

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.18 - phase: 0
(387 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
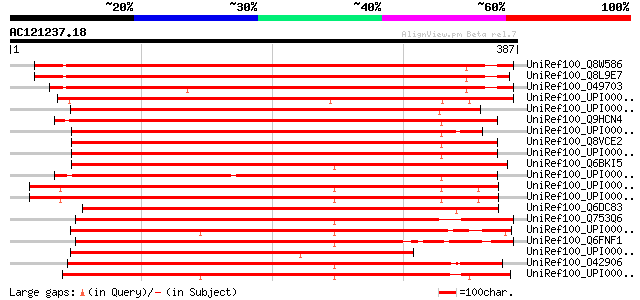
UniRef100_Q8W586 AT4g21800/F17L22_260 [Arabidopsis thaliana] 516 e-145
UniRef100_Q8L9E7 Hypothetical protein [Arabidopsis thaliana] 514 e-144
UniRef100_O49703 Hypothetical protein T8O5.10 [Arabidopsis thali... 499 e-140
UniRef100_UPI000042CFF9 UPI000042CFF9 UniRef100 entry 377 e-103
UniRef100_UPI0000360750 UPI0000360750 UniRef100 entry 363 4e-99
UniRef100_Q9HCN4 XPA-binding protein 1 [Homo sapiens] 362 1e-98
UniRef100_UPI00003AD1B1 UPI00003AD1B1 UniRef100 entry 359 8e-98
UniRef100_Q8VCE2 XPA-binding protein 1 [Mus musculus] 358 2e-97
UniRef100_UPI000036926A UPI000036926A UniRef100 entry 357 2e-97
UniRef100_Q6BKI5 Debaryomyces hansenii chromosome F of strain CB... 355 1e-96
UniRef100_UPI000021D85D UPI000021D85D UniRef100 entry 353 6e-96
UniRef100_UPI000042F8C7 UPI000042F8C7 UniRef100 entry 348 1e-94
UniRef100_UPI000042C294 UPI000042C294 UniRef100 entry 348 1e-94
UniRef100_Q6DC83 Zgc:100927 protein [Brachydanio rerio] 346 6e-94
UniRef100_Q753Q6 AFR428Cp [Ashbya gossypii] 343 6e-93
UniRef100_UPI000021BEF2 UPI000021BEF2 UniRef100 entry 342 1e-92
UniRef100_Q6FNF1 Similar to sp|P47122 Saccharomyces cerevisiae Y... 340 5e-92
UniRef100_UPI00002D0F68 UPI00002D0F68 UniRef100 entry 339 9e-92
UniRef100_O42906 SPBC119.15 protein [Schizosaccharomyces pombe] 338 2e-91
UniRef100_UPI000023E707 UPI000023E707 UniRef100 entry 337 3e-91
>UniRef100_Q8W586 AT4g21800/F17L22_260 [Arabidopsis thaliana]
Length = 379
Score = 516 bits (1328), Expect = e-145
Identities = 259/371 (69%), Positives = 313/371 (83%), Gaps = 15/371 (4%)
Query: 20 KQKEELSESMKKLDIEGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRG 79
++ ++L +S+ KL + +SS S NFK+KP+IIIVVGMAGSGKT+ +HRLV HT S G
Sbjct: 14 EESQKLVDSLDKLRVSAASSSS-NFKKKPIIIIVVGMAGSGKTSFLHRLVCHTFDSKSHG 72
Query: 80 YVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIE 139
YV+NLDPAVM+LP+ +NIDIRDTVKYKEVMKQ+NLGPNGGILTSLNLFATKFDEV+SVIE
Sbjct: 73 YVVNLDPAVMSLPFGANIDIRDTVKYKEVMKQYNLGPNGGILTSLNLFATKFDEVVSVIE 132
Query: 140 KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNM 199
KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVV YVVDTPR+ +P TFMSNM
Sbjct: 133 KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVTYVVDTPRSSSPITFMSNM 192
Query: 200 LYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDFEVFQAAASSDQSYTSNLTQSLSLA 259
LYACSILYKTRLPL+LAFNK DVA H+FALEWM+DFEVFQAA SD SYT+ L SLSL+
Sbjct: 193 LYACSILYKTRLPLVLAFNKTDVADHKFALEWMEDFEVFQAAIQSDNSYTATLANSLSLS 252
Query: 260 LDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKE 319
L EFY N+RS GVSA++GAG++GFFKA+EASAEEYMETYKADLD R+ +K+ LEE R+K
Sbjct: 253 LYEFYRNIRSVGVSAISGAGMDGFFKAIEASAEEYMETYKADLDMRKADKERLEEERKKH 312
Query: 320 NMDKLRREMEKSGGETVVLSTGLKNKEE------DEDEEDEEMDDDDNVDFGTYTEDEDA 373
M+KLR++ME S G TVVL+TGLK+++ +ED+ED +++D+++ D DA
Sbjct: 313 EMEKLRKDMESSQGGTVVLNTGLKDRDATEKMMLEEDDEDFQVEDEEDSD--------DA 364
Query: 374 IDEDEDEEVDK 384
IDED++++ K
Sbjct: 365 IDEDDEDDETK 375
>UniRef100_Q8L9E7 Hypothetical protein [Arabidopsis thaliana]
Length = 379
Score = 514 bits (1325), Expect = e-144
Identities = 258/368 (70%), Positives = 312/368 (84%), Gaps = 15/368 (4%)
Query: 20 KQKEELSESMKKLDIEGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRG 79
++ ++L +S+ KL + +SS S NFK+KP+IIIVVGMAGSGKT+ +HRLV HT S G
Sbjct: 14 EESQKLVDSLDKLRVSAASSSS-NFKKKPIIIIVVGMAGSGKTSFLHRLVCHTFDSKSHG 72
Query: 80 YVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIE 139
YV+NLDPAVM+LP+ +NIDIRDTVKYKEVMKQ+NLGPNGGILTSLNLFATKFDEV+SVIE
Sbjct: 73 YVVNLDPAVMSLPFGANIDIRDTVKYKEVMKQYNLGPNGGILTSLNLFATKFDEVVSVIE 132
Query: 140 KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNM 199
KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVV YVVDTPR+ +P TFMSNM
Sbjct: 133 KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVTYVVDTPRSSSPITFMSNM 192
Query: 200 LYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDFEVFQAAASSDQSYTSNLTQSLSLA 259
LYACSILYKTRLPL+LAFNK DVA H+FALEWM+DFEVFQAA SD SYT+ L SLSL+
Sbjct: 193 LYACSILYKTRLPLVLAFNKTDVADHKFALEWMEDFEVFQAAIQSDXSYTATLANSLSLS 252
Query: 260 LDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKE 319
L EFY N+RS GVSA++GAG++GFFKA+EASAEEYMETYKADLD R+ +K+ LEE R+K
Sbjct: 253 LYEFYRNIRSVGVSAISGAGMDGFFKAIEASAEEYMETYKADLDMRKADKERLEEERKKH 312
Query: 320 NMDKLRREMEKSGGETVVLSTGLKNKEE------DEDEEDEEMDDDDNVDFGTYTEDEDA 373
M+KLR++ME S G TVVL+TGLK+++ +ED+ED +++D+++ D DA
Sbjct: 313 EMEKLRKDMESSQGGTVVLNTGLKDRDATEKMMLEEDDEDFQVEDEEDSD--------DA 364
Query: 374 IDEDEDEE 381
IDED++++
Sbjct: 365 IDEDDEDD 372
>UniRef100_O49703 Hypothetical protein T8O5.10 [Arabidopsis thaliana]
Length = 373
Score = 499 bits (1284), Expect = e-140
Identities = 257/378 (67%), Positives = 305/378 (79%), Gaps = 33/378 (8%)
Query: 31 KLDIEGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMT 90
KL + +SS S NFK+KP+IIIVVGMAGSGKT+ +HRLV HT S GYV+NLDPAVM+
Sbjct: 1 KLRVSAASSSS-NFKKKPIIIIVVGMAGSGKTSFLHRLVCHTFDSKSHGYVVNLDPAVMS 59
Query: 91 LPYASNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEV---------------- 134
LP+ +NIDIRDTVKYKEVMKQ+NLGPNGGILTSLNLFATKFDEV
Sbjct: 60 LPFGANIDIRDTVKYKEVMKQYNLGPNGGILTSLNLFATKFDEVNIPLELVQMITDFFSH 119
Query: 135 --ISVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENP 192
+SVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVV YVVDTPR+ +P
Sbjct: 120 CVVSVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVTYVVDTPRSSSP 179
Query: 193 TTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDFEVFQAAASSDQSYTSNL 252
TFMSNMLYACSILYKTRLPL+LAFNK DVA H+FALEWM+DFEVFQAA SD SYT+ L
Sbjct: 180 ITFMSNMLYACSILYKTRLPLVLAFNKTDVADHKFALEWMEDFEVFQAAIQSDNSYTATL 239
Query: 253 TQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLL 312
SLSL+L EFY N+RS GVSA++GAG++GFFKA+EASAEEYMETYKADLD R+ +K+ L
Sbjct: 240 ANSLSLSLYEFYRNIRSVGVSAISGAGMDGFFKAIEASAEEYMETYKADLDMRKADKERL 299
Query: 313 EENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE------DEDEEDEEMDDDDNVDFGT 366
EE R+K M+KLR++ME S G TVVL+TGLK+++ +ED+ED +++D+++ D
Sbjct: 300 EEERKKHEMEKLRKDMESSQGGTVVLNTGLKDRDATEKMMLEEDDEDFQVEDEEDSD--- 356
Query: 367 YTEDEDAIDEDEDEEVDK 384
DAIDED++++ K
Sbjct: 357 -----DAIDEDDEDDETK 369
>UniRef100_UPI000042CFF9 UPI000042CFF9 UniRef100 entry
Length = 405
Score = 377 bits (969), Expect = e-103
Identities = 194/364 (53%), Positives = 255/364 (69%), Gaps = 16/364 (4%)
Query: 37 SSSGSPNF--KRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYA 94
SS+ P+ K++PV+I+ +GMAGSGKTTLM RL +H H N Y++NLDPAV +PY+
Sbjct: 12 SSTNPPSVEGKKQPVVILCIGMAGSGKTTLMQRLNSHLHSKNTPPYILNLDPAVTHMPYS 71
Query: 95 SNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPG 154
+NIDIRDTV YKEVMKQ+ LGPNGGILT+LNLF TKFD+V+ +EKRA+ +DY+LVDTPG
Sbjct: 72 ANIDIRDTVDYKEVMKQYKLGPNGGILTALNLFTTKFDQVLGYVEKRAETVDYILVDTPG 131
Query: 155 QIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLI 214
QIEIFTWSASGAIIT+A AS+ PTVVAY+VDTPR +P TFMSNMLYACSILYKT+LP I
Sbjct: 132 QIEIFTWSASGAIITDAIASSLPTVVAYIVDTPRTASPVTFMSNMLYACSILYKTKLPFI 191
Query: 215 LAFNKVDVAQHEFALEWMKDFEVFQAAAS-------SDQSYTSNLTQSLSLALDEFYSNL 267
+ FNK+DV HEFAL+WM DFE +Q A + + SY ++L S++L L+EFY+NL
Sbjct: 192 IVFNKIDVQPHEFALDWMTDFEKYQEALNDKSRDEHGEGSYVNSLMSSMNLVLEEFYNNL 251
Query: 268 RSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRRE 327
R+ GVSA+TG G++ FF AVE + +EY YK +LD+ E+ E +K +++L R+
Sbjct: 252 RAVGVSAMTGEGMKAFFSAVEEARKEYETDYKPELDRLAAERAAQTEADKKAQLERLMRD 311
Query: 328 ME-----KSGGETVVLSTGLKNKEEDE--DEEDEEMDDDDNVDFGTYTEDEDAIDEDEDE 380
M +SG +N ED D+E E D D+ + E+ ++ E E
Sbjct: 312 MNISDSPRSGPGGNPFGPHARNDREDRYYDDEGEASDIDEQEQEAIRRQMEEEEEDAEAE 371
Query: 381 EVDK 384
E+ K
Sbjct: 372 ELGK 375
>UniRef100_UPI0000360750 UPI0000360750 UniRef100 entry
Length = 357
Score = 363 bits (932), Expect = 4e-99
Identities = 179/327 (54%), Positives = 234/327 (70%), Gaps = 14/327 (4%)
Query: 47 KPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYK 106
KPV +IV+GMAGSGKTT + RL H H YV+NLDPAV +P+ +NIDIRDTV YK
Sbjct: 14 KPVCLIVLGMAGSGKTTFVQRLTAHLHSVKSPPYVINLDPAVHEVPFPANIDIRDTVNYK 73
Query: 107 EVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGA 166
EVMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEK+ YVL+DTPGQIE+FTWSASG
Sbjct: 74 EVMKQYGLGPNGGIVTSLNLFATRFDQVMQFIEKKQQNHRYVLIDTPGQIEVFTWSASGT 133
Query: 167 IITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHE 226
IITEA AS+FP VV YV+DT R+ NP TFMSNMLYACSILYKT+LP I+ NK D+ H
Sbjct: 134 IITEALASSFPCVVIYVMDTSRSVNPVTFMSNMLYACSILYKTKLPFIVVMNKTDIIDHS 193
Query: 227 FALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKA 286
FA+EWM+DFEVFQ A + + SY SNLT+S+SL LDEFY+NLR GVSAVTG+G++ F
Sbjct: 194 FAVEWMQDFEVFQDALNQETSYVSNLTRSMSLVLDEFYANLRVVGVSAVTGSGLDKLFVQ 253
Query: 287 VEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRR--------------EMEKSG 332
VE +A+EY Y+ + ++ R+E + +++E +++LR+ E + +G
Sbjct: 254 VEDAAQEYERDYRPEYERLRKELVEAQNRKQQEQLERLRKDLGAVDMATTALTEEADVAG 313
Query: 333 GETVVLSTGLKNKEEDEDEEDEEMDDD 359
+++ G ++ EE+ D + DDD
Sbjct: 314 PSDFIMTRGNRDDEEEAAASDSDTDDD 340
>UniRef100_Q9HCN4 XPA-binding protein 1 [Homo sapiens]
Length = 374
Score = 362 bits (929), Expect = 1e-98
Identities = 186/348 (53%), Positives = 240/348 (68%), Gaps = 13/348 (3%)
Query: 35 EGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYA 94
E +SG P R PV ++V+GMAGSGKTT + RL H H YV+NLDPAV +P+
Sbjct: 9 ELQASGGP---RHPVCLLVLGMAGSGKTTFVQRLTGHLHAQGTPPYVINLDPAVHEVPFP 65
Query: 95 SNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPG 154
+NIDIRDTVKYKEVMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEK + YVL+DTPG
Sbjct: 66 ANIDIRDTVKYKEVMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNMSKYVLIDTPG 125
Query: 155 QIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLI 214
QIE+FTWSASG IITEA AS+FPTVV YV+DT R+ NP TFMSNMLYACSILYKT+LP I
Sbjct: 126 QIEVFTWSASGTIITEALASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFI 185
Query: 215 LAFNKVDVAQHEFALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSA 274
+ NK D+ H FA+EWM+DFE FQ A + + +Y SNLT+S+SL LDEFYS+LR GVSA
Sbjct: 186 VVMNKTDIIDHSFAVEWMQDFEAFQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSA 245
Query: 275 VTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREM------ 328
V G G++ F V ++AEEY Y+ + ++ ++ E +++E +++LR++M
Sbjct: 246 VLGTGLDELFVQVTSAAEEYEREYRPEYERLKKSLANAESQQQREQLERLRKDMGSVALD 305
Query: 329 ---EKSGGETVVLSTGLKNKEEDEDEEDEEMDDD-DNVDFGTYTEDED 372
K V+ + L DEEDEE D D D++D E +
Sbjct: 306 AGTAKDSLSPVLHPSDLILTRGTLDEEDEEADSDTDDIDHRVTEESHE 353
>UniRef100_UPI00003AD1B1 UPI00003AD1B1 UniRef100 entry
Length = 364
Score = 359 bits (921), Expect = 8e-98
Identities = 176/330 (53%), Positives = 234/330 (70%), Gaps = 18/330 (5%)
Query: 48 PVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKE 107
PV ++V+GMAGSGKTT + L H H YV+NLDPAV LP+ +NIDIRDTVKYKE
Sbjct: 8 PVCVLVLGMAGSGKTTFVQCLAAHLHGQRCPPYVINLDPAVHELPFPANIDIRDTVKYKE 67
Query: 108 VMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAI 167
VMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEKR + YV++DTPGQIE+FTWSASG I
Sbjct: 68 VMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKRQNASKYVIIDTPGQIEVFTWSASGTI 127
Query: 168 ITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEF 227
ITEA AS+FP+VV YV+DT R+ NP TFMSNMLYACSILYKT+LP I+ NK D+ H F
Sbjct: 128 ITEALASSFPSVVVYVMDTSRSTNPITFMSNMLYACSILYKTKLPFIVVMNKTDIIDHSF 187
Query: 228 ALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAV 287
A+EWM+DFE FQ A + + SY SNLT+S+SL LDEFYS+L+ GVSAV G G++ FF +
Sbjct: 188 AVEWMQDFETFQDALNQETSYVSNLTRSMSLVLDEFYSSLKVVGVSAVLGTGLDDFFVQL 247
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREM----------------EKS 331
+ EEY Y+ + ++ R+ + + ++K+ +++LR++M
Sbjct: 248 SKAVEEYEREYRPEYERLRKTLEKAQNKQKKDQLERLRKDMGPVCMQGNTLAGSDDASAM 307
Query: 332 GGETVVLSTGLKNKEEDEDEEDEEMDDDDN 361
G ++L+ G N +EDE+E + + DD D+
Sbjct: 308 GPSELILTRG--NLDEDEEERESDTDDIDH 335
>UniRef100_Q8VCE2 XPA-binding protein 1 [Mus musculus]
Length = 372
Score = 358 bits (918), Expect = 2e-97
Identities = 181/335 (54%), Positives = 233/335 (69%), Gaps = 10/335 (2%)
Query: 48 PVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKE 107
PV ++V+GMAGSGKTT + RL H H YV+NLDPAV +P+ +NIDIRDTVKYKE
Sbjct: 19 PVCLLVLGMAGSGKTTFVQRLTGHLHNKGCPPYVINLDPAVHEVPFPANIDIRDTVKYKE 78
Query: 108 VMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAI 167
VMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEK + YVL+DTPGQIE+FTWSASG I
Sbjct: 79 VMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNTFRYVLIDTPGQIEVFTWSASGTI 138
Query: 168 ITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEF 227
ITEA AS+FPTVV YV+DT R+ NP TFMSNMLYACSILYKT+LP I+ NK D+ H F
Sbjct: 139 ITEALASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFIVVMNKTDIIDHSF 198
Query: 228 ALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAV 287
A+EWM+DFE FQ A + + +Y SNLT+S+SL LDEFYS+LR GVSAV G G + V
Sbjct: 199 AVEWMQDFEAFQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSAVVGTGFDELCTQV 258
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREM---------EKSGGETVVL 338
++AEEY Y+ + ++ ++ + N++KE +++LR++M K V+
Sbjct: 259 TSAAEEYEREYRPEYERLKKSLANAQSNQQKEQLERLRKDMGSVALDPEAGKGNASPVLD 318
Query: 339 STGLKNKEEDEDEEDEEMDDD-DNVDFGTYTEDED 372
+ L DEEDEE D D D++D E +
Sbjct: 319 PSDLILTRGTLDEEDEEADSDTDDIDHRVTEESRE 353
>UniRef100_UPI000036926A UPI000036926A UniRef100 entry
Length = 374
Score = 357 bits (917), Expect = 2e-97
Identities = 180/335 (53%), Positives = 234/335 (69%), Gaps = 10/335 (2%)
Query: 48 PVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKE 107
PV ++V+GMAGSGKTT + RL H H YV+NLDPAV +P+ +NIDIRDTVKYKE
Sbjct: 19 PVCLLVLGMAGSGKTTFVQRLTGHLHAQGTPPYVINLDPAVHEVPFPANIDIRDTVKYKE 78
Query: 108 VMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAI 167
VMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEK + YVL+DTPGQIE+FTWSASG I
Sbjct: 79 VMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNMSKYVLIDTPGQIEVFTWSASGTI 138
Query: 168 ITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEF 227
ITEA AS+FPTVV YV+DT R+ NP TFMSNMLYACSILYKT+LP I+ NK D+ H F
Sbjct: 139 ITEALASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFIVVMNKTDIIDHSF 198
Query: 228 ALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAV 287
A+EWM+DFE FQ A + + +Y SNLT+S+SL LDEFYS+LR GVSAV G G++ F V
Sbjct: 199 AVEWMQDFEAFQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSAVLGTGLDELFVQV 258
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREM---------EKSGGETVVL 338
++AEEY Y+ + ++ ++ E +++E +++LR++M + V+
Sbjct: 259 TSAAEEYEREYRPEYERLKKSLANAESQQQREQLERLRKDMGSVALDAGTARDSLSPVLH 318
Query: 339 STGLKNKEEDEDEEDEEMDDD-DNVDFGTYTEDED 372
+ L DEEDEE D D D++D E +
Sbjct: 319 PSDLILTRGTLDEEDEEADSDTDDIDHRVTEESHE 353
>UniRef100_Q6BKI5 Debaryomyces hansenii chromosome F of strain CBS767 of Debaryomyces
hansenii [Debaryomyces hansenii]
Length = 380
Score = 355 bits (911), Expect = 1e-96
Identities = 182/344 (52%), Positives = 241/344 (69%), Gaps = 11/344 (3%)
Query: 48 PVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKE 107
P +I +GMAGSGKTT M RL +H H YV+NLDPAV+ +P+ +NIDIRD+VKYK+
Sbjct: 3 PSTVICIGMAGSGKTTFMQRLNSHLHAKKSPPYVINLDPAVLKVPFGANIDIRDSVKYKK 62
Query: 108 VMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAI 167
VM+++NLGPNG I+TSLNLFATK D+VI ++EKR+D+++ V++DTPGQIE F WSASGAI
Sbjct: 63 VMEEYNLGPNGAIVTSLNLFATKIDQVIKLVEKRSDKVENVIIDTPGQIECFIWSASGAI 122
Query: 168 ITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEF 227
ITEAFASTFPTV+AY+VDTPR +PTTFMSNMLYACSILYKT+LP+I+ FNK DV +F
Sbjct: 123 ITEAFASTFPTVIAYIVDTPRNSSPTTFMSNMLYACSILYKTKLPMIIVFNKSDVKNADF 182
Query: 228 ALEWMKDFEVFQAAASSDQ----------SYTSNLTQSLSLALDEFYSNLRSAGVSAVTG 277
A EWM+DFE FQ A S DQ Y S+L S+SL L+EFYS L GVS+ TG
Sbjct: 183 AKEWMQDFETFQRALSEDQELNGEDGTSSGYMSSLVNSMSLMLEEFYSQLDVVGVSSYTG 242
Query: 278 AGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREME-KSGGETV 336
G + F AV+ +EY E YKA+ ++ +EK+ E R+ +++KL ++++ K +
Sbjct: 243 EGFDEFLGAVDNKVDEYNEYYKAERERILKEKEEKENKRQSSSLNKLMKDLDIKDKNQEG 302
Query: 337 VLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDE 380
S L + E ++DE E+ D++ YT ED E D+
Sbjct: 303 KDSEVLSDYESEDDEYQGELPRDEDEVEREYTFPEDRNSEVNDK 346
>UniRef100_UPI000021D85D UPI000021D85D UniRef100 entry
Length = 370
Score = 353 bits (905), Expect = 6e-96
Identities = 183/348 (52%), Positives = 236/348 (67%), Gaps = 16/348 (4%)
Query: 35 EGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYA 94
E +SG+P PV ++V+GMAGSGKTT + RL H H YV+NLDPAV +P+
Sbjct: 9 EPQASGAPEL---PVCLLVLGMAGSGKTTFVQRLTGHLHNKGCPPYVINLDPAVHEVPFP 65
Query: 95 SNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPG 154
+NIDIRDTVKYKEVMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEK + YVL+DTPG
Sbjct: 66 ANIDIRDTVKYKEVMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNTFRYVLIDTPG 125
Query: 155 QIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLI 214
QIE+FTWSASG IIT AS+FPTVV YV+DT R+ NP TFMSNMLYACSILYKT+LP I
Sbjct: 126 QIEVFTWSASGTIIT---ASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFI 182
Query: 215 LAFNKVDVAQHEFALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSA 274
+ NK D+ H FA+EWM+DFE FQ A + + +Y SNLT+S+SL LDEFYS+LR GVSA
Sbjct: 183 VVMNKTDIIDHSFAVEWMQDFEAFQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSA 242
Query: 275 VTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREM------ 328
V G G + V ++AEEY Y+ + ++ ++ + N +KE +++LR++M
Sbjct: 243 VVGTGFDELCAQVTSAAEEYEREYRPEYERLKKSLASAQSNEQKEQLERLRKDMGSIALD 302
Query: 329 ---EKSGGETVVLSTGLKNKEEDEDEEDEEMDDD-DNVDFGTYTEDED 372
G V+ + L DEEDEE D D D++D E +
Sbjct: 303 PGASTDGASPVLDPSDLILTRGTLDEEDEEADSDTDDIDHRVTEESRE 350
>UniRef100_UPI000042F8C7 UPI000042F8C7 UniRef100 entry
Length = 425
Score = 348 bits (894), Expect = 1e-94
Identities = 181/379 (47%), Positives = 259/379 (67%), Gaps = 21/379 (5%)
Query: 16 KEKNKQKEELSESMKKLDIEGS----SSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTH 71
+ K+K +++ S+ D++ S ++ P II +GMAGSGKTT + RL +H
Sbjct: 6 RRTEKKKYKINHSLLLFDLKPSPILYTANHHTMPTPPPTIICIGMAGSGKTTFVQRLNSH 65
Query: 72 THLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKF 131
H Y++NLDPAV+ +P+ +NIDIRD+VKYK+VM+++NLGPNG I+TSLNLF+TK
Sbjct: 66 LHSKKTPPYLINLDPAVLKIPFGANIDIRDSVKYKKVMEEYNLGPNGAIVTSLNLFSTKI 125
Query: 132 DEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAEN 191
D+VI +I+K+ D+++ V++DTPGQIE F WSASG+IITE+FAS FPTV+AY+VDTPR +
Sbjct: 126 DQVIKLIDKKQDKINNVVIDTPGQIECFIWSASGSIITESFASEFPTVIAYIVDTPRNTS 185
Query: 192 PTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDFEVFQAAASSDQ----- 246
PTTFMSNMLYACSILYKT+LP+I+ FNK DV + +FA EWM DFE FQ A D+
Sbjct: 186 PTTFMSNMLYACSILYKTKLPMIVVFNKTDVTKDDFAKEWMTDFESFQMAIQKDKDLNNE 245
Query: 247 ---SYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLD 303
Y S+L S+SL L+EFYSNL GVS+ TG G + F +AV+ +EY E YKA+ +
Sbjct: 246 QGSGYMSSLINSMSLMLEEFYSNLDVVGVSSYTGQGFDKFMEAVDNKVDEYNEFYKAEKE 305
Query: 304 KRREEKQLLEENRRKENMDKLRREM---EKSGGETVVLSTGLKNKEEDEDEEDEEM---- 356
+ ++K+ E+ R+ ++++KL ++M + G T S L + EED++E D+E+
Sbjct: 306 RILKQKEEDEKKRQTKSLNKLMKDMKMKDTKGDHTKKDSEVLSDYEEDDNEIDDEIQGEV 365
Query: 357 --DDDDNVDFGTYTEDEDA 373
D+D+ T+ ED +
Sbjct: 366 LRDEDEPEREYTFPEDRQS 384
>UniRef100_UPI000042C294 UPI000042C294 UniRef100 entry
Length = 425
Score = 348 bits (894), Expect = 1e-94
Identities = 181/379 (47%), Positives = 259/379 (67%), Gaps = 21/379 (5%)
Query: 16 KEKNKQKEELSESMKKLDIEGS----SSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTH 71
+ K+K +++ S+ D++ S ++ P II +GMAGSGKTT + RL +H
Sbjct: 6 RRTEKKKYKINHSLLLFDLKPSPILYTANHHTMPTPPPTIICIGMAGSGKTTFVQRLNSH 65
Query: 72 THLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKF 131
H Y++NLDPAV+ +P+ +NIDIRD+VKYK+VM+++NLGPNG I+TSLNLF+TK
Sbjct: 66 LHSKKTPPYLINLDPAVLKIPFGANIDIRDSVKYKKVMEEYNLGPNGAIVTSLNLFSTKI 125
Query: 132 DEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAEN 191
D+VI +I+K+ D+++ V++DTPGQIE F WSASG+IITE+FAS FPTV+AY+VDTPR +
Sbjct: 126 DQVIKLIDKKQDKINNVVIDTPGQIECFIWSASGSIITESFASEFPTVIAYIVDTPRNTS 185
Query: 192 PTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDFEVFQAAASSDQ----- 246
PTTFMSNMLYACSILYKT+LP+I+ FNK DV + +FA EWM DFE FQ A D+
Sbjct: 186 PTTFMSNMLYACSILYKTKLPMIVVFNKTDVTKDDFAKEWMTDFESFQMAIQKDKDLNNE 245
Query: 247 ---SYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLD 303
Y S+L S+SL L+EFYSNL GVS+ TG G + F +AV+ +EY E YKA+ +
Sbjct: 246 QGSGYMSSLINSMSLMLEEFYSNLDVVGVSSYTGQGFDKFMEAVDNKVDEYNEFYKAERE 305
Query: 304 KRREEKQLLEENRRKENMDKLRREM---EKSGGETVVLSTGLKNKEEDEDEEDEEM---- 356
+ ++K+ E+ R+ ++++KL ++M + G T S L + EED++E D+E+
Sbjct: 306 RILKQKEEDEKKRQTKSLNKLMKDMKMKDTKGDHTKKDSEVLSDYEEDDNEIDDEIQGEV 365
Query: 357 --DDDDNVDFGTYTEDEDA 373
D+D+ T+ ED +
Sbjct: 366 LRDEDEPEREYTFPEDRQS 384
>UniRef100_Q6DC83 Zgc:100927 protein [Brachydanio rerio]
Length = 349
Score = 346 bits (888), Expect = 6e-94
Identities = 175/327 (53%), Positives = 230/327 (69%), Gaps = 9/327 (2%)
Query: 56 MAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLG 115
MAGSGKTT + RL + H YV+NLDPAV +P+ +NIDIRDTV YKEVMKQ+ LG
Sbjct: 1 MAGSGKTTFVQRLTAYLHSKKTPPYVINLDPAVHEVPFPANIDIRDTVNYKEVMKQYGLG 60
Query: 116 PNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITEAFAST 175
PNGGI+TSLNLFAT+FD+V+ IEK+ +YVL+DTPGQIE+FTWSASG IITEA AS+
Sbjct: 61 PNGGIVTSLNLFATRFDQVMKFIEKKQSNHEYVLIDTPGQIEVFTWSASGTIITEALASS 120
Query: 176 FPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDF 235
FP VV YV+DT R+ NP TFMSNMLYACSILYKT+LP I+ NK D+ H FA+EWM+DF
Sbjct: 121 FPCVVIYVMDTSRSVNPVTFMSNMLYACSILYKTKLPFIVVMNKTDIIDHSFAVEWMQDF 180
Query: 236 EVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYM 295
EVFQ A + + SY SNLT+S+SL LDEFY+NLR GVSAVTG+G++ F V +A+EY
Sbjct: 181 EVFQDALNQETSYISNLTRSMSLVLDEFYTNLRVVGVSAVTGSGLDELFVQVADAAKEYE 240
Query: 296 ETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLS--------TGLKNKEE 347
Y+ + ++ E + +++E +++LR++M ET + + L
Sbjct: 241 TEYRPEYERLHRELAEAQSQKQQEQLERLRKDMGAVPMETAAPTEAPDLGGPSDLIFTRG 300
Query: 348 DEDEEDEEMDDD-DNVDFGTYTEDEDA 373
+ DE +EE+D D D++D E ++A
Sbjct: 301 NPDEAEEEIDSDTDDIDHTVTEESKEA 327
>UniRef100_Q753Q6 AFR428Cp [Ashbya gossypii]
Length = 383
Score = 343 bits (879), Expect = 6e-93
Identities = 175/345 (50%), Positives = 232/345 (66%), Gaps = 27/345 (7%)
Query: 51 IIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMK 110
I+ +GMAGSGKTT M RL +H H S YV+NLDPAV+ +PY +NIDIRD++KYK+VM
Sbjct: 4 ILCIGMAGSGKTTFMQRLNSHLHASKEPPYVINLDPAVLKVPYGANIDIRDSIKYKKVMS 63
Query: 111 QFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITE 170
+NLGPNG I+TSLNLF+TK D+VI ++EK+ D+ + +VDTPGQIE F WSASG+IITE
Sbjct: 64 NYNLGPNGAIVTSLNLFSTKIDQVIGLVEKKQDKYQHCIVDTPGQIECFVWSASGSIITE 123
Query: 171 AFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFALE 230
+FASTFPTV+AY+VDTPR +PTTFMSNMLYACSILYKT+LP+++ FNK DV FA E
Sbjct: 124 SFASTFPTVIAYIVDTPRNTSPTTFMSNMLYACSILYKTKLPMVVVFNKTDVQDASFAKE 183
Query: 231 WMKDFEVFQAAASSDQ----------SYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGI 280
WM DFE FQ A +DQ Y +L S+SL L+EFYS L GVS+ TG G+
Sbjct: 184 WMTDFEAFQEALRADQELNGEAGLGSGYMGSLINSMSLVLEEFYSQLDMVGVSSFTGEGV 243
Query: 281 EGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLST 340
F +AV+ AEEY YKA+ + + EK EE R++++++KL +++
Sbjct: 244 SDFLRAVDKKAEEYETDYKAEYQRLQAEKAQKEEARKEKSLEKLMKDL------------ 291
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAI-DEDEDEEVDK 384
+D ++ + DD NV + D I DEDE+V++
Sbjct: 292 ----GVQDATKKPADTDDGVNVVSDIEGREHDGIVPRDEDEDVER 332
>UniRef100_UPI000021BEF2 UPI000021BEF2 UniRef100 entry
Length = 408
Score = 342 bits (876), Expect = 1e-92
Identities = 181/368 (49%), Positives = 239/368 (64%), Gaps = 43/368 (11%)
Query: 47 KPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYK 106
KPV ++ VGMAGSGKTT M R+ H H YV+NLDPAV+ +P+ SNIDIRD+V YK
Sbjct: 17 KPVAVVCVGMAGSGKTTFMQRINAHLHGKKDPPYVINLDPAVLNVPFDSNIDIRDSVNYK 76
Query: 107 EVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQ----------LDYVLVDTPGQI 156
EVMKQ+NLGPNGGILTSLNLFATK D++++++EKR + ++++LVDTPGQI
Sbjct: 77 EVMKQYNLGPNGGILTSLNLFATKVDQILNLLEKRTARDTPENAGRKPIEHILVDTPGQI 136
Query: 157 EIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILA 216
E F WSASG I+ E+ AS+FPTV+AYVVDTPR + +TFMSNMLYACSILYKT+LP+IL
Sbjct: 137 EAFVWSASGTILLESLASSFPTVIAYVVDTPRTRSTSTFMSNMLYACSILYKTKLPMILV 196
Query: 217 FNKVDVAQHEFALEWMKDFEVFQAAASSDQ-----------------SYTSNLTQSLSLA 259
FNK DV FA EWM DF+ FQAA D+ Y S+L S+S+
Sbjct: 197 FNKTDVQDASFAKEWMTDFDAFQAALREDEERNAFGSAEGESGSGGSGYMSSLLNSMSMM 256
Query: 260 LDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKE 319
LDEFY++L GVS++TG G++ FF AV AEE+ Y+ +L++RREE+ ++ R++
Sbjct: 257 LDEFYAHLSVVGVSSMTGHGVDDFFTAVAEKAEEFRRDYQPELERRREERDEAKKLIREQ 316
Query: 320 NMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDED-- 377
+DK+ + M +G + TG + EDE N D DAID D
Sbjct: 317 ELDKMMKGMSFAGDKD---GTGASGSNDAEDE---------NPDIAALGRRVDAIDSDDS 364
Query: 378 --EDEEVD 383
+D+E+D
Sbjct: 365 VADDDEMD 372
>UniRef100_Q6FNF1 Similar to sp|P47122 Saccharomyces cerevisiae YJR072c [Candida
glabrata]
Length = 382
Score = 340 bits (871), Expect = 5e-92
Identities = 175/344 (50%), Positives = 236/344 (67%), Gaps = 28/344 (8%)
Query: 51 IIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMK 110
+I +GMAGSGKTT M RL +H YV+NLDPAV+ +PY +NIDIRD++KYK+VM+
Sbjct: 6 VICIGMAGSGKTTFMQRLNSHIRSKKEVPYVINLDPAVLRVPYGANIDIRDSIKYKKVME 65
Query: 111 QFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITE 170
+ LGPNG I+TSLNLF+TK D+VI ++EK+ D D+ ++DTPGQIE F WSASG+IITE
Sbjct: 66 NYQLGPNGAIVTSLNLFSTKIDQVIKLVEKKRDTHDFCIIDTPGQIECFVWSASGSIITE 125
Query: 171 AFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFALE 230
+FASTFPTV+AY+VDTPR +PTTFMSNMLYACSILYKT+LP+I+ FNK DV + +FA E
Sbjct: 126 SFASTFPTVIAYIVDTPRNSSPTTFMSNMLYACSILYKTKLPMIIVFNKTDVKKSDFAKE 185
Query: 231 WMKDFEVFQAAASSDQ----------SYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGI 280
WM DFE FQ A DQ Y S+L S+SL L+EFYS L GVS+ TG G
Sbjct: 186 WMTDFEAFQQAVREDQDANGDFGMGSGYMSSLVNSMSLMLEEFYSTLDVVGVSSFTGEGF 245
Query: 281 EGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLST 340
+ F AV+ +EY E YKA E +++L ++KE +KLR++ +G ++
Sbjct: 246 DDFLDAVDKKVDEYEEFYKA------ERERIL---KQKEEEEKLRKDKSLNG---LMKDL 293
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
GL +K++ ED+ + + D + E++ +D DEDE V++
Sbjct: 294 GLNDKKDKEDDSADVISDLEE------GENDGLVDRDEDEGVER 331
>UniRef100_UPI00002D0F68 UPI00002D0F68 UniRef100 entry
Length = 442
Score = 339 bits (869), Expect = 9e-92
Identities = 166/266 (62%), Positives = 205/266 (76%), Gaps = 4/266 (1%)
Query: 47 KPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYK 106
KPV +IV+GMAGSGKTT + RL H H YV+NLDPAV +P+ +NIDIRDTV YK
Sbjct: 37 KPVCLIVLGMAGSGKTTFVQRLTAHLHSIEAPPYVINLDPAVHQVPFPANIDIRDTVNYK 96
Query: 107 EVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGA 166
EVMKQF LGPNGGI+TSLNLFAT+FD+V+ IEK+ +VL+DTPGQIE+FTWSASG
Sbjct: 97 EVMKQFGLGPNGGIVTSLNLFATRFDQVMQFIEKKQQNHRFVLIDTPGQIEVFTWSASGT 156
Query: 167 IITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKV----DV 222
IITEA AS+FP VV YV+DT R+ NP TFMSNMLYACSILYKT+LP I+ NK+ D+
Sbjct: 157 IITEALASSFPCVVVYVMDTSRSVNPVTFMSNMLYACSILYKTKLPFIVVMNKLLLQTDI 216
Query: 223 AQHEFALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEG 282
H FA+EWM+DFEVFQ A + + SY SNLT+S+SL LDEFY++LR GVSAVTG+G++
Sbjct: 217 IDHSFAVEWMQDFEVFQDALNQETSYVSNLTRSMSLVLDEFYTSLRVVGVSAVTGSGLDQ 276
Query: 283 FFKAVEASAEEYMETYKADLDKRREE 308
F+ VE +A EY Y+ + ++ R E
Sbjct: 277 LFRQVEDAAAEYERDYRPEYERLRRE 302
>UniRef100_O42906 SPBC119.15 protein [Schizosaccharomyces pombe]
Length = 367
Score = 338 bits (866), Expect = 2e-91
Identities = 173/341 (50%), Positives = 236/341 (68%), Gaps = 13/341 (3%)
Query: 45 KRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVK 104
++KP IIVVGMAGSGKTT M +L H H N Y++NLDPAV LPY +NIDIRDT+
Sbjct: 5 EKKPCAIIVVGMAGSGKTTFMQQLNAHLHSKNKPPYILNLDPAVRNLPYEANIDIRDTIN 64
Query: 105 YKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSAS 164
YKEVMKQ+NLGPNGGI+TSLNLF TKFD+V+ ++EKRA +D++L+DTPGQIEIF WSAS
Sbjct: 65 YKEVMKQYNLGPNGGIMTSLNLFVTKFDQVLKILEKRAPTVDHILIDTPGQIEIFQWSAS 124
Query: 165 GAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQ 224
G+II + AS++PT +AYVVDTPRA + +T+MS+MLYACS+LYK +LPLI+ +NK DV
Sbjct: 125 GSIICDTLASSWPTCIAYVVDTPRATSTSTWMSSMLYACSMLYKAKLPLIIVYNKCDVQD 184
Query: 225 HEFALEWMKDFEVFQAAASSDQ---------SYTSNLTQSLSLALDEFYSNLRSAGVSAV 275
EFA +WM DFE FQ A + D+ Y +L S+SL L+EFY +L S+V
Sbjct: 185 SEFAKKWMTDFEEFQQAVTKDEGMSSEGATSGYMGSLVNSMSLMLEEFYRHLDFVSCSSV 244
Query: 276 TGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGET 335
TG G++ F +AV+A +EY E Y ++++ +E ++ +E +++ + KL ++M S +
Sbjct: 245 TGEGMDDFLEAVKAKVKEYEEEYVPEMERMKEIQRQTKERQKEAQLSKLMKDMHVSKDKE 304
Query: 336 VVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDE 376
V GL + EDE + E+ D D D T + ED I +
Sbjct: 305 DV---GL-TVSDAEDEYNGELVDPDEDDGLTAEDREDMIKQ 341
>UniRef100_UPI000023E707 UPI000023E707 UniRef100 entry
Length = 388
Score = 337 bits (865), Expect = 3e-91
Identities = 174/372 (46%), Positives = 242/372 (64%), Gaps = 38/372 (10%)
Query: 41 SPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIR 100
+P PV I+ VGMAGSGKTT M R+ H H ++ YV+NLDPAV+++P+ SNIDIR
Sbjct: 11 APKASSPPVAIVCVGMAGSGKTTFMRRINAHLHQNDTPPYVINLDPAVLSVPFESNIDIR 70
Query: 101 DTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQ---------LDYVLVD 151
D+V Y+EVMKQ+NLGPNGGILTSLNLFATK D++++++EKRA +D +LVD
Sbjct: 71 DSVNYEEVMKQYNLGPNGGILTSLNLFATKVDQIVNLLEKRAQPDPEKPDRKPIDRILVD 130
Query: 152 TPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRL 211
TPGQIE+F WSASG I+ E+ AS+FPTV+AYV+DTPR + +TFMSNMLYACSILYKT+L
Sbjct: 131 TPGQIEVFVWSASGTILLESLASSFPTVIAYVIDTPRTASTSTFMSNMLYACSILYKTKL 190
Query: 212 PLILAFNKVDVAQHEFALEWMKDFEVFQAAASSDQ----------------SYTSNLTQS 255
P+IL FNK DV EFA EWM DFE FQ A D+ Y +L S
Sbjct: 191 PMILVFNKTDVKDAEFAKEWMTDFEAFQEALRKDEESDELGGVEGGGHGGSGYMGSLLNS 250
Query: 256 LSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEEN 315
+SL L+EFY++L GVS+ G G++ FF+AVE +E++ Y+ +L+++R E+ ++
Sbjct: 251 MSLMLEEFYAHLSVVGVSSRLGTGVDEFFEAVEEKRQEFLRDYQPELERKRAERDEQKKA 310
Query: 316 RRKENMDKLRREMEKSGGETVVLSTGLKNKEEDE-----DEEDEEMDDDDNVDFGTYTED 370
R++ +DK+ + M G KEED+ D++ ++ D D+ + +
Sbjct: 311 TREKELDKMMQGMSVGGLPVA--------KEEDDADVASDDDGDDTDSDEEIQKQGLQDR 362
Query: 371 EDAIDEDEDEEV 382
+A D+D+ V
Sbjct: 363 YEAAMGDQDDSV 374
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 646,562,192
Number of Sequences: 2790947
Number of extensions: 28911588
Number of successful extensions: 452244
Number of sequences better than 10.0: 5207
Number of HSP's better than 10.0 without gapping: 1958
Number of HSP's successfully gapped in prelim test: 3534
Number of HSP's that attempted gapping in prelim test: 333464
Number of HSP's gapped (non-prelim): 43896
length of query: 387
length of database: 848,049,833
effective HSP length: 129
effective length of query: 258
effective length of database: 488,017,670
effective search space: 125908558860
effective search space used: 125908558860
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC121237.18


