

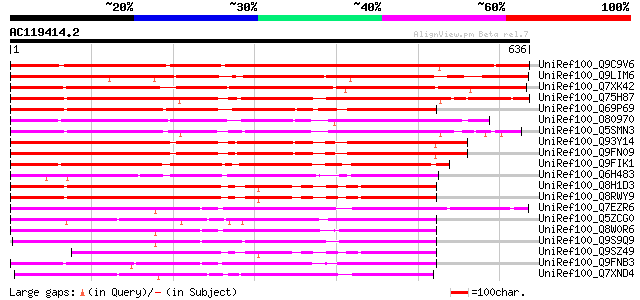
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119414.2 + phase: 0
(636 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C9V6 Hypothetical protein T23K23.25 [Arabidopsis tha... 889 0.0
UniRef100_Q9LIM6 Gb|AAF16571.1 [Arabidopsis thaliana] 549 e-154
UniRef100_Q7XK42 OSJNBa0044K18.3 protein [Oryza sativa] 516 e-145
UniRef100_Q75H87 Putative NPH3 family protein [Oryza sativa] 471 e-131
UniRef100_Q69P69 Putative RPT2 [Oryza sativa] 436 e-120
UniRef100_O80970 Hypothetical protein At2g14820 [Arabidopsis tha... 436 e-120
UniRef100_Q5SMN3 Non-phototropic hypocotyl 3-like [Oryza sativa] 435 e-120
UniRef100_Q93Y14 Photoreceptor-interacting protein-like [Arabido... 429 e-119
UniRef100_Q9FN09 Photoreceptor-interacting protein-like [Arabido... 429 e-118
UniRef100_Q9FIK1 Putative photoreceptor-interacting protein [Ara... 428 e-118
UniRef100_Q6H483 Putative non-phototropic hypocotyl 3 [Oryza sat... 387 e-106
UniRef100_Q8H1D3 Hypothetical protein At4g31820 [Arabidopsis tha... 385 e-105
UniRef100_Q8RWY9 Hypothetical protein At4g31820 [Arabidopsis tha... 383 e-104
UniRef100_Q7EZR6 Putative non-phototropic hypocotyl 3 (NPH3)/pho... 363 1e-98
UniRef100_Q5ZCG0 Putative non-phototropic hypocotyl 3 [Oryza sat... 358 3e-97
UniRef100_Q8W0R6 Putative photoreceptor-interacting protein [Sor... 348 4e-94
UniRef100_Q9S9Q9 F26G16.2 protein [Arabidopsis thaliana] 344 5e-93
UniRef100_Q9SZ49 Hypothetical protein AT4g31820 [Arabidopsis tha... 338 3e-91
UniRef100_Q9FNB3 Photoreceptor-interacting protein-like; non-pho... 338 4e-91
UniRef100_Q7XND4 OSJNBb0034I13.5 protein [Oryza sativa] 329 1e-88
>UniRef100_Q9C9V6 Hypothetical protein T23K23.25 [Arabidopsis thaliana]
Length = 631
Score = 889 bits (2297), Expect = 0.0
Identities = 468/645 (72%), Positives = 534/645 (82%), Gaps = 24/645 (3%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSES 60
MKFMKLGSRPDTFYTSE +R +SSEVSSD I+V GSRYLLHKFPLLSKCL LQR+CSES
Sbjct: 1 MKFMKLGSRPDTFYTSEDLRCVSSEVSSDFTIEVSGSRYLLHKFPLLSKCLRLQRMCSES 60
Query: 61 PPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKG 120
P I+QLP+FPGGVEAFELCAKFCYGI IT+S YNIVAARCAAEYLQM+EEVEKG
Sbjct: 61 P-----ESIIQLPEFPGGVEAFELCAKFCYGITITISAYNIVAARCAAEYLQMSEEVEKG 115
Query: 121 NLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKVS 180
NL++KLEVFFNSCIL+GW+D+IV+LQTTKA LWSEDL ITSRCIE IASKVLSHP+KVS
Sbjct: 116 NLVYKLEVFFNSCILNGWRDSIVTLQTTKAFPLWSEDLAITSRCIEAIASKVLSHPSKVS 175
Query: 181 LSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPSNL 240
LSHSHSRRVRDD D S R + S+GWWAED+AEL IDLYWRTMIAIKSGGKVP++L
Sbjct: 176 LSHSHSRRVRDD----DMSSNRAAASSRGWWAEDIAELGIDLYWRTMIAIKSGGKVPASL 231
Query: 241 IGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLPTEK 300
IGDAL++YAS+WLP + +N + V + +S+SDS ++ +SKHRLLLESI+SLLP EK
Sbjct: 232 IGDALRVYASKWLPTLQRN----RKVVKKKEDSDSDSDTDTSSKHRLLLESIISLLPAEK 287
Query: 301 GAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTLYDVE 360
GAVSCSFLLKLLKA+NILNAS+SSKMELARRV LQLEEATV+DLLIP +SY ++ LYDV+
Sbjct: 288 GAVSCSFLLKLLKAANILNASTSSKMELARRVALQLEEATVSDLLIPPMSYKSELLYDVD 347
Query: 361 LVMTILEQFMLQGQ-SPPTSPPRSLKTF--ERRRSRSAENINFELQESRRSSSASHSSKL 417
+V TILEQFM+QGQ SPPTSP R K RRRSRSAENI+ E QESRRSSSASHSSKL
Sbjct: 348 IVATILEQFMVQGQTSPPTSPLRGKKGMMDRRRRSRSAENIDLEFQESRRSSSASHSSKL 407
Query: 418 KVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKSE 477
KVAKLVD YLQ++ARDVN LSKF+ LA+ +PEF+R DHDDLYRAIDIYLKAH LNKSE
Sbjct: 408 KVAKLVDGYLQQIARDVNLPLSKFVTLAESVPEFSRLDHDDLYRAIDIYLKAHKNLNKSE 467
Query: 478 RKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSGG----KVTELQSN 533
RKR+CR+LDCKKLSMEACMHAAQNE+LPLRVVVQVLF+EQARAA++ TEL SN
Sbjct: 468 RKRVCRVLDCKKLSMEACMHAAQNEMLPLRVVVQVLFYEQARAAAATNNGEKNTTELPSN 527
Query: 534 IKALLTTHGIDPSKHTTTQLSSTTSIQG-EDNWSVSGFKSPKSK-SSTLRMKLAEEHDEF 591
IKALL H IDPS S+TTSI ED WSVSG KSPKSK S TLR KLAE+ +E
Sbjct: 528 IKALLAAHNIDPSNPNAAAFSTTTSIAAPEDRWSVSGLKSPKSKLSGTLRSKLAED-EEV 586
Query: 592 DENGLVNDDGIGRNSKFKSICALPTQPKKMLSKLWSTNRTATEKN 636
DE +N DG S+FK+ CA+P +PKKM SKL S NR +++KN
Sbjct: 587 DER-FMNGDGGRTPSRFKAFCAIPGRPKKMFSKLLSINRNSSDKN 630
>UniRef100_Q9LIM6 Gb|AAF16571.1 [Arabidopsis thaliana]
Length = 588
Score = 549 bits (1414), Expect = e-154
Identities = 326/651 (50%), Positives = 418/651 (64%), Gaps = 80/651 (12%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSES 60
MKFM+LG RPDTFYT E++R++SS++ +DL+IQV+ ++YLLHKFP+LSKCL L+ L S
Sbjct: 1 MKFMELGFRPDTFYTVESVRSVSSDLLNDLVIQVKSTKYLLHKFPMLSKCLRLKNLVSSQ 60
Query: 61 PPDSSHHQ-IVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEK 119
++S Q ++QL DFPG EAFELCAKFCYGI ITL +N+VA RCAAEYL MTEEVE
Sbjct: 61 ETETSQEQQVIQLVDFPGETEAFELCAKFCYGITITLCAHNVVAVRCAAEYLGMTEEVEL 120
Query: 120 G---NLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHP 176
G NL+ +LE+F +C+ W+D+ V+LQTTK L LWSEDLGIT+RCIE IA+ V P
Sbjct: 121 GETENLVQRLELFLTTCVFKSWRDSYVTLQTTKVLPLWSEDLGITNRCIEAIANGVTVSP 180
Query: 177 -------TKVSLSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIA 229
+ L + SR RD+I CN K+ S WW EDLAEL +DLY RTM+A
Sbjct: 181 GEDFSTQLETGLLRNRSRIRRDEILCNGGGGS--KAESLRWWGEDLAELGLDLYRRTMVA 238
Query: 230 IKSGG-KVPSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLL 288
IKS K+ LIG+AL+IYAS+WLP+I ES++DS L+
Sbjct: 239 IKSSHRKISPRLIGNALRIYASKWLPSIQ--------------ESSADS--------NLV 276
Query: 289 LESIVSLLPTEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPS 348
LES++SLLP EK +V CSFLL+LLK +N++N S SSKMELA + G QL++ATV++LLIP
Sbjct: 277 LESVISLLPEEKSSVPCSFLLQLLKMANVMNVSHSSKMELAIKAGNQLDKATVSELLIP- 335
Query: 349 LSYTNDTLYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFE-LQESRR 407
LS + LYDV++V +++QF L SP P R+ E RRSRS ENIN E +QE R
Sbjct: 336 LSDKSGMLYDVDVVKMMVKQF-LSHISPEIRPTRTRT--EHRRSRSEENINLEEIQEVRG 392
Query: 408 SSSASHSSK---LKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAID 464
S S S S KVAKLVD YLQE+ARDVN ++SKF+ LA+ IP+ +R HDDLY AID
Sbjct: 393 SLSTSSSPPPLLSKVAKLVDSYLQEIARDVNLTVSKFVELAETIPDTSRICHDDLYNAID 452
Query: 465 IYLKAHPELNKSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSG 524
+YL+ H ++ K ERKRLCRILDCKKLS+EA AAQNELLPLRV+VQ+LF EQARA
Sbjct: 453 VYLQVHKKIEKCERKRLCRILDCKKLSVEASKKAAQNELLPLRVIVQILFVEQARA---- 508
Query: 525 GKVTELQSNIKALLTTHGIDPSKHTTTQLSSTTSIQGEDNWSVSGFKSPKSKSSTLRMKL 584
L TT + T S T T R +
Sbjct: 509 -----------TLATTRNNITTNETAVLKRSFT---------------------TRREEG 536
Query: 585 AEEHDEFDENGLVNDDGIGRNSKFKSICALPTQPKKMLSKLWSTNRTATEK 635
+E +E DE S+F ++CA+P QPKK+L KL S +R+ +++
Sbjct: 537 GQEEEERDETKPSGGFLQSTPSRFMALCAIPRQPKKLLCKLLSISRSLSQR 587
>UniRef100_Q7XK42 OSJNBa0044K18.3 protein [Oryza sativa]
Length = 627
Score = 516 bits (1330), Expect = e-145
Identities = 298/655 (45%), Positives = 405/655 (61%), Gaps = 50/655 (7%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSES 60
M+ MKLG+RPDTF++S +R++S+++++D+ I V G + LHKFPLLSKC+ LQ LC ES
Sbjct: 1 MRVMKLGNRPDTFFSSGPVRSVSTDLATDMQILVDGCLFRLHKFPLLSKCMWLQALCVES 60
Query: 61 PPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKG 120
V+LP FPGG EAFE CAKFCYG+ +T+ P+N+VA RCAA L M+E ++G
Sbjct: 61 GDGGG---AVELPAFPGGAEAFEACAKFCYGVAVTIGPHNVVAVRCAAARLGMSEAADRG 117
Query: 121 NLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKVS 180
NL KL+ F +SC+L WKD + L +T+ E+LG+TSRC++ +A+ + +
Sbjct: 118 NLAAKLDAFLSSCLLRRWKDALAVLHSTRRYAALCEELGVTSRCVDAVAALAVGDASGAV 177
Query: 181 LSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPSNL 240
+ S S S WWA D++EL +DLYWR M+A+K+ G V +
Sbjct: 178 PAGS-------------------SSSSPPWWARDISELGVDLYWRVMVAVKATGTVHAKA 218
Query: 241 IGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLPTEK 300
IGD LK YA RWLP +KN + + + + +A T HRLL+E IVSLLP E+
Sbjct: 219 IGDTLKAYARRWLPIAAKNHHAAERTA-GGGGGGAANAERATKNHRLLVEKIVSLLPAER 277
Query: 301 GAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLS-YTNDTLYDV 359
AVSC FLLKLLKA+NIL AS +SK EL RRV QLE+A V+DLLIP+ LYDV
Sbjct: 278 NAVSCGFLLKLLKAANILGASPASKAELTRRVASQLEDANVSDLLIPAPPPCAGGVLYDV 337
Query: 360 ELVMTILEQFMLQGQSPPTSPPRS-LKTFERRRSRSAENINFELQESRRSSS---ASHSS 415
+ V+TILE+F L+ + SP S + RRS SAE+ EL+ +RRS+S SH +
Sbjct: 338 DAVVTILEEFALRQAAASGSPKGSPARAGRHRRSMSAES--GELEGARRSTSMAAVSHGA 395
Query: 416 KLKVAKLVDRYLQEVA-RDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELN 474
++V +LVD +L VA +D L K IA+A+ +P+FAR +HDDLYRAID YL+AHPE++
Sbjct: 396 MVRVGRLVDGFLAMVATKDARTPLDKMIAVAEAVPDFARPEHDDLYRAIDTYLRAHPEMD 455
Query: 475 KSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSGGKVTELQSNI 534
KS RK+LCR+L+C+KLS +A MHAAQNELLPLRVVVQVLFFE ARAA G + + +
Sbjct: 456 KSSRKKLCRVLNCRKLSEKASMHAAQNELLPLRVVVQVLFFENARAA---GLSSGHGNRV 512
Query: 535 KALLTTHGIDPSKHTTTQLSSTTSIQGED--------------NWSVSGFKSPKSKSSTL 580
A G D S T TT G+D +WSV G + S+ +TL
Sbjct: 513 AARFPGDGGDVSALLGTGRPRTTEENGKDGQSPAAAGSVAADGDWSVEGLRRAASRVATL 572
Query: 581 RMKLAEEH-DEFDENGLVNDDGIG-RNSKFKSICALPTQPKKMLSKLWSTNRTAT 633
+M+L EE ++ + V+ G S + A + K+MLS+LW T+RT T
Sbjct: 573 KMRLEEEDGEDAGDEAFVHRTRAGLARSASSRVTAAAGRSKRMLSRLWPTSRTFT 627
>UniRef100_Q75H87 Putative NPH3 family protein [Oryza sativa]
Length = 634
Score = 471 bits (1213), Expect = e-131
Identities = 273/643 (42%), Positives = 401/643 (61%), Gaps = 45/643 (6%)
Query: 1 MKFMKLGSRPDTFYTSEA-IRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE 59
MK MKLGS+PD F T IR +++E+++D++I + ++ LHKFPLLSK CLQRL +
Sbjct: 1 MKHMKLGSKPDLFQTEGGNIRFVATELATDIVISIGDVKFCLHKFPLLSKSSCLQRLAAS 60
Query: 60 SPPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEK 119
S + + + + D PGG AFE+CAKFCYG+ +TL+ YN++AARCAAEYL+M E +EK
Sbjct: 61 SNVEGNEE--LDISDIPGGPSAFEICAKFCYGMIVTLNAYNVLAARCAAEYLEMFETIEK 118
Query: 120 GNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKV 179
GNLI+K++VF S I WKD+I+ LQ+TK+L WSE+L + + CI++IASK L P++V
Sbjct: 119 GNLIYKIDVFLTSSIFRAWKDSIIVLQSTKSLLPWSENLKVINHCIDSIASKALIDPSEV 178
Query: 180 SLSHSHSRRVRDDISCNDTESLRIKSG---SKGWWAEDLAELSIDLYWRTMIAIKSGGKV 236
S++++R+ + +D+ ++ K WW EDL +L +DLY R ++ IK+ G+
Sbjct: 179 EWSYTYNRKKLPSENGHDSHWNGVRKQLIVPKDWWVEDLCDLEMDLYKRVIMMIKAKGRT 238
Query: 237 PSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLL 296
+IG+AL+ YA R L SL ++ SN + K R +LE+I+ LL
Sbjct: 239 SPIVIGEALRAYAYRRLLG-----------SLEDAVSNGVDCT----KRRAVLETIIFLL 283
Query: 297 PTEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTL 356
PTEKG+VSC FLLKLLKA+ +L A S L +R+G QL+ A+V+DLLIP+ + N TL
Sbjct: 284 PTEKGSVSCGFLLKLLKAACLLEAGESCHDILIKRIGTQLDGASVSDLLIPANTSEN-TL 342
Query: 357 YDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSSK 416
Y+V L++ I+E+F+ + + + + E+ E + S +S
Sbjct: 343 YNVNLIIAIVEEFVSR---------------QSDTGKMKFQDDDEIVEVENLTPVSSTSN 387
Query: 417 LKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKS 476
L VA L+D YL E+A+D N LSKFIA+A+++P +R +HD LYRAID+YLK HP L+KS
Sbjct: 388 LAVANLIDGYLAEIAKDTNLPLSKFIAIAEMVPPASRKNHDGLYRAIDMYLKEHPSLSKS 447
Query: 477 ERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSGGK---VTELQSN 533
E+K LCR++DCKKLS +AC+HA QNE LPLRVVVQVLFFEQ RA+++ G+ EL S
Sbjct: 448 EKKALCRLMDCKKLSQDACLHAVQNERLPLRVVVQVLFFEQIRASAASGRTDAAAELTSA 507
Query: 534 IKALLTTHGIDPSKHTTTQLSSTTSIQGEDNWSVSGFKSPKSKSSTLRMKLAEEHDEFDE 593
+ +LL + + + +++ ++TT+ + ED V S SS + +
Sbjct: 508 VHSLLPRE--NGNSYGSSRSAATTTTE-EDGTGVPTSSDINSFSSLRLANNSGGSERSSG 564
Query: 594 NGLVNDDGIGRNSKFKSICALPTQPKKMLSKLWSTNRTATEKN 636
+ +N+ S KS +L PKK+LSKLWS A+E +
Sbjct: 565 SSDINNKSCDDKSSSKSKGSL--MPKKILSKLWSGKTNASENS 605
>UniRef100_Q69P69 Putative RPT2 [Oryza sativa]
Length = 618
Score = 436 bits (1121), Expect = e-120
Identities = 245/527 (46%), Positives = 342/527 (64%), Gaps = 43/527 (8%)
Query: 1 MKFMKLGSRPDTFYTSEA-IRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE 59
MKFMKLGS+PD F + A +R + S++++D+I+ V ++ LHKFPLLSK LQRL +
Sbjct: 1 MKFMKLGSKPDAFQSDGADVRYVISDLATDVIVHVSEVKFYLHKFPLLSKSSKLQRLVIK 60
Query: 60 SPPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEK 119
+ + + V + FPGGV AFE+CAKFCYG+ +TLSP+N+VAARCAAEYL+MTE+V+K
Sbjct: 61 ATEEGTDE--VHIDGFPGGVTAFEICAKFCYGMVVTLSPHNVVAARCAAEYLEMTEDVDK 118
Query: 120 GNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKV 179
GNLI K++VF NS IL WKD+I+ LQ+TKAL WSE+L + RCI+ IASK P V
Sbjct: 119 GNLIFKIDVFINSSILRSWKDSIIVLQSTKALLPWSEELKVIGRCIDAIASKTSVDPANV 178
Query: 180 SLSHSHSRRVRDDISCND-TESL-RIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVP 237
+ S+SHSR+ +SC + ES R K WW EDL EL +DLY R M+A+KS G++
Sbjct: 179 TWSYSHSRK---GMSCTEIVESTGRTSIAPKDWWVEDLCELDVDLYKRVMVAVKSKGRMS 235
Query: 238 SNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLP 297
LIG+ALK YA RWLP+ ++ A + +++ L+E+I+ LLP
Sbjct: 236 PELIGEALKAYAVRWLPD----------------SYDALVAEDYMRRNQCLVETIIWLLP 279
Query: 298 TEK-GAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTL 356
++K SC FLLKLLK + ++ A K EL RR+ QL +A+V DLL+P+ S +D
Sbjct: 280 SDKTSGCSCRFLLKLLKVAILVGAGQHVKEELMRRISFQLHKASVKDLLLPAAS-PSDGA 338
Query: 357 YDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSSK 416
+DV+LV ++++F+ + T+ + E+ + E +NFE S
Sbjct: 339 HDVKLVHNLVQRFVAR-----TAMSHNGGFVEKSDDKMIE-LNFE-----------QEST 381
Query: 417 LKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKS 476
L + +LVD YL EVA D + SLS F+ LA +PE AR HD LY A+D YLK HP ++K+
Sbjct: 382 LALGELVDGYLSEVASDPDLSLSTFVELATAVPEAARPVHDSLYSAVDAYLKEHPNISKA 441
Query: 477 ERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASS 523
++K++C ++D KKLS +A MHA QN+ LPLR+VVQVLFF+Q RA SS
Sbjct: 442 DKKKICGLIDVKKLSTDASMHATQNDRLPLRLVVQVLFFQQLRAGSS 488
>UniRef100_O80970 Hypothetical protein At2g14820 [Arabidopsis thaliana]
Length = 634
Score = 436 bits (1120), Expect = e-120
Identities = 255/598 (42%), Positives = 359/598 (59%), Gaps = 57/598 (9%)
Query: 1 MKFMKLGSRPDTFYTS-EAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE 59
MKFMK+GS+ D+F T +R + +E++SD+ + V GSR+ LHKFPLLSKC CLQ+L S
Sbjct: 1 MKFMKIGSKLDSFKTDGNNVRYVENELASDISVDVEGSRFCLHKFPLLSKCACLQKLLSS 60
Query: 60 SPPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEK 119
+ D ++ + + PGG AFE CAKFCYG+ +TLS YN+VA RCAAEYL M E VEK
Sbjct: 61 T--DKNNIDDIDISGIPGGPTAFETCAKFCYGMTVTLSAYNVVATRCAAEYLGMHETVEK 118
Query: 120 GNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKV 179
GNLI+K++VF +S + WKD+I+ LQTTK SEDL + S CI+ IA+K + V
Sbjct: 119 GNLIYKIDVFLSSSLFRSWKDSIIVLQTTKPFLPLSEDLKLVSLCIDAIATKACVDVSHV 178
Query: 180 SLSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPSN 239
S++++++ + + N +S++ + WW EDL EL ID Y R ++ IK+ +
Sbjct: 179 EWSYTYNKKKLAEEN-NGADSIKARDVPHDWWVEDLCELEIDYYKRVIMNIKTKCILGGE 237
Query: 240 LIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLPTE 299
+IG+ALK Y R L +K + ++ KH+ ++E++V LLP E
Sbjct: 238 VIGEALKAYGYRRLSGFNKGVMEQGDLV----------------KHKTIIETLVWLLPAE 281
Query: 300 KGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTLYDV 359
K +VSC FLLKLLKA ++N+ K +L RR+G QLEEA++ +LLI S ++TLYDV
Sbjct: 282 KNSVSCGFLLKLLKAVTMVNSGEVVKEQLVRRIGQQLEEASMAELLIKS-HQGSETLYDV 340
Query: 360 ELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAE------NINFELQESRRSSSA-S 412
+LV I+ +FM RR +++E FE+QE R+ S
Sbjct: 341 DLVQKIVMEFM-------------------RRDKNSEIEVQDDEDGFEVQEVRKLPGILS 381
Query: 413 HSSKLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPE 472
+SKL VAK++D YL E+A+D N SKFI +A+ + R HD LYRAID++LK HP
Sbjct: 382 EASKLMVAKVIDSYLTEIAKDPNLPASKFIDVAESVTSIPRPAHDALYRAIDMFLKEHPG 441
Query: 473 LNKSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSGGKVT-ELQ 531
+ K E+KR+C+++DC+KLS+EACMHA QN+ LPLRVVVQVLFFEQ RAA+S G T +L
Sbjct: 442 ITKGEKKRMCKLMDCRKLSVEACMHAVQNDRLPLRVVVQVLFFEQVRAAASSGSSTPDLP 501
Query: 532 SNIKALLTTHGIDPSKHTTTQLSSTTSIQGEDNWSVSGFKSPKS--KSSTLRMKLAEE 587
+ L + G S + + ED W + KS +KL EE
Sbjct: 502 RGMGRELRSCGTYGSSRSVPTVM-------EDEWEAVATEEEMRALKSEIAALKLQEE 552
>UniRef100_Q5SMN3 Non-phototropic hypocotyl 3-like [Oryza sativa]
Length = 644
Score = 435 bits (1118), Expect = e-120
Identities = 273/651 (41%), Positives = 376/651 (56%), Gaps = 78/651 (11%)
Query: 1 MKFMKLGSRPDTFYTSEA--IRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCS 58
MKFMKLGS+PD F T R + SE+ SD++I V +R+ LHKFPLLSK LQRL
Sbjct: 1 MKFMKLGSKPDAFQTDGGGDSRYVLSELPSDIVIHVEEARFYLHKFPLLSKSSLLQRLII 60
Query: 59 ESPPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVE 118
E+ + + V + D PGGV+ FE+CAKFCYG+ +TL+ YN+VAARCAAE L MTE+V+
Sbjct: 61 EASQNGTDE--VYIHDIPGGVKIFEICAKFCYGMVVTLNAYNVVAARCAAELLGMTEDVD 118
Query: 119 KGNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTK 178
K NL+ K+EVF NS I WKD+I++LQTT AL WSE L + +RCI++IASK S+P
Sbjct: 119 KSNLVFKIEVFLNSGIFRSWKDSIIALQTTDALLPWSEQLKLAARCIDSIASKATSNPCN 178
Query: 179 VSLSHSHSRRVRDDISCNDTESLRIKSGS----KGWWAEDLAELSIDLYWRTMIAIKSGG 234
V S++++R+ S + E + + S K WW EDL EL +DLY R M+A+KS G
Sbjct: 179 VVWSYTYNRK-----SASSDEIVEARKNSQPVPKDWWVEDLCELDVDLYKRVMVAVKSRG 233
Query: 235 KVPSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVS 294
++ S+++G+ALK YASRWLP ++ + D A + H LLE+IV
Sbjct: 234 RITSDVVGEALKAYASRWLP------------ECFDTAAIDDDAYSMAYNH--LLETIVW 279
Query: 295 LLPTEKGA--VSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYT 352
LLP++KG+ SC F LKLLK + ++ + K EL RV LQL +A+V DLLIP+
Sbjct: 280 LLPSDKGSSCCSCRFFLKLLKVAVLIGSGEMLKEELMDRVILQLHKASVCDLLIPARPPA 339
Query: 353 NDTLYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINF--ELQESRRSSS 410
T YD++LV+T++ +FM RR+ E+ F L + +
Sbjct: 340 L-TTYDIQLVLTLVGRFM-------------------RRAGVTEDGIFLNNLDQEMFETD 379
Query: 411 ASHSSKLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAH 470
S L ++K+VD YL EVA D N S+S F+A+A +P+ AR HD LY AID++LK H
Sbjct: 380 VDDESLLALSKIVDGYLAEVASDPNLSVSSFVAVATSMPDAARATHDGLYTAIDVFLKLH 439
Query: 471 PELNKSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSGGK---- 526
P L K+E++++ ++D KKLS EAC+HAAQN+ LPLRVVVQVLFFEQ RAA+ G
Sbjct: 440 PNLPKAEKRKISSLMDVKKLSKEACIHAAQNDRLPLRVVVQVLFFEQLRAAAGGNNPAAA 499
Query: 527 VTELQSNIKALLTTHGIDPSKHTTTQLSSTTSIQGEDNWSVSGFKSPKSKSSTLR----- 581
I LL D G +WS S P S L+
Sbjct: 500 AAAASGGIARLLVEEEDDD--------DDDVGGGGGGDWSKSR-ALPTPTPSLLKKQLGS 550
Query: 582 MKLAEEHDEFDENGLVNDDG-----IGRNSKFKSICALPTQPKKMLSKLWS 627
+KLA DE G DDG + + S +L ++ ++M +LW+
Sbjct: 551 LKLAAAGDE----GGGGDDGRQLARVSSVANQSSRLSLSSRSRRMFDRLWA 597
>UniRef100_Q93Y14 Photoreceptor-interacting protein-like [Arabidopsis thaliana]
Length = 579
Score = 429 bits (1104), Expect = e-119
Identities = 238/573 (41%), Positives = 352/573 (60%), Gaps = 58/573 (10%)
Query: 1 MKFMKLGSRPDTFYTSE-AIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE 59
MKFMKLGS+PD+F + E +R +++E+++D+++ V ++ LHKFPLLSK LQ+L +
Sbjct: 1 MKFMKLGSKPDSFQSDEDCVRYVATELATDVVVIVGDVKFHLHKFPLLSKSARLQKLIAT 60
Query: 60 SPPDS-SHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVE 118
+ D S +++PD PGG AFE+CAKFCYG+ +TL+ YN+VA RCAAEYL+M E +E
Sbjct: 61 TTTDEQSDDDEIRIPDIPGGPPAFEICAKFCYGMAVTLNAYNVVAVRCAAEYLEMYESIE 120
Query: 119 KGNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTK 178
GNL++K+EVF NS +L WKD+I+ LQTT++ + WSED+ + RC+E+IA K P +
Sbjct: 121 NGNLVYKMEVFLNSSVLRSWKDSIIVLQTTRSFYPWSEDVKLDVRCLESIALKAAMDPAR 180
Query: 179 VSLSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPS 238
V S++++RR N+ S + WW EDLAELSIDL+ R + I+ G V
Sbjct: 181 VDWSYTYNRRKLLPPEMNN------NSVPRDWWVEDLAELSIDLFKRVVSTIRRKGGVLP 234
Query: 239 NLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLPT 298
+IG+AL++YA++ +P + ++++N D E + R LLE++VS+LP+
Sbjct: 235 EVIGEALEVYAAKRIPGF-----------MIQNDNNDD--EEDVMEQRSLLETLVSMLPS 281
Query: 299 EKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTLYD 358
EK +VSC FL+KLLK+S + EL+RR+G +LEEA V DLLI + +T+YD
Sbjct: 282 EKQSVSCGFLIKLLKSSVSFECGEEERKELSRRIGEKLEEANVGDLLIRA-PEGGETVYD 340
Query: 359 VELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSSKLK 418
+++V T++++F+ Q T +R +++IN SSK
Sbjct: 341 IDIVETLIDEFVTQ-------------TEKRDELDCSDDIN-------------DSSKAN 374
Query: 419 VAKLVDRYLQEVAR-DVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKSE 477
VAKL+D YL E++R + N S +KFI +A+ + F R HD +YRAID++LK HP + KSE
Sbjct: 375 VAKLIDGYLAEISRIETNLSTTKFITIAEKVSTFPRQSHDGVYRAIDMFLKQHPGITKSE 434
Query: 478 RKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARA---------ASSGGKVT 528
+K +++DC+KLS EAC HA QNE LPLRVVVQ+LFFEQ RA S G T
Sbjct: 435 KKSSSKLMDCRKLSPEACAHAVQNERLPLRVVVQILFFEQVRATTKPSLPPSGSHGSSRT 494
Query: 529 ELQSNIKALLTTHGIDPSKHTTTQLSSTTSIQG 561
+ +++ T + T S T +G
Sbjct: 495 TTEEECESVTATEETTTTTRDKTSSSEKTKAKG 527
>UniRef100_Q9FN09 Photoreceptor-interacting protein-like [Arabidopsis thaliana]
Length = 579
Score = 429 bits (1103), Expect = e-118
Identities = 238/573 (41%), Positives = 351/573 (60%), Gaps = 58/573 (10%)
Query: 1 MKFMKLGSRPDTFYTSE-AIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE 59
MKFMKLGS+PD+F + E +R +++E+++D+++ V ++ LHKFPLLSK LQ+L +
Sbjct: 1 MKFMKLGSKPDSFQSDEDCVRYVATELATDVVVIVGDVKFHLHKFPLLSKSARLQKLIAT 60
Query: 60 SPPDS-SHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVE 118
+ D S +++PD PGG AFE+CAKFCYG+ +TL+ YN+VA RCAAEYL+M E +E
Sbjct: 61 TTTDEQSDDDEIRIPDIPGGPPAFEICAKFCYGMAVTLNAYNVVAVRCAAEYLEMYESIE 120
Query: 119 KGNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTK 178
GNL++K+EVF NS +L WKD+I+ LQTT++ + WSED+ + RC+E+IA K P +
Sbjct: 121 NGNLVYKMEVFLNSSVLRSWKDSIIVLQTTRSFYPWSEDVKLDVRCLESIALKAAMDPAR 180
Query: 179 VSLSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPS 238
V S++++RR N+ S + WW EDLAELSIDL+ R + I+ G V
Sbjct: 181 VDWSYTYNRRKLLPPEMNN------NSVPRDWWVEDLAELSIDLFKRVVSTIRRKGGVLP 234
Query: 239 NLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLPT 298
+IG+AL++YA++ +P + +++ N D E + R LLE++VS+LP+
Sbjct: 235 EVIGEALEVYAAKRIPGF-----------MIQNDDNDD--EEDVMEQRSLLETLVSMLPS 281
Query: 299 EKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTLYD 358
EK +VSC FL+KLLK+S + EL+RR+G +LEEA V DLLI + +T+YD
Sbjct: 282 EKQSVSCGFLIKLLKSSVSFECGEEERKELSRRIGEKLEEANVGDLLIRA-PEGGETVYD 340
Query: 359 VELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSSKLK 418
+++V T++++F+ Q T +R +++IN SSK
Sbjct: 341 IDIVETLIDEFVTQ-------------TEKRDELDCSDDIN-------------DSSKAN 374
Query: 419 VAKLVDRYLQEVAR-DVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKSE 477
VAKL+D YL E++R + N S +KFI +A+ + F R HD +YRAID++LK HP + KSE
Sbjct: 375 VAKLIDGYLAEISRIETNLSTTKFITIAEKVSTFPRQSHDGVYRAIDMFLKQHPGITKSE 434
Query: 478 RKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARA---------ASSGGKVT 528
+K +++DC+KLS EAC HA QNE LPLRVVVQ+LFFEQ RA S G T
Sbjct: 435 KKSSSKLMDCRKLSPEACAHAVQNERLPLRVVVQILFFEQVRATTKPSLPPSGSHGSSRT 494
Query: 529 ELQSNIKALLTTHGIDPSKHTTTQLSSTTSIQG 561
+ +++ T + T S T +G
Sbjct: 495 TTEEECESVTATEETTTTTRDKTSSSEKTKAKG 527
>UniRef100_Q9FIK1 Putative photoreceptor-interacting protein [Arabidopsis thaliana]
Length = 559
Score = 428 bits (1100), Expect = e-118
Identities = 230/539 (42%), Positives = 343/539 (62%), Gaps = 47/539 (8%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSES 60
MKFMK+G++PDTFYT EA R + ++ +DL+I++ + Y LH+ L+ KC L+RLC++
Sbjct: 1 MKFMKIGTKPDTFYTQEASRILITDTPNDLVIRINNTTYHLHRSCLVPKCGLLRRLCTDL 60
Query: 61 PPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKG 120
+ S ++L D PGG +AFELCAKFCY I I LS +N+V A CA+++L+M++ V+KG
Sbjct: 61 --EESDTVTIELNDIPGGADAFELCAKFCYDITINLSAHNLVNALCASKFLRMSDSVDKG 118
Query: 121 NLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKVS 180
NL+ KLE FF+SCIL GWKD+IV+LQ+T L W E+LGI +CI++I K+L+ ++VS
Sbjct: 119 NLLPKLEAFFHSCILQGWKDSIVTLQSTTKLPEWCENLGIVRKCIDSIVEKILTPTSEVS 178
Query: 181 LSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPSNL 240
SH+++R + S + WW ED+++L +DL+ + A +S +P L
Sbjct: 179 WSHTYTR--------PGYAKRQHHSVPRDWWTEDISDLDLDLFRCVITAARSTFTLPPQL 230
Query: 241 IGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLPTEK 300
IG+AL +Y RWLP N + S+ E+E+ +HR L+ ++V+++P +K
Sbjct: 231 IGEALHVYTCRWLPYFKSNSH--SGFSVKENEA-------ALERHRRLVNTVVNMIPADK 281
Query: 301 GAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTLYDVE 360
G+VS FLL+L+ ++ + AS ++K EL R+ LQLEEAT+ DLL+PS S ++ YD +
Sbjct: 282 GSVSEGFLLRLVSIASYVRASLTTKTELIRKSSLQLEEATLEDLLLPSHSSSHLHRYDTD 341
Query: 361 LVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSSKLKVA 420
LV T+LE F++ + R+ S + N +L S R KVA
Sbjct: 342 LVATVLESFLM--------------LWRRQSSAHLSSNNTQLLHSIR----------KVA 377
Query: 421 KLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKSERKR 480
KL+D YLQ VA+DV+ +SKF++L++ +P+ AR HD LY+AI+I+LK HPE++K E+KR
Sbjct: 378 KLIDSYLQAVAQDVHMPVSKFVSLSEAVPDIARQSHDRLYKAINIFLKVHPEISKEEKKR 437
Query: 481 LCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSGGKVTELQSNIKALLT 539
LCR LDC+KLS + HA +NE +PLR VVQ LFF+Q SG K +S + L T
Sbjct: 438 LCRSLDCQKLSAQVRAHAVKNERMPLRTVVQALFFDQ----ESGSKGASSRSESQELFT 492
>UniRef100_Q6H483 Putative non-phototropic hypocotyl 3 [Oryza sativa]
Length = 594
Score = 387 bits (993), Expect = e-106
Identities = 224/550 (40%), Positives = 314/550 (56%), Gaps = 66/550 (12%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHK----------------- 43
MK+MKLG++PDTFYT EA+R++ S+V +DLII V ++Y LHK
Sbjct: 1 MKYMKLGTKPDTFYTEEAVRSVLSDVPADLIIHVNNTKYQLHKLKFCSHSENADTEGTGC 60
Query: 44 -----FPLLSKCLCLQRLCSESPPDSSHHQI---VQLPDFPGGVEAFELCAKFCYGIQIT 95
FPLL KC LQRLCS+ V L D PGG EAFELCAKFCYGI I
Sbjct: 61 HPYYQFPLLLKCGLLQRLCSDDDGVDGVDAAPVPVALHDIPGGEEAFELCAKFCYGISIN 120
Query: 96 LSPYNIVAARCAAEYLQMTEEVEKGNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWS 155
+ N VAA AA +L+MTE V KGNL+ KL+ FF+SCIL GWKD I +L + WS
Sbjct: 121 IGAGNFVAAALAARFLRMTEAVAKGNLVAKLDSFFDSCILQGWKDPIAALTAAWRISGWS 180
Query: 156 EDLGITSRCIETIASKVLSHPTKVSLSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDL 215
E I C++ I K+L+ P+KV+ S++++R +S K WW ED+
Sbjct: 181 ESR-IVQPCVDAIVEKILTPPSKVTWSYTYTRP--------GYAKKAHQSVPKDWWTEDV 231
Query: 216 AELSIDLYWRTMIAIKSGGKVPSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNS 275
+EL ID++ + + + +P LIG+AL +YA + LP+ + + A + S
Sbjct: 232 SELDIDVFRSLLSTVCAARLLPPPLIGEALHVYACKHLPDPLNHA-----AAAATANGQS 286
Query: 276 DSASEITSKHRLLLESIVSLLPTEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQ 335
+K R +LE+IV+++P + GAV+ FLL+LL+ ++ + AS S++ +L R+ G Q
Sbjct: 287 SELETAAAKQRRVLETIVTMIPGDAGAVTGRFLLRLLRVASYVGASPSTRAQLVRQAGAQ 346
Query: 336 LEEATVNDLLIPSLSYTNDTLYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSA 395
L+EA DLL+P S ++ YDV +LE F+ Q P +PP
Sbjct: 347 LDEARAVDLLVPMPSSSDPPAYDVGAAEAVLEHFLALFQRP--APP-------------- 390
Query: 396 ENINFELQESRRSSSASHSSKLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYD 455
E RR S+A KVA+ D YL+ VA +F + KF+ LA+ +P+ AR D
Sbjct: 391 -------DERRRMSAAME----KVARTFDEYLRAVALHADFPVGKFVDLAECLPDIARND 439
Query: 456 HDDLYRAIDIYLKAHPELNKSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFF 515
HD LY AID YLK HPEL+K+++KRLCR++DC+KLS + A N+ +PLR +VQ+LF
Sbjct: 440 HDGLYHAIDTYLKEHPELSKADKKRLCRMIDCRKLSPDVRAQAISNDRMPLRTIVQLLFV 499
Query: 516 EQARAASSGG 525
EQ RA GG
Sbjct: 500 EQERAMGGGG 509
>UniRef100_Q8H1D3 Hypothetical protein At4g31820 [Arabidopsis thaliana]
Length = 571
Score = 385 bits (990), Expect = e-105
Identities = 225/529 (42%), Positives = 321/529 (60%), Gaps = 63/529 (11%)
Query: 1 MKFMKLGSRPDTFYTS-EAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE 59
MKFMKLGS+PDTF + + ++ S++ SD+ I V + LHKFPLLSK +QRL E
Sbjct: 1 MKFMKLGSKPDTFESDGKFVKYAVSDLDSDVTIHVGEVTFHLHKFPLLSKSNRMQRLVFE 60
Query: 60 SPPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEK 119
+ + + + + D PGG +AFE+CAKFCYG+ +TL+ YNI A RCAAEYL+MTE+ ++
Sbjct: 61 ASEEKTDE--ITILDMPGGYKAFEICAKFCYGMTVTLNAYNITAVRCAAEYLEMTEDADR 118
Query: 120 GNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKV 179
GNLI+K+EVF NS I WKD+I+ LQTT++L WSEDL + RCI+++++K+L +P +
Sbjct: 119 GNLIYKIEVFLNSGIFRSWKDSIIVLQTTRSLLPWSEDLKLVGRCIDSVSAKILVNPETI 178
Query: 180 SLSHSHSRRVR--DDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVP 237
+ S++ +R++ D I E K WW ED+ EL ID++ R + +KS G++
Sbjct: 179 TWSYTFNRKLSGPDKIVEYHREKREENVIPKDWWVEDVCELEIDMFKRVISVVKSSGRMN 238
Query: 238 SNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLP 297
+ +I +AL+ Y +RWLP ++ +E+ SN D L+E++V LLP
Sbjct: 239 NGVIAEALRYYVARWLPESMES-------LTSEASSNKD-----------LVETVVFLLP 280
Query: 298 TEKGAV---SCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTND 354
A+ SCSFLLKLLK S ++ A + + +L V L+L EA+V DLLI
Sbjct: 281 KVNRAMSYSSCSFLLKLLKVSILVGADETVREDLVENVSLKLHEASVKDLLI-------- 332
Query: 355 TLYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHS 414
++VELV I++QFM E+R S F L +
Sbjct: 333 --HEVELVHRIVDQFMAD---------------EKRVSEDDRYKEFVL---------GNG 366
Query: 415 SKLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELN 474
L V +L+D YL A + +LS F+ L++++PE AR HD LY+AID ++K HPEL
Sbjct: 367 ILLSVGRLIDAYL---ALNSELTLSSFVELSELVPESARPIHDGLYKAIDTFMKEHPELT 423
Query: 475 KSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASS 523
KSE+KRLC ++D +KL+ EA HAAQNE LPLRVVVQVL+FEQ RA S
Sbjct: 424 KSEKKRLCGLMDVRKLTNEASTHAAQNERLPLRVVVQVLYFEQLRANHS 472
>UniRef100_Q8RWY9 Hypothetical protein At4g31820 [Arabidopsis thaliana]
Length = 571
Score = 383 bits (983), Expect = e-104
Identities = 224/529 (42%), Positives = 320/529 (60%), Gaps = 63/529 (11%)
Query: 1 MKFMKLGSRPDTFYTS-EAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE 59
MKFMKLGS+PDTF + + ++ S++ SD+ I V + LHKFPLLSK +QRL E
Sbjct: 1 MKFMKLGSKPDTFESDGKFVKYAVSDLDSDVTIHVGEVTFHLHKFPLLSKSNRMQRLVFE 60
Query: 60 SPPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEK 119
+ + + + + D PGG +AFE+CAKFCYG+ +TL+ YNI A RCAAEYL+MTE+ ++
Sbjct: 61 ASEEKTDE--ITILDMPGGYKAFEICAKFCYGMTVTLNAYNITAVRCAAEYLEMTEDADR 118
Query: 120 GNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKV 179
GNLI+K+EVF NS I WKD+I+ LQTT++L WSEDL + RCI+++++K+L +P +
Sbjct: 119 GNLIYKIEVFLNSGIFRSWKDSIIVLQTTRSLLPWSEDLKLVGRCIDSVSAKILVNPETI 178
Query: 180 SLSHSHSRRVR--DDISCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVP 237
+ S++ +R++ D I E K WW ED+ EL ID++ + +KS G++
Sbjct: 179 TWSYTFNRKLSGPDKIVEYHREKREENVIPKDWWVEDVCELEIDMFKGVISVVKSSGRMN 238
Query: 238 SNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLP 297
+ +I +AL+ Y +RWLP ++ +E+ SN D L+E++V LLP
Sbjct: 239 NGVIAEALRYYVARWLPESMES-------LTSEASSNKD-----------LVETVVFLLP 280
Query: 298 TEKGAV---SCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTND 354
A+ SCSFLLKLLK S ++ A + + +L V L+L EA+V DLLI
Sbjct: 281 KVNRAMSYSSCSFLLKLLKVSILVGADETVREDLVENVSLKLHEASVKDLLI-------- 332
Query: 355 TLYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHS 414
++VELV I++QFM E+R S F L +
Sbjct: 333 --HEVELVHRIVDQFMAD---------------EKRVSEDDRYKEFVL---------GNG 366
Query: 415 SKLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELN 474
L V +L+D YL A + +LS F+ L++++PE AR HD LY+AID ++K HPEL
Sbjct: 367 ILLSVGRLIDAYL---ALNSELTLSSFVELSELVPESARPIHDGLYKAIDTFMKEHPELT 423
Query: 475 KSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASS 523
KSE+KRLC ++D +KL+ EA HAAQNE LPLRVVVQVL+FEQ RA S
Sbjct: 424 KSEKKRLCGLMDVRKLTNEASTHAAQNERLPLRVVVQVLYFEQLRANHS 472
>UniRef100_Q7EZR6 Putative non-phototropic hypocotyl 3 (NPH3)/phototropic response
protein [Oryza sativa]
Length = 641
Score = 363 bits (931), Expect = 1e-98
Identities = 237/644 (36%), Positives = 347/644 (53%), Gaps = 38/644 (5%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSES 60
M +KLGSR D F ++ + SD+ + V + LHKFPLLSK L+R E
Sbjct: 1 MACLKLGSRADVFRKQGQDWYCTTGLPSDITVTVGEQSFHLHKFPLLSKSGLLERCIREK 60
Query: 61 PPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKG 120
+ + ++ L D PGG +AFEL AKFCYG++ ++ N+V RCAAEYL+MTEE+ +G
Sbjct: 61 IENGDNSCVIDLSDIPGGAKAFELTAKFCYGVKFEMTASNVVHLRCAAEYLEMTEEIAEG 120
Query: 121 NLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPT--- 177
NLI + E F +L WKD+I +L T + +E L I RCI+++A++ + P
Sbjct: 121 NLIAQTENFLTQTVLRSWKDSIKALHTCDDIIDLAEKLQIVKRCIDSVATRSCTDPDLFG 180
Query: 178 -KVSLSHSHSRRVRDDISCNDTES-LRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGK 235
V + + N + R + S WW +D++ LS+ LY + + A++ G
Sbjct: 181 WPVVQYGGPMQSPGGSVLWNGISTGARPRHSSPDWWYDDVSCLSLPLYKKVISAMEYRG- 239
Query: 236 VPSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKH--RLLLESIV 293
+ ++I +L YA R LP + NR+K++S S + S + I S+ + LLE I
Sbjct: 240 INQDIIVGSLNHYAKRRLPGL----NRRKSISDVSSCLSISSLTSIPSEEEQKYLLEEID 295
Query: 294 SLLPTEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTN 353
LLP ++G SC L LL+ + IL AS S L RR+G+QL++AT+ DLLIP++S +
Sbjct: 296 RLLPFQRGVTSCKLLFGLLRTAIILKASPSCVSNLERRIGMQLDKATLEDLLIPNISESV 355
Query: 354 DTLYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASH 413
+TLYDV+ V IL+ F+ Q + P + + S S I
Sbjct: 356 ETLYDVDCVHKILDHFLAMDQETGGASPGLGEDAQMLASPSLMPITM------------- 402
Query: 414 SSKLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPEL 473
VAKL+D YL EVA DVN L KF +LA IP++AR D LYRAIDIYLKAHP L
Sbjct: 403 -----VAKLIDGYLAEVAPDVNLKLPKFRSLAAAIPDYARPIDDGLYRAIDIYLKAHPHL 457
Query: 474 NKSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASSGGKVTELQSN 533
++SE++ LCR++DC+KLS+EAC HAAQNE LPLRV+VQVLFFEQ + SS + + N
Sbjct: 458 SESEKEELCRVMDCQKLSLEACTHAAQNERLPLRVIVQVLFFEQLQLRSSIAECLMVSEN 517
Query: 534 IKALLTTHGIDPSKHTTTQLSSTTSIQGEDNWSVSGFKSPKSKSSTLRMKLAEEHDEFDE 593
++ + + P+ E+ G + K + + L + + DE +
Sbjct: 518 LEG--GSRQLIPTISGEQYRPGWPLASRENQALREGMDNMKQRVADLEKECSTMRDEIER 575
Query: 594 NGLVNDDGIGRNS--KFKSICALPTQPKKMLSKLWSTNRTATEK 635
G G GR S IC+ K+ + +T TA+E+
Sbjct: 576 LGRSRSTGKGRFSLNMKPQICS----TKEAIPTTATTTATASEE 615
>UniRef100_Q5ZCG0 Putative non-phototropic hypocotyl 3 [Oryza sativa]
Length = 659
Score = 358 bits (919), Expect = 3e-97
Identities = 224/560 (40%), Positives = 324/560 (57%), Gaps = 50/560 (8%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSE- 59
M MKLGS+P+ F +E+ SD++++V + LHKFPLLS+ LQR+ SE
Sbjct: 1 MATMKLGSKPEIFVLEGLTWRCMTELESDVVVEVGEMSFYLHKFPLLSRSGVLQRMISEY 60
Query: 60 -SPPDSSHHQ----IVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMT 114
+P + +QL D PGG +AFEL A+FCY ++I L+ +N+V RCAAEYL+MT
Sbjct: 61 QAPQEDGGGGGGMCTLQLDDIPGGAKAFELAARFCYDVKIELNAHNVVCLRCAAEYLRMT 120
Query: 115 EEVEKGNLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVL- 173
++ +GNLI + E F +L WKD+I +L+T + + +EDL I SRCI +ASK
Sbjct: 121 DDYAEGNLITQAESFLAD-VLANWKDSIKALETCEGVLPTAEDLHIVSRCITALASKACA 179
Query: 174 SHPTKVSLSHSHSRRVRDDISCNDT--ESLRIKSGSKG-------------------WWA 212
S S H+ + ++ + ++L GS G WW
Sbjct: 180 SDAAACSTGHAAASASAVAVAAKNASYDALWNGIGSGGTPRGGGGAAGAAAGCSGMDWWY 239
Query: 213 EDLAELSIDLYWRTMIAIKSGGKVPSNLIGDALKIYASRWLPNISKNGNRKKNVSL---- 268
ED++ LS+ ++ R + A++ G P ++ G A+ YA R+LP + +N + N S
Sbjct: 240 EDVSFLSLPMFKRLIQAMEGKGMRPESIAG-AIMFYAGRFLPGLKRNTSFS-NASFGGDC 297
Query: 269 -AESESNSDSASEITSK----HRLLLESIVSLLPTEKGAVSCSFLLKLLKASNILNASSS 323
A S S + A+ +++ R LE IV+LLP +KG S FLL +L+ + +L+AS
Sbjct: 298 GAGSRSITPRAANVSAPSEGDQRYFLEEIVALLPAKKGVASTRFLLGMLRTAMLLHASPL 357
Query: 324 SKMELARRVGLQLEEATVNDLLIPSLSYTNDTLYDVELVMTILEQFMLQGQSPPTSPPRS 383
+ L RR+G QLE+A ++DLL+P+L YT +TLYD++ V IL+ FM T
Sbjct: 358 CRENLERRIGAQLEDACLDDLLVPNLGYTVETLYDIDCVQRILDYFMSSTDGLGTG---- 413
Query: 384 LKTFERRRSRSAENINFELQESRRSSSASHSSKLKVAKLVDRYLQEVARDVNFSLSKFIA 443
S + +L S +S S VAKL+D YL EVA D N L KF A
Sbjct: 414 ------YTSPAVVEEGSQLGALHAGSPSSLSPITMVAKLMDGYLAEVAPDTNLKLPKFQA 467
Query: 444 LADIIPEFARYDHDDLYRAIDIYLKAHPELNKSERKRLCRILDCKKLSMEACMHAAQNEL 503
LA ++P++AR D +YRA+DIYLK+HP L++SER++LCR+++C+KLS+EAC HAAQNE
Sbjct: 468 LAAVVPDYARPVDDGIYRAMDIYLKSHPWLSESEREQLCRLMNCQKLSLEACTHAAQNER 527
Query: 504 LPLRVVVQVLFFEQARAASS 523
LPLRVVVQVLFFEQ R +S
Sbjct: 528 LPLRVVVQVLFFEQLRLRTS 547
>UniRef100_Q8W0R6 Putative photoreceptor-interacting protein [Sorghum bicolor]
Length = 658
Score = 348 bits (892), Expect = 4e-94
Identities = 209/530 (39%), Positives = 305/530 (57%), Gaps = 30/530 (5%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSES 60
M +KLGSR D F +S + SD+ + V + LHKFPLLSK L++ E
Sbjct: 1 MACLKLGSRADVFRKQGQEWYCTSGLPSDITVVVGEQSFHLHKFPLLSKSGLLEKRIREK 60
Query: 61 PPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKG 120
+ L D PGG +AFEL AKFCYG++ ++ N+V RCAA+YL+MTEE+ +
Sbjct: 61 IDKGDDSCTIDLSDIPGGAKAFELAAKFCYGVKFEMTASNVVHLRCAADYLEMTEEISEE 120
Query: 121 NLIHKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPT--- 177
NLI + E F +L WKD++ +LQT + +E L I RC+++IA++ S P
Sbjct: 121 NLIAQTEYFLTQTVLRSWKDSVKALQTCDNVLEVAERLQIVKRCVDSIATRSCSDPDLFG 180
Query: 178 -KVSLSHSHSRRVRDDISCNDTES-LRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGK 235
V+ + + N + R ++ S WW +D++ LS+ LY + + A++ G
Sbjct: 181 WPVAQYGGPMQSPGGSLLWNGISTGARPRNSSPDWWYDDVSCLSLPLYKKLISAMEYRG- 239
Query: 236 VPSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSK--HRLLLESIV 293
+ +I +L YA R LP + NR+K++ + + S + I S+ + LLE I
Sbjct: 240 ISQEIIVGSLNHYAKRRLPGL----NRRKSIGDVSNCLSMTSLTSIPSEDDQKYLLEEID 295
Query: 294 SLLPTEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTN 353
LLP ++G SC L LL+ + L AS S L RR+G+QL++A++ DLLIP++S +
Sbjct: 296 RLLPFQRGVTSCKLLFGLLRTAIFLKASPSCVSNLERRIGMQLDKASLEDLLIPNVSDSV 355
Query: 354 DTLYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASH 413
+TLYDV+ IL+ F+ Q + P I ++ + +S S
Sbjct: 356 ETLYDVDCAQRILDHFLAMDQETGGASP---------------GIG---EDGQMLASPSL 397
Query: 414 SSKLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPEL 473
+ VAKL+D YL EVA D+N L KF +LA IP++AR D LYRAIDIYLKAHP L
Sbjct: 398 MPIMMVAKLIDGYLAEVAPDMNLKLPKFRSLAAAIPDYARPIDDGLYRAIDIYLKAHPHL 457
Query: 474 NKSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASS 523
++SE++ LCR++DC+KLS+EAC HAAQNE LPLR++VQVLFFEQ + SS
Sbjct: 458 SESEKEELCRVMDCQKLSLEACTHAAQNERLPLRIIVQVLFFEQLQLRSS 507
>UniRef100_Q9S9Q9 F26G16.2 protein [Arabidopsis thaliana]
Length = 662
Score = 344 bits (882), Expect = 5e-93
Identities = 203/526 (38%), Positives = 301/526 (56%), Gaps = 23/526 (4%)
Query: 4 MKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSESPPD 63
MKLGS+ D F ++ + SD++++V + LHKFPLLS+ ++R +E+ +
Sbjct: 1 MKLGSKSDAFQRQGQAWFCTTGLPSDIVVEVGEMSFHLHKFPLLSRSGVMERRIAEASKE 60
Query: 64 SSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKGNLI 123
++++ D PGG + FEL AKFCYG+++ L+ N+V RCAAE+L+MTEE +GNLI
Sbjct: 61 GDDKCLIEISDLPGGDKTFELVAKFCYGVKLELTASNVVYLRCAAEHLEMTEEHGEGNLI 120
Query: 124 HKLEVFFNSCILHGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPT----KV 179
+ E FFN +L WKD+I +L + + ++++L IT +CIE++A + + P V
Sbjct: 121 SQTETFFNQVVLKSWKDSIKALHSCDEVLEYADELNITKKCIESLAMRASTDPNLFGWPV 180
Query: 180 SLSHSHSRRVRDDISCND-TESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPS 238
+ + N + R K S WW ED + LS L+ R + ++S G +
Sbjct: 181 VEHGGPMQSPGGSVLWNGISTGARPKHTSSDWWYEDASMLSFPLFKRLITVMESRG-IRE 239
Query: 239 NLIGDALKIYASRWLPNIS-KNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLP 297
++I +L Y + LP + + G + + + + + SE K+ LLE I LL
Sbjct: 240 DIIAGSLTYYTRKHLPGLKRRRGGPESSGRFSTPLGSGNVLSEEEQKN--LLEEIQELLR 297
Query: 298 TEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTLY 357
+KG V F + +L+ + IL AS L +R+G+QL++A + DL++PS S+T +TLY
Sbjct: 298 MQKGLVPTKFFVDMLRIAKILKASPDCIANLEKRIGMQLDQAALEDLVMPSFSHTMETLY 357
Query: 358 DVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSSKL 417
DV+ V IL+ F+ Q P + + + S S +
Sbjct: 358 DVDSVQRILDHFLGTDQIMPGGVGSPCSSVD--------------DGNLIGSPQSITPMT 403
Query: 418 KVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKSE 477
VAKL+D YL EVA DVN L KF ALA IPE+AR D LYRAIDIYLK HP L ++E
Sbjct: 404 AVAKLIDGYLAEVAPDVNLKLPKFQALAASIPEYARLLDDGLYRAIDIYLKHHPWLAETE 463
Query: 478 RKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASS 523
R+ LCR+LDC+KLS+EAC HAAQNE LPLR++VQVLFFEQ + +S
Sbjct: 464 RENLCRLLDCQKLSLEACTHAAQNERLPLRIIVQVLFFEQLQLRTS 509
>UniRef100_Q9SZ49 Hypothetical protein AT4g31820 [Arabidopsis thaliana]
Length = 498
Score = 338 bits (867), Expect = 3e-91
Identities = 194/453 (42%), Positives = 275/453 (59%), Gaps = 60/453 (13%)
Query: 76 PGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKGNLIHKLEVFFNSCIL 135
PGG +AFE+CAKFCYG+ +TL+ YNI A RCAAEYL+MTE+ ++GNLI+K+EVF NS I
Sbjct: 2 PGGYKAFEICAKFCYGMTVTLNAYNITAVRCAAEYLEMTEDADRGNLIYKIEVFLNSGIF 61
Query: 136 HGWKDTIVSLQTTKALHLWSEDLGITSRCIETIASKVLSHPTKVSLSHSHSRRVR--DDI 193
WKD+I+ LQTT++L WSEDL + RCI+++++K+L +P ++ S++ +R++ D I
Sbjct: 62 RSWKDSIIVLQTTRSLLPWSEDLKLVGRCIDSVSAKILVNPETITWSYTFNRKLSGPDKI 121
Query: 194 SCNDTESLRIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGKVPSNLIGDALKIYASRWL 253
E K WW ED+ EL ID++ R + +KS G++ + +I +AL+ Y +RWL
Sbjct: 122 VEYHREKREENVIPKDWWVEDVCELEIDMFKRVISVVKSSGRMNNGVIAEALRYYVARWL 181
Query: 254 PNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSLLPTEKGAV---SCSFLLK 310
P ++ +E+ SN D L+E++V LLP A+ SCSFLLK
Sbjct: 182 PESMES-------LTSEASSNKD-----------LVETVVFLLPKVNRAMSYSSCSFLLK 223
Query: 311 LLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDTLYDVELVMTILEQFM 370
LLK S ++ A + + +L V L+L EA+V DLLI ++VELV I++QFM
Sbjct: 224 LLKVSILVGADETVREDLVENVSLKLHEASVKDLLI----------HEVELVHRIVDQFM 273
Query: 371 LQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSSKLKVAKLVDRYLQEV 430
E+R S F L + L V +L+D YL
Sbjct: 274 AD---------------EKRVSEDDRYKEFVL---------GNGILLSVGRLIDAYL--- 306
Query: 431 ARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNKSERKRLCRILDCKKL 490
A + +LS F+ L++++PE AR HD LY+AID ++K HPEL KSE+KRLC ++D +KL
Sbjct: 307 ALNSELTLSSFVELSELVPESARPIHDGLYKAIDTFMKEHPELTKSEKKRLCGLMDVRKL 366
Query: 491 SMEACMHAAQNELLPLRVVVQVLFFEQARAASS 523
+ EA HAAQNE LPLRVVVQVL+FEQ RA S
Sbjct: 367 TNEASTHAAQNERLPLRVVVQVLYFEQLRANHS 399
>UniRef100_Q9FNB3 Photoreceptor-interacting protein-like; non-phototropic
hypocotyl-like protein [Arabidopsis thaliana]
Length = 591
Score = 338 bits (866), Expect = 4e-91
Identities = 214/530 (40%), Positives = 306/530 (57%), Gaps = 40/530 (7%)
Query: 1 MKFMKLGSRPDTFYTSEAIRTISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSES 60
M +KLGS+ + F+ S + + D++IQV + LHKFPLLS+ L+ L S++
Sbjct: 1 MASLKLGSKSEVFHLSGHTWLCKTGLKPDVMIQVVDESFHLHKFPLLSRSGYLETLFSKA 60
Query: 61 PPDSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVEKG 120
+ + QL D PGG E F L AKFCYG++I ++P N V+ RCAAEYLQM+E
Sbjct: 61 SETTC---VAQLHDIPGGPETFLLVAKFCYGVRIEVTPENAVSLRCAAEYLQMSENYGDA 117
Query: 121 NLIHKLEVFFNSCILHGWKDTIVSLQTT---KALHLWSEDLGITSRCIETIASKVLSHPT 177
NLI+ E F N + W+D+I +L+ + K L L +E+L I SRCI ++A K +
Sbjct: 118 NLIYLTESFLNDHVFVNWEDSIKALEKSCEPKVLPL-AEELHIVSRCIGSLAMKACAEDN 176
Query: 178 KVSLSHSHSRRVRDDISCNDTESLRIKSGSKGWWAEDLAE-LSIDLYWRTMIAIKSGGKV 236
+ S + ++ K+ S+ WW D++ L + +Y R + ++S G V
Sbjct: 177 TSFFNWPISLPEGTTTTTIYWNGIQTKATSENWWFNDVSSFLDLPMYKRFIKTVESRG-V 235
Query: 237 PSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESN-SDSASEITSKHRLLLESIVSL 295
+ +I ++ YA R LP + G +K+ S +E +N D + R LLE IV L
Sbjct: 236 NAGIIAASVTHYAKRNLPLL---GCSRKSGSPSEEGTNYGDDMYYSHEEQRSLLEEIVEL 292
Query: 296 LPTEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDT 355
LP +K S FLL+LL+ S +L+AS ++ L +R+G+QL+EA + DLLIP++ Y+ +T
Sbjct: 293 LPGKKCVTSTKFLLRLLRTSMVLHASQVTQETLEKRIGMQLDEAALEDLLIPNMKYSGET 352
Query: 356 LYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSS 415
LYD + V IL+ FML TF+ + E ++ SH
Sbjct: 353 LYDTDSVQRILDHFML--------------TFDS-----------SIVEEKQMMGDSHPL 387
Query: 416 K--LKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPEL 473
K KVA L+D YL EVA D N LSKF AL +IPE R D +YRAIDIY+KAHP L
Sbjct: 388 KSITKVASLIDGYLAEVASDENLKLSKFQALGALIPEDVRPMDDGIYRAIDIYIKAHPWL 447
Query: 474 NKSERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQARAASS 523
+SER++LC +++C+KLS+EAC HAAQNE LPLRV+VQVLFFEQ R +S
Sbjct: 448 TESEREQLCLLMNCQKLSLEACTHAAQNERLPLRVIVQVLFFEQMRLRTS 497
>UniRef100_Q7XND4 OSJNBb0034I13.5 protein [Oryza sativa]
Length = 644
Score = 329 bits (844), Expect = 1e-88
Identities = 208/524 (39%), Positives = 302/524 (56%), Gaps = 35/524 (6%)
Query: 6 LGSRP-DTFYTSEAIR-TISSEVSSDLIIQVRGSRYLLHKFPLLSKCLCLQRLCSESPP- 62
LGS+P D F + T +E+ SD++++V + LHKFPL+S+ LQ+L SE+
Sbjct: 7 LGSKPADCFQFQDPNSWTCMTELVSDVVVEVGDFSFHLHKFPLMSRSGTLQKLISEAAAG 66
Query: 63 -DSSHHQIVQLPDFPGGVEAFELCAKFCYGIQITLSPYNIVAARCAAEYLQMTEEVE-KG 120
D V+L D PGG AFEL A+FCY ++ L N+VA RCAAE+L MTE+ +G
Sbjct: 67 ADDGEPCSVKLHDVPGGAAAFELAARFCYDVRAELDAGNVVALRCAAEHLGMTEDHGGEG 126
Query: 121 NLIHKLEVFFNSCILHGWKDTIVSLQTTK-ALHLWSEDLGITSRCIETIASKVLSHPTKV 179
NL+ + E F +L W D + +L++ AL +E+L + RCI+ +ASK + PT
Sbjct: 127 NLVEQAEAFLRD-VLGSWDDALRALRSCDGALLPLAEELLVVPRCIDALASKACADPTLF 185
Query: 180 S---LSHSHSRRVRDDISCNDTESL-RIKSGSKGWWAEDLAELSIDLYWRTMIAIKSGGK 235
+ + +R + + + N + + +S WW + + L + +Y R + A++S G
Sbjct: 186 GWPMVEYYTARGLEETVMWNGIATAGKPRSPGPDWWYKQASSLKLPVYKRLITAMRSKGM 245
Query: 236 VPSNLIGDALKIYASRWLPNISKNGNRKKNVSLAESESNSDSASEITSKHRLLLESIVSL 295
P N+ G +L YA R L ++++ + ++ S++ + R LLE IV+L
Sbjct: 246 SPENIAG-SLTHYAKRHLSGLTRHSGY-----VGGGGASGTVLSDV--EQRALLEEIVAL 297
Query: 296 LPTEKGAVSCSFLLKLLKASNILNASSSSKMELARRVGLQLEEATVNDLLIPSLSYTNDT 355
LP E+G + FLL LL+ + ILNA ++ + L R G QLEEA + DLLIP+ Y +T
Sbjct: 298 LPVERGVATTRFLLGLLRTATILNAGAACRDALERMAGNQLEEAALEDLLIPNTGYAVET 357
Query: 356 LYDVELVMTILEQFMLQGQSPPTSPPRSLKTFERRRSRSAENINFELQESRRSSSASHSS 415
LYDV+ V +LEQF+ S + P + S E +
Sbjct: 358 LYDVDCVQRMLEQFVAANTSAFAASPEITDEAQLVDGPSGELMPIST------------- 404
Query: 416 KLKVAKLVDRYLQEVARDVNFSLSKFIALADIIPEFARYDHDDLYRAIDIYLKAHPELNK 475
VAKLVD YL EVA D N LSKF ++A+++P++AR D +YRAIDIYLKAH L
Sbjct: 405 ---VAKLVDGYLAEVATDTNVKLSKFQSIAELVPDYARAIDDGIYRAIDIYLKAHSWLTA 461
Query: 476 SERKRLCRILDCKKLSMEACMHAAQNELLPLRVVVQVLFFEQAR 519
SER++LCR+++C+KLS+EAC HAAQNE LPLRVVVQVLFFEQ R
Sbjct: 462 SEREQLCRLMNCQKLSLEACTHAAQNERLPLRVVVQVLFFEQLR 505
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 981,793,009
Number of Sequences: 2790947
Number of extensions: 38719653
Number of successful extensions: 114605
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 80
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 114158
Number of HSP's gapped (non-prelim): 197
length of query: 636
length of database: 848,049,833
effective HSP length: 134
effective length of query: 502
effective length of database: 474,062,935
effective search space: 237979593370
effective search space used: 237979593370
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC119414.2


