISNPSCFPVALHFLKQLRPKIVVTL 405
+ PSC + D EA+GVNF +S + L +K L PK+V +
Sbjct: 223 SRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMVKSLSPKVVTLV 282
Query: 406 DKNCDRMDVPLPTNVVHVLQCYSALLESLDA-VNVNLDVLQKIERHYIQPTINKIVLSHH 464
++ C+ P + L Y+A+ ES+D + N IE+H + + I+
Sbjct: 283 EQECNTNTSPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMARDVVNIMACEG 342
Query: 465 NQR----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCW 520
+R + L W++ F +GF P+ S+ A L+ R G+ +E + +L L W
Sbjct: 343 AERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALL-RDYSNGYAIEERDGALYLGW 401
Query: 521 QRKELISVSTWR 532
+ L+S W+
Sbjct: 402 MDRILVSSCAWK 413
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 888,022,547
Number of Sequences: 2790947
Number of extensions: 37587769
Number of successful extensions: 126739
Number of sequences better than 10.0: 328
Number of HSP's better than 10.0 without gapping: 170
Number of HSP's successfully gapped in prelim test: 171
Number of HSP's that attempted gapping in prelim test: 123353
Number of HSP's gapped (non-prelim): 1061
length of query: 533
length of database: 848,049,833
effective HSP length: 132
effective length of query: 401
effective length of database: 479,644,829
effective search space: 192337576429
effective search space used: 192337576429
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC121238.1


